Abstract
High-level induction of fetal (γ) globin gene expression for therapy of β-hemoglobinopathies likely requires local chromatin modification and dissociation of repressor complexes for γ-globin promoter activation. A novel γ-globin–inducing short-chain fatty acid derivative (SCFAD), RB7, which was identified through computational modeling, produced a 6-fold induction in a reporter assay that detects only strong inducers of the γ-globin gene promoter and in cultured human erythroid progenitors. To elucidate the molecular mechanisms used by high-potency SCFADs, chromatin immunoprecipitation (ChIP) assays performed at the human γ- and β-globin gene promoters in GM979 cells and in erythroid progenitors demonstrate that RB7 and butyrate induce dissociation of HDAC3 (but not HDAC1 or HDAC2) and its adaptor protein NCoR, specifically from the γ-globin gene promoter. A coincident and proportional recruitment of RNA polymerase II to the γ-globin gene promoter was observed with exposure to these γ-globin inducers. Knockdown of HDAC3 by siRNA induced transcription of the γ-globin gene promoter, demonstrating that displacement of HDAC3 from the γ-globin gene promoter by the SCFAD is sufficient to induce γ-globin gene expression. These studies demonstrate new dynamic alterations in transcriptional regulatory complexes associated with SCFAD-induced activation of the γ-globin gene and provide a specific molecular target for potential therapeutic intervention.
Introduction
Discerning molecular mechanisms to reactivate expression of fetal (γ) globin has been the subject of intense investigation for more than 2 decades, with potential application to treatment of the β hemoglobin disorders, sickle cell disease and β-thalassemia. A number of molecular events associated with the developmental activation of the adult β-globin genes, and reciprocal suppression of the fetal globin genes, have been elucidated, including chromatin modifications that may promote interaction between the β-globin locus control region (LCR) and the β- and γ-globin gene promoters, and binding of certain transcription factors, such as EKLF, NF-E2, and GATA-1, during erythroid cell development.1-6 Transcription factors that activate the γ-globin gene promoter include the fetal Kruppel-like factor (FKLF), which binds to the CACCC element of the γ-globin promoter,7-9 and the stage selector complex (SSC), composed of the transcription factor CP2 and an erythroid-specific factor NF-E4.10,11 More recently, additional data implicate p14 NF-E4, a shorter form of NF-E4 generated by an alternate start site in the NF-E4 open-reading frame, in competing with, and sequestering CP2 away from, the active γ-globin promoter, thereby silencing it.12 Alternatively, persistent activation of the γ-globin gene is observed when repressor complexes cannot silence the promoter, as in some hereditary persistence of fetal hemoglobin (HPFH) point mutations in the region between –114 and –202 upstream of Gγ- or Aγ-globin transcription start.13 The repressor proteins that normally bind this and other upstream regions, and are presumably disrupted in HPFH, have not been established definitively.14-16
Potent therapeutic agents that specifically induce high-level fetal globin expression would be beneficial for treating the β-globin diseases.1,17 The short-chain fatty acid (SCFA) arginine butyrate (AB) stimulates fetal globin gene expression in cultured erythroid cells, animal models, and treated patients through induction of the proximal γ-globin gene promoter and, in some cases, by increasing translational efficiency.18-21 In vivo footprinting studies performed in nucleated erythroid cells of patients in whom γ-globin gene expression was induced with butyrate treatment revealed alterations in DNA-binding proteins in 4 regions of the proximal γ-globin gene promoter, designated butyrate response elements γ 1 to 4 (BRE-G 1-4).22 New binding of one component of the stage selector complex, CP2, to the γ-globin promoter was identified in nucleated erythroid cells in which γ-globin was being induced.23 The identity of 2 other putative activators, and a putative repressor that disassociated with butyrate induction, was not established. Although generalized acetylation of histones to confer chromatin accessibility was initially considered a mechanism of γ-globin gene activation by the butyrates, other SCFAs induce fetal globin expression without causing generalized histone acetylation.24-26 Thus, specific molecular events underlying γ-globin gene induction in response to SCFAs have not been entirely elucidated, and a better understanding would be useful for the development of high-potency therapies.
Using computational modeling and screening of a chemical library, we recently identified novel SCFA derivative (SCFAD) fetal globin–inducing compounds, of which, a few demonstrate higher potency than the prototype SCFA, butyrate.27 These compounds were tested in a validated model system composed of a reporter construct containing the μLCRβprRlucAγprFluc cassette, which detects only potent inducers of the upstream Aγ-globin gene promoter.28 A novel SCFAD, designated RB7 (3-2ox-2H chromen-3yl benzoic acid), was found to have significantly higher activity than AB in this assay and acts at a concentration that is a log lower than the concentration necessary for butyrate. Investigation of the molecular effects of this high-potency inducer of the γ-globin gene promoter demonstrated a displacement of the histone deacetylase 3/nuclear receptor corepressor (HDAC3/NCoR) complex and a coordinate association of RNA polymerase II concurrent with γ-globin transcription, both in the integrated reporter construct–carrying cell line and in human erythroid progenitor cells. The magnitude of these effects was greater with RB7 exposure than with AB and was proportional to the degree of induction observed. Furthermore, specific knockdown of endogenous HDAC3 by siRNA resulted in increased transcription at the γ-globin gene promoter. These studies suggest a pivotal role of HDAC3 displacement from the γ-globin gene promoter as a mechanism of γ-globin gene reactivation by SCFADs.
Materials and methods
Cell culture and reagents
Human K562 cells and GM979 cells stably transfected with the human μLCRβprRlucAγprFluc construct were studied as previously described.28 Cells were cultured with and without the candidate SCFAD compounds and lysed in Jurkat lysis buffer (JLB) (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, 0.5% Triton-X 100) as described.29 Antibodies to HDAC1 and HDAC3 were purchased from Abcam (Cambridge, MA); antibodies against HDAC2 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); anti–acetyl histone H3 and anti–hyperacetylated histone H4 antibodies were purchased from Upstate Cell Signaling Solutions (Waltham, MA). Coimmunoprecipitation was carried out using the Seize Primary Mammalian Immunoprecipitation Kit (Pierce, Rockford, IL) according to the manufacturer's recommendations.
Human cord blood cells were studied with approval of the institutional review board of the Boston University School of Medicine. Erythroid progenitors were cultured from unidentified cord blood for the large quantities of erythroid cells necessary for chromatin immunoprecipitation (ChIP) assays. Mononuclear cells were isolated by Ficoll Paque PLUS (Amersham Biosciences AB, Uppsala, Sweden) and density separated and cultured in liquid culture conditions that provide sequential differentiation and enrichment of erythroid progenitors cells, as previously described.30,31 Briefly, cells were cultured in Iscove modified Dulbecco media (Cellgro, Herndon, VA), 30% charcoal-absorbed fetal bovine serum (Atlanta Biotechnologies, Lawrenceville, GA.), 1% deionized bovine serum albumin, 10–4 M β mercaptoethanol, 10–6 M hydrocortisone, 100 U/mL penicillin streptomycin (all from Sigma, St Louis, MO), 2 mM/l glutamine (Gibco BRL Life Technologies, Gaithersburg, MD), with 10 U/mL erythropoietin (Amgen, Thousand Oaks, CA), and low concentrations of GM-CSF (0.001 ng/mL) and IL-3 0.01 U/mL (Stem Cell Technologies, Vancouver, BC) as previously described.31 Cells were cultured alone (control conditions) or with one of the test compounds (RB7 [3-2ox-2H chromen-3yl benzoic acid]) at 20 μM (SPECS, Delft, the Netherlands) or arginine butyrate (AB) at 100-μM concentrations (Gene Regulation Laboratories, Newton, MA). Test compounds RB7 and RB25 were added on days 1, 7, 8, 9, 10, and 11 of culture, and cells were harvested for analysis of globin mRNA and chromatin immunoprecipitation assay on day 11, when erythroblasts predominate by 90%.30
Erythroid cell culture and globin mRNA analysis from adult peripheral blood
Erythroid progenitor cells were also studied from normal adult peripheral blood by culturing purified CD34+ cells (obtained from the NHLBI Program of Excellence in Gene Therapy, National Hematopoietic Cell Processing Core, Fred Hutchinson Cancer Center, Seattle, WA) with 100 ng/mL Flt-3 ligand, 100 ng/mL stem cell factor, and 50 ng/mL rhu-IL-3 (R & D Systems, Minneapolis, MN) for 7 days, followed by a second phase culture initiated on day 8 in fresh DMEM (GIBCO), with 30% FBS (GIBCO), 1% BSA, 0.3% saturated transferrin, 1 μM dexamethasone, 10 μM beta mercaptoethanol (all from Sigma), and 4 U/mL EPO (Ortho Biotech, Raritan, NJ) for 14 days.30,31 RB7 or arginine butyrate (AB) was added to cultures after 2 days of culture with EPO. Every 2 days thereafter, cells were enumerated and RNA was harvested from 106 cells using an RNeasy Plus Mini Kit (Qiagen, Valencia, CA). Quantitative reverse-transcription–polymerase chain reaction (RT-PCR) for γ-globin mRNA was performed using IQ SybrGreen Supermix on an Opticon Monitor instrument (MJ Research, Watertown, MA). Samples were analyzed in triplicate, and raw data from the instrument were normalized to β-actin and G3PD. Primers used for γ-globin were 5′ AGACGCCATGGGTCATTTCCA 3′ and 5′ GCCTATCCTTGAAAGCTCTGCT 3′.
Luciferase reporter assays
Luciferase reporter assay was performed using GM979 cells stably transfected with the dual luciferase construct (μLCRβprRlucAγprFluc).28 Signals were detected using the Dual Luciferase Reporter System (Promega, Madison, WI) as previously described.28 Results were calculated as a ratio of firefly luciferase to renilla luciferase normalized to untreated control.
Immunoprecipitation and in vitro HDAC assay
K562 cells were washed 3 times with PBS and lysed with JLB containing a protease inhibitor cocktail (10 μg/mL aprotinin, 10 μg/mL leupeptin, 1 mM PMSF) at 4°C. The HDAC antibody was incubated with cell extract for 2 hours at 4°C on a rotating platform. Immunocomplexes were collected using protein A/G agarose beads (Santa Cruz Biotechnology). The beads were collected by gentle centrifugation, and washed first with JLB, and then with histone deacetylase buffer (Upstate Cell Signaling Solutions). The histone deacetylase assay was performed using the fluorometric HDAC assay kit (Upstate Cell Signaling Solutions) as per the manufacturer's recommendations. The immunoprecipitates from each reaction were incubated with 100 μM substrate and test compounds for 1 hour at 37°C. Fluorescence was read using a Millipore Cytofluor 2300 Fluorescence Plate Reader (Millipore, Billerica, MA).
Chromatin immunoprecipitation assays
GM979 cells were cultured in the presence of either RB7 (3-2ox-2H chromen-3yl benzoic acid, 200 μM), RB25 (1-(2-oxo-2-thien-2-ylethyl) cyclohexanecarboxylic acid, 200 μM) (SPECS), or arginine butyrate (2500 μM). Untreated cells cultured in the media alone were used as a control. GM979 or erythroid-enriched human cord blood cells were cross-linked by adding formaldehyde to a final concentration of 1%, and incubated for 10 minutes at 37°C. The reaction was quenched by adding glycine to a final concentration of 0.125 M. Cells were washed twice with ice-cold PBS containing the protease inhibitor cocktail (1 mM PMSF, 1 μg/mL aprotinin, 1 μg/mL pepstatinA), and then lysed with SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-Cl, pH 8.0) containing protease inhibitors at 4°C. The lysate was sonicated on ice to obtain DNA fragments in the range of 200 to 1000 bases (10% power, 4 10-second pulses) using the 550 Sonic Dismembrator Probe Sonicator (Fisher Scientific, Swanee, GA). Tissue debris was removed by centrifugation, and the supernatant was diluted in ChIP dilution buffer (0.01% SDS, 1.1% TritonX-100, 1.2 mM EDTA, 16.7 mM Tris-HCL, pH 8.1, 167 mM NaCl). Samples were immunoprecipitated by incubating them with the appropriate antibody for 2 hours on a rotating platform at room temperature. The immunocomplexes were collected using protein A/G agarose beads (Santa Cruz Biotechnology) and washed twice with low-salt wash buffer (0.01% SDS, 1% TritonX-100, 2 mM EDTA, 20 mM Tris-HCL, pH 8.1, 150 mM NaCl). The antibody-DNA complex was eluted from the beads using freshly prepared elution buffer (0.1 M NaHCO3, 1% SDS). Cross-links were reversed by adding NaCl to a final concentration of 0.2 M and incubating for 6 to 8 hours at 70°C. The samples were treated with proteinase K for 1 hour at 37°C, and the DNA was purified by phenol-chloroform extraction. Ethanol precipitation was facilitated using glycogen (Roche Diagnostics, Indianapolis, IN) as a coprecipitant. Primers used for amplifying the human β-globin gene promoter were as follows: 5′ GACAGGTACGGCTGTCATCA 3′ and 5′GTGTCTGTTTGAGGTTGCTA 3′. Primers used for amplifying the γ-globin gene promoter were as follows: 5′ AAACGGTCCCTGGCTAAACT 3′ and 5′ GACGTTCCAGAAGCGAGTGT 3′.
HDAC3 knockdown
Endogenous HDAC3 RNA knockdown was performed in the GM979 cell line carrying the μLCRβprRlucAγprFluc cassette, using predesigned siH-DAC3 RNA targeted against exons 8 and 9 of the HDAC3 open-reading frame (Ambion, Austin, TX). Transfection conditions for the cell line were optimized using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA) in 24-well plates. Cells (1 × 105) were incubated with siRNA (HDAC3 or a scrambled siRNA control) at a concentration of 200 to 500 nM. Twenty-four hours after transfection, the cells were washed and lysed to obtain either protein extracts or total RNA.
RNA preparation, cDNA synthesis, and RT-PCR
Total RNA was extracted from the cells using TRIzol reagent (Gibco BRL Life Technologies, Gaithersburg, MD) following directions of the manufacturer. First-strand synthesis was done using Superscript III (Invitrogen Life Technologies) using oligo dT for priming. For each primer pair used, homogeneity of the product was confirmed on an agarose gel prior to real-time PCR. Relative quantification PCR was performed using the ABI 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA) by the ΔΔCt method, with α-globin as the internal control to normalize for RNA levels. The following primers were used for PCR: human α-globin gene, 5′ CAAGACCTACTTCCCGCACT 3′ and 5′ AGGCAGTGGCTTAGGAGCTT 3′; human β-globin gene, 5′ CTGAGGAGAAGTCTGCCGTTA 3′ and 5′ GAGGTTGTCCAGGTGAGCCA 3′; and human γ-globin gene, 5′ TCACAGAGGAGGACAAGGCTA 3′ and 5′ GAGATCATCCAGGTGCTTT3′.
Evaluation of RB7 in an in vivo primate model
Globin mRNA was quantitated in blood samples obtained from chronically catheterized anemic baboons prior to and following administration of RB7 (50 mg/kg, 2 doses given 5 days apart) (administered on days 0 and 5), as previously described.28 Animal studies were approved by the IACUC of the University of Oklahoma. Globin mRNA was quantitated by RNase protection using a BD RiboQuant kit (BD Biosciences, San Diego, CA). Briefly, a γ-globin probe (kind gift of Dr Qiliang Li) and 18S RNA probe (Ambion) were radiolabeled with [α-32P] UTP (PerkinElmer Life and Analytical Sciences, Boston, MA) using the in vitro transcription kit (BD Biosciences) according to the manufacturer's directions. The radiolabeled probes and total RNA were hybridized overnight at 52°C. Following digestion of the single-stranded mRNA fragments, the protected fragments were precipitated and resolved on a denaturing 5% polyacrylamide gel. The density of each band on an autoradiograph was calculated using the NIH 1.62 software (http://rsb.info.nih.gov/nih-image).
Results
RB7 is a potent inducer of γ-globin gene transcription in vitro and in vivo
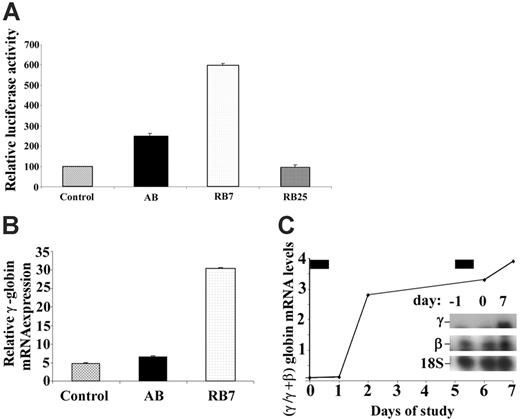
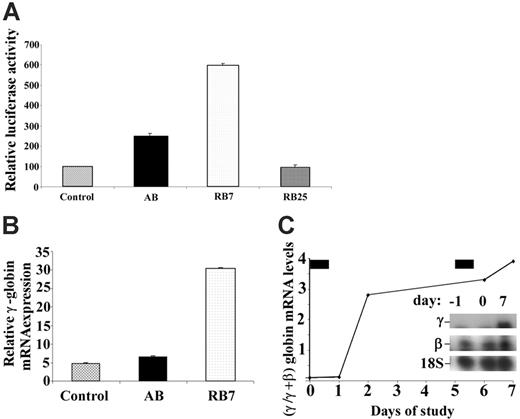
A dual luciferase construct containing the LCR and the β-globin promoter linked to renilla luciferase and the Aγ-globin promoter linked to firefly luciferase stably transfected into GM979 cells28 was used to determine the relative activity of new SCFADs in inducing γ-globin gene transcription. The orientation of the promoters and μLCR allows detection of only strong inducers of the γ-globin gene promoter. The compound RB7, at a concentration of 200 μM, induced expression of the γ-globin gene promoter, producing a 5.9-fold increase (P < .001) in the firefly–renilla luciferase signal ratio compared with untreated controls after a 24-hour exposure (Figure 1A). In the same experiment, AB, at a concentration of 2500 μM, produced a 2.5-fold increase in γ-globin gene promoter activity over control. Another SCFAD, RB25, which does not activate the γ-globin gene, was used as a negative control in this assay and had no significant effect. In erythroid progenitors cultured from adult peripheral blood, expression of all globin mRNAs was maximal on day 13 of culture, 5 days after EPO was added and 4 days after addition of the test compounds. Relative γ-globin transcript expression was 4.7 (relative units) in untreated cells, 6.4 with exposure to butyrate (at 100 μM), and 30.4 with exposure to RB7 (at 20 μM) (Figure 1B). There were no significant differences in cell numbers between control culture conditions and cultures treated with RB7. Exposure to butyrate at 100 μM caused a slight decrease in relative cell numbers, consistent with previous reports. Treatment with RB7 also induced γ-globin transcript expression in vivo in an anemic baboon model (Figure 1C). The γ/γ+β-globin mRNA ratio increased 4-fold relative to baseline levels, following 2 doses (50 mg/kg) of RB7.
Relative induction of the γ-globin gene promoter by test compounds. (A) GM979 cells containing the integrated cassette (μLCRβprRlucAγprFluc) were assayed for luciferase activity. The ratio of firefly to renilla luciferase is shown for each compound tested: control, arginine butyrate (AB, 2500 μM), RB7 (200 μM), and RB25 (200 μM). Results are reported as relative induction and represent the mean of 4 experiments. Error bars indicate the standard errors of the mean. (B) γ-Globin mRNA induction in adult peripheral blood. Relative γ-globin mRNA expression levels (normalized to β-actin and G3PD transcripts) from cultured adult erythroid cells are shown in untreated control cells (C), or cells cultured with 100 μM arginine butyrate (AB) or 20 μM RB7. γ-Globin mRNA was induced 1.4-fold above control levels in cells cultured with AB, and 6.5-fold above control levels in cells treated with RB7. (C) Induction of fetal globin mRNA by RB7 treatment in an anemic baboon in vivo. Ratios of γ/γ+β-globin mRNA were induced by 2.8-fold and 4-fold above the animal's baseline level after 1 and 2 doses of RB7, respectively. The drug was administered on day 0 and day 5, as indicated by the solid bars above the graph. Insert shows baseline and on-treatment levels of γ-globin mRNA and 18S RNA.
Relative induction of the γ-globin gene promoter by test compounds. (A) GM979 cells containing the integrated cassette (μLCRβprRlucAγprFluc) were assayed for luciferase activity. The ratio of firefly to renilla luciferase is shown for each compound tested: control, arginine butyrate (AB, 2500 μM), RB7 (200 μM), and RB25 (200 μM). Results are reported as relative induction and represent the mean of 4 experiments. Error bars indicate the standard errors of the mean. (B) γ-Globin mRNA induction in adult peripheral blood. Relative γ-globin mRNA expression levels (normalized to β-actin and G3PD transcripts) from cultured adult erythroid cells are shown in untreated control cells (C), or cells cultured with 100 μM arginine butyrate (AB) or 20 μM RB7. γ-Globin mRNA was induced 1.4-fold above control levels in cells cultured with AB, and 6.5-fold above control levels in cells treated with RB7. (C) Induction of fetal globin mRNA by RB7 treatment in an anemic baboon in vivo. Ratios of γ/γ+β-globin mRNA were induced by 2.8-fold and 4-fold above the animal's baseline level after 1 and 2 doses of RB7, respectively. The drug was administered on day 0 and day 5, as indicated by the solid bars above the graph. Insert shows baseline and on-treatment levels of γ-globin mRNA and 18S RNA.
RB7 does not inhibit class I histone deacetylases
The prototypic γ-globin–inducing SCFADs phenylbutyrate and butyrate are established inhibitors of histone deacetylases.19,32-34 To determine whether RB7 and related compounds shared this property with butyrate, and whether they exhibit any HDAC isozyme specificity, class I histone deacetylases 1, 2, and 3 were immunoprecipitated from K562 cell extract, and their activities were assayed in vitro. As shown in Figure 2A, the effects of candidate SCFAD compounds (ST-20, RB7, RB25)35 on HDAC activity were compared with established HDAC inhibitors such as AB, SAHA, butyl hydroxamic acid (BHA), and trichostatin A (TSA). RB25 did not induce the γ-globin gene promoter and was used as a negative control. While the known HDAC inhibitors suppressed the activity of all 3 class I HDAC isozymes by more than 50% at the concentrations tested, with TSA demonstrating the highest potency, ST-20, RB7, and RB25 demonstrated no significant inhibition of class I HDAC isozymes. None of the compounds tested displayed HDAC subclass-specific inhibition. RB7 displayed an unexpected and specific 2-fold increase in HDAC3 activity, while HDAC1 and HDAC2 activities remained unaffected by the compound. The deacetylase activities of HDAC1, HDAC2, and HDAC3 were assayed at RB7 concentrations of 10, 100, and 200 μM. While the relative activity of HDAC1 and HDAC2 remained largely unaffected at the concentrations tested, RB7 increased HDAC3 activity in a concentration-dependent fashion (Figure 2B).
In vitro HDAC assay. (A) In vitro assay of HDAC1, HDAC2, and HDAC3 isolated by immunoprecipitation from K562 cells. HDACs were assayed in the presence of control (C), AB (arginine butyrate, 2.5 mM), SAHA (suberoylanilide hydroxamic acid, 10 μM), BHA (butyryl hydroxamic acid, 2.5 mM), TSA (trichostatin A, 1 μM), ST-20 (1 mM), RB7 (200 μM), and RB25 (200 μM). For each enzyme assayed, the values represented are relative to the control reaction. Values are a mean of 3 independent experiments; error bars indicate the standard errors of the mean. (B) RB7 dose-response curve of immunoprecipitated HDAC1, HDAC2, and HDAC3 activity. Activity was assayed at RB7 concentrations of 10, 100, and 200 μM. Values reported are relative to the control reaction and are an average of 2 independent experiments.
In vitro HDAC assay. (A) In vitro assay of HDAC1, HDAC2, and HDAC3 isolated by immunoprecipitation from K562 cells. HDACs were assayed in the presence of control (C), AB (arginine butyrate, 2.5 mM), SAHA (suberoylanilide hydroxamic acid, 10 μM), BHA (butyryl hydroxamic acid, 2.5 mM), TSA (trichostatin A, 1 μM), ST-20 (1 mM), RB7 (200 μM), and RB25 (200 μM). For each enzyme assayed, the values represented are relative to the control reaction. Values are a mean of 3 independent experiments; error bars indicate the standard errors of the mean. (B) RB7 dose-response curve of immunoprecipitated HDAC1, HDAC2, and HDAC3 activity. Activity was assayed at RB7 concentrations of 10, 100, and 200 μM. Values reported are relative to the control reaction and are an average of 2 independent experiments.
RB7 displaces HDAC3/NCoR from the γ-globin gene promoter
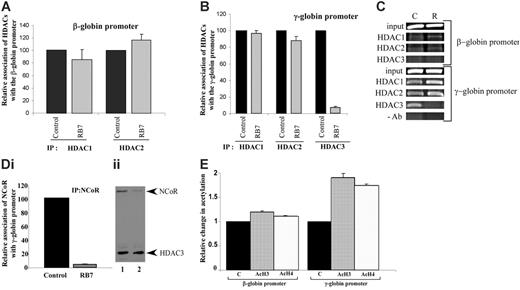
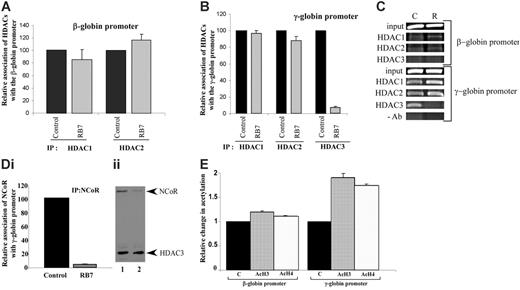
To elucidate the mechanisms underlying RB7-mediated γ-globin gene induction, chromatin immunoprecipitation was performed on the integrated globin promoters in the GM979 cells using antibodies to HDAC1, HDAC2, and HDAC3. At the human β-globin gene promoter, only minor and nonsignificant changes were observed in HDAC1 and HDAC2 associations after exposure to RB7 for 24 hours; HDAC1 levels were reduced to 85% of the control conditions, while HDAC2 increased to 116% of the control (Figure 3A). HDAC3 occupancy was not observed at the human β-globin gene promoter (Figure 3C). In cells treated with RB7, only minor changes in the levels of HDAC1 and HDAC2 were observed at the human γ-globin gene promoter; HDAC1 levels remained at 97% of control, while HDAC2 presence was reduced to 88% of the untreated control. In contrast, HDAC3 detected at the γ-globin gene promoter was significantly reduced to 7.8% of the control (P = .007) in cells exposed to RB7 (Figure 3B-C).
NCoR has been shown to associate with, and thereby modulate, the histone deacetylase activity of HDAC3 in multiprotein complexes, possibly acting as an adaptor to recruit HDAC3 to the complex, as well as an activator of HDAC3 activity.36-39 IP of the γ-globin gene promoter using an NCoR antibody to precipitate the associated chromatin demonstrated a 95.3% decrease (P = .001) in NCoR occupancy at the promoter within 24 hours of exposure to RB7 (Figure 3Di). The displacement of NCoR paralleled the pattern observed for HDAC3, both temporally and in magnitude, suggesting simultaneous displacement of a HDAC3/NCoR repressor complex from the γ-globin gene promoter. Coimmunoprecipitation studies were consistent with this observation, as the amount of NCoR associated with HDAC3 decreased after 24 hours of treatment with RB7 (Figure 3Dii). We next investigated the local acetylation status of histones at the integrated β- and γ-globin gene promoters 24 hours after treatment with RB7. Chromatin IP at the γ-globin promoter using antibodies to acetylated histones indicated that there was a corresponding 1.9-fold and 1.7-fold increase (P ≤ .008) in the acetylation of histones H3 and H4, respectively, at the γ-globin promoter (Figure 3E), at a time when dissociation of HDAC3 had already occurred, while only a minor change (1.1-fold baseline) was detected at the β-globin gene promoter following RB7 treatment. These reciprocal changes in HDAC3 occupancy and local histone acetylation strongly suggest that the increase in acetylation is due to HDAC3 displacement from the γ-globin gene promoter.
Extent of HDAC3 displacement from the γ-globin promoter correlates with the level of gene induction
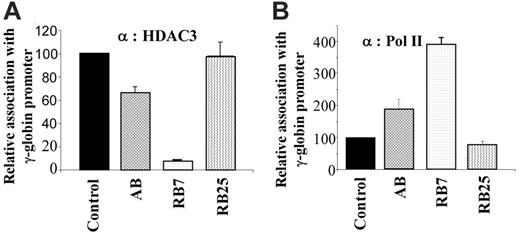
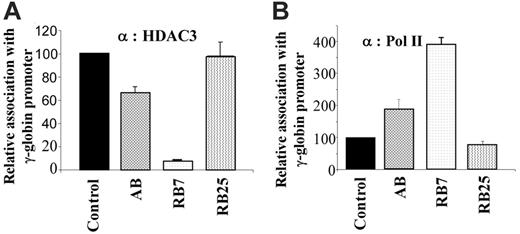
We further examined whether there was a correlation between the extent of HDAC3 displacement and the efficiency of induction by the γ-globin–inducing compounds in the dual luciferase assay. For comparison, we evaluated the γ-globin gene inducers RB7 (which had higher potency) and arginine butyrate (AB), and used the inactive SCFAD RB25 as a negative control. Chromatin IP using antibodies to HDAC3 and RNA polymerase II demonstrated an inverse correlation when cells were cultured in the presence of the γ-globin gene–inducing compounds RB7 and AB. Quantitative ChIP assays demonstrated that while RB7 resulted in a more than 90% decrease in HDAC3 levels at the γ-globin gene promoter, and AB treatment resulted in approximately 30% reduction in HDAC3 binding compared with untreated control conditions, exposure to the inactive SCFAD RB25 produced no change in HDAC3 binding at the γ-globin gene promoter (Figure 4A). These findings correlated with the simultaneous and inversely proportional recruitment of RNA polymerase II to the promoter in response to both active compounds. Treatment with RB7 and AB resulted in a 4-fold and 2-fold greater recruitment of RNA polymerase II to the γ-globin gene promoter, respectively, while RB25 exposure did not alter recruitment of RNA polymerase II to the promoter (Figure 4B).
Relative association of class I HDACs at the integrated β- and γ-globin gene promoters. Antibodies to HDAC1, HDAC2, and HDAC3 were used to immunoprecipitate chromatin isolated from GM979 cells cultured in either control conditions or 200 μM RB7. Precipitated DNA was amplified and quantitated by real-time PCR using primers flanking either the β-globin gene promoter (A) or the γ-globin gene promoter (B). For each HDAC antibody used, the presence of the HDAC protein bound at the promoter is represented relative to its control (arbitrarily set at 100). Data represent average of 3 independent experiments; error bars represent the standard error of the mean. (C) Agarose gel showing relative association of HDAC1, HDAC2, and HDAC3 with the beta- and gamma-globin promoters in GM979 cells cultured in either control conditions (C), or in the presence of 200 μM RB7 (R). Results are representative of 3 independent experiments. (Di) ChIP of NCoR at the γ-globin gene promoter. NCoR association with the human γ-globin gene promoter in control and RB7 (200 μM)–treated cells; values are reported relative to the control and are an average of 3 experiments. Error bar indicates standard error of the mean. (Dii) Dissociation of HDAC3/NCoR complex after RB7 treatment. Immunoprecipitation was carried out using HDAC3 antibodies. The immunoblot was performed using antibodies to HDAC3 and NCoR. Lane 1: control cells; lane 2: 200 μM RB7 treatment. (E) Acetylation status of β- and γ-globin gene promoters. Change in acetylation status of histones H3 and H4 at the β- and γ-globin gene promoter 24 hours after GM979 cells were treated with RB7. Values are represented as a percentage of the untreated control (C) and are an average of 3 independent experiments. Error bars represent the standard errors of the mean.
Relative association of class I HDACs at the integrated β- and γ-globin gene promoters. Antibodies to HDAC1, HDAC2, and HDAC3 were used to immunoprecipitate chromatin isolated from GM979 cells cultured in either control conditions or 200 μM RB7. Precipitated DNA was amplified and quantitated by real-time PCR using primers flanking either the β-globin gene promoter (A) or the γ-globin gene promoter (B). For each HDAC antibody used, the presence of the HDAC protein bound at the promoter is represented relative to its control (arbitrarily set at 100). Data represent average of 3 independent experiments; error bars represent the standard error of the mean. (C) Agarose gel showing relative association of HDAC1, HDAC2, and HDAC3 with the beta- and gamma-globin promoters in GM979 cells cultured in either control conditions (C), or in the presence of 200 μM RB7 (R). Results are representative of 3 independent experiments. (Di) ChIP of NCoR at the γ-globin gene promoter. NCoR association with the human γ-globin gene promoter in control and RB7 (200 μM)–treated cells; values are reported relative to the control and are an average of 3 experiments. Error bar indicates standard error of the mean. (Dii) Dissociation of HDAC3/NCoR complex after RB7 treatment. Immunoprecipitation was carried out using HDAC3 antibodies. The immunoblot was performed using antibodies to HDAC3 and NCoR. Lane 1: control cells; lane 2: 200 μM RB7 treatment. (E) Acetylation status of β- and γ-globin gene promoters. Change in acetylation status of histones H3 and H4 at the β- and γ-globin gene promoter 24 hours after GM979 cells were treated with RB7. Values are represented as a percentage of the untreated control (C) and are an average of 3 independent experiments. Error bars represent the standard errors of the mean.
SCFAD(s) induce γ-globin gene expression in cord blood erythroid progenitor cells
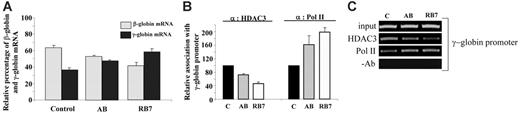
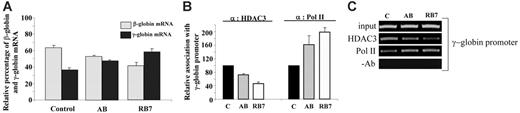
To determine whether the observed molecular effects of RB7 and AB on the human γ-globin gene were operable in primary erythroid cells, we investigated the effects of these SCFADs on erythroid progenitors cultured from cord blood. Following exposure to AB (100 μM) or RB7 (20 μM), γ-globin mRNA increased 30% and 60%, respectively, above levels observed in untreated control cells (Figure 5A). Chromatin IP performed in these primary cells at the same time point produced the molecular effects observed in the GM979 cells. HDAC3 occupancy at the γ-globin gene promoter in primary erythroid cells cultured in the presence of AB was reduced to 72% of HDAC3 occupancy in control cells, while HDAC3 binding in cells cultured in RB7 was reduced to 47% of control at this promoter. A reciprocal and proportional increase in RNA polymerase II association with the γ-globin gene promoter was observed following treatment with the test compounds; a 63% increase was found in AB-treated cells, and a 99% increase was found in cells cultured with RB7 (Figure 5B-C).
Chromatin immunoprecipitation and quantitative PCR of the integrated γ-globin gene promoter. GM979 cells were treated with active SCFADs (RB7 or AB) and an inactive SCFAD, RB25. Antibodies against HDAC3 (A) or RNA polymerase II (B) were used for chromatin IP. The results are following a 24-hour exposure to the drugs: control, RB7 (200 μM), arginine butyrate (2500 μM), and RB25 (200 μM). Values are an average of 3 independent experiments and are reported relative to the control. Error bars indicate the standard errors of the mean.
Chromatin immunoprecipitation and quantitative PCR of the integrated γ-globin gene promoter. GM979 cells were treated with active SCFADs (RB7 or AB) and an inactive SCFAD, RB25. Antibodies against HDAC3 (A) or RNA polymerase II (B) were used for chromatin IP. The results are following a 24-hour exposure to the drugs: control, RB7 (200 μM), arginine butyrate (2500 μM), and RB25 (200 μM). Values are an average of 3 independent experiments and are reported relative to the control. Error bars indicate the standard errors of the mean.
Knockdown of endogenous HDAC3 is sufficient to activate transcription from the γ-globin gene promoter
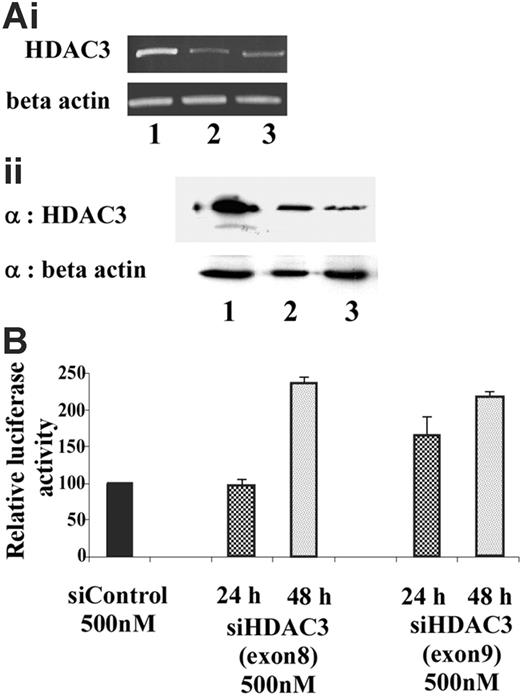
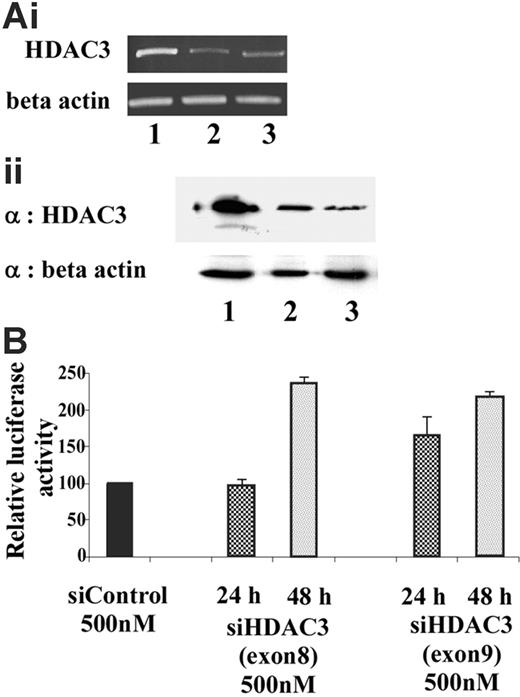
To determine whether HDAC3 plays a critical role in maintaining the γ-globin promoter in a transcriptionally silent state, siRNA was used to selectively target and knockdown HDAC3 levels. Endogenous HDAC3 levels in GM979 cells were suppressed using siRNA directed toward exons 8 and 9 of the open-reading frame of murine HDAC3. A scrambled siRNA was used as a negative control to exclude nonspecific effects of the siRNA oligomer or the transfection protocol. HDAC3-specific siRNA treatment resulted in a reduction in HDAC3 mRNA and protein levels within 24 hours (Figure 6A). Reciprocal induction of γ-globin promoter activity, detected by an increase in the firefly–renilla luciferase ratio, was observed following treatment with the HDAC3 siRNA (Figure 6B). While approximately 1.5-fold induction was observed 24 hours after the siRNA treatment, a significant effect (2.5-fold, P ≤ .03) was observed 48 hours following the treatment.
Relative expression of β-globin and γ-globin mRNA in human primary erythroid cells. (A) Globin mRNA analysis by RT-PCR. Cord blood erythroid progenitors were cultured either in control conditions, in the presence of AB (100 μM), or in the presence of RB7 (20 μM), and globin expression was assayed by RT-PCR. Values represent an average of 3 experiments, from independent cord blood specimens; error bars represent the standard errors of the mean. An increase in γ-globin mRNA, and corresponding decrease in the β-globin mRNA, was observed with SCFAD treatment compared with untreated controls. (B) Chromatin immunoprecipitation and quantitative PCR at the endogenous γ-globin gene promoter. Relative association of HDAC3 and RNA polymerase II at the γ-globin gene promoter in cord blood erythroid progenitors determined under control conditions, or with RB7 or AB treatment; error bars represent the standard errors of the mean. (C) Agarose gel showing relative association of HDAC3 and RNA polymerase II with the γ-globin promoter in cord blood erythroid progenitors cultured in either control conditions, AB (100 μM), or RB7 (20 μM). Results are representative of 3 independent experiments.
Relative expression of β-globin and γ-globin mRNA in human primary erythroid cells. (A) Globin mRNA analysis by RT-PCR. Cord blood erythroid progenitors were cultured either in control conditions, in the presence of AB (100 μM), or in the presence of RB7 (20 μM), and globin expression was assayed by RT-PCR. Values represent an average of 3 experiments, from independent cord blood specimens; error bars represent the standard errors of the mean. An increase in γ-globin mRNA, and corresponding decrease in the β-globin mRNA, was observed with SCFAD treatment compared with untreated controls. (B) Chromatin immunoprecipitation and quantitative PCR at the endogenous γ-globin gene promoter. Relative association of HDAC3 and RNA polymerase II at the γ-globin gene promoter in cord blood erythroid progenitors determined under control conditions, or with RB7 or AB treatment; error bars represent the standard errors of the mean. (C) Agarose gel showing relative association of HDAC3 and RNA polymerase II with the γ-globin promoter in cord blood erythroid progenitors cultured in either control conditions, AB (100 μM), or RB7 (20 μM). Results are representative of 3 independent experiments.
Discussion
Mechanisms that up-regulate fetal globin gene expression for potential application to therapy of the β-globin disorders have been elusive, except for the induction of γ-globin gene expression through binding of the stage selector complex in the proximal γ-globin promoter and FKLFs in fetal stage erythroblasts.7,8,10,11 As HDAC inhibitors often also augment expression of the fetal globin genes, creation of an “accessible” chromatin structure was initially considered to be required for γ-globin gene induction.25,26 HDAC inhibitors typically induce acetylation of bulk histones and result in multiple cellular effects, including cell growth arrest, which is counterproductive and undesirable in the β-hemoglobinopathies, particularly in the β-thalassemias wherein accelerated apoptosis occurs during early erythroid differentiation.27 However, we and others have reported that the magnitude of HDAC inhibition does not correlate directly with the magnitude of induction of γ-globin expression by these compounds, and several SCFADs activate the γ-globin gene promoter but do not induce generalized histone acetylation.24-26 These observations have suggested that a specific HDAC subtype, or a specific site of acetylation, may be more important for γ-globin gene promoter activation than general effects on histone acetylation.
Knockdown of endogenous HDAC3 in GM979 cells. (Ai) RT-PCR analysis of HDAC3 mRNA levels 24 hours after transfection of the siRNA. (Top panel) HDAC3 levels. (Bottom panel) β-actin levels. Lane 1: control (nonspecific siRNA); lane 2: siHDAC3 (exon 8); lane 3: siHDAC3 (exon 9). (Aii) (Top panel) Immunoblot showing HDAC3 levels after cells were treated with siRNA. Lane 1: control (nonspecific siRNA); lane 2: siHDAC3 (exon 8), lane 3: siHDAC3 (exon 9). (Bottom panel) Immunoblot using β-actin as a loading control. (B) Relative induction of the Aγ-globin gene following siHDAC3 treatment. Ratios of firefly luciferase–renilla luciferase of cells treated with siHDAC3 (exons 8 and 9), represented as a percentage of the control transfection. Results shown were an average of 2 independent experiments and were recorded either at 24 hours (checked) or 48 hours (dotted) following siRNA treatment; error bars represent the standard errors of the mean.
Knockdown of endogenous HDAC3 in GM979 cells. (Ai) RT-PCR analysis of HDAC3 mRNA levels 24 hours after transfection of the siRNA. (Top panel) HDAC3 levels. (Bottom panel) β-actin levels. Lane 1: control (nonspecific siRNA); lane 2: siHDAC3 (exon 8); lane 3: siHDAC3 (exon 9). (Aii) (Top panel) Immunoblot showing HDAC3 levels after cells were treated with siRNA. Lane 1: control (nonspecific siRNA); lane 2: siHDAC3 (exon 8), lane 3: siHDAC3 (exon 9). (Bottom panel) Immunoblot using β-actin as a loading control. (B) Relative induction of the Aγ-globin gene following siHDAC3 treatment. Ratios of firefly luciferase–renilla luciferase of cells treated with siHDAC3 (exons 8 and 9), represented as a percentage of the control transfection. Results shown were an average of 2 independent experiments and were recorded either at 24 hours (checked) or 48 hours (dotted) following siRNA treatment; error bars represent the standard errors of the mean.
To further elucidate potential mechanisms of SCFAD-inducing activity and any correlation with HDAC inhibition, we compared the activities of several structurally distinct SCFAD compounds, identified through computational modeling, to the activity of the HDAC-inhibitor butyrate, regarding isozyme-specific inhibition of immunoprecipitated HDAC1, HDAC2, or HDAC3, in vitro. No general or isozyme-specific HDAC-inhibitory activity of any of the new SCFADs was observed, and arginine butyrate showed no isozyme specificity. Yet, in the dual luciferase reporter assay, as well as in the in vivo model, RB7 significantly enhanced γ-globin expression 5-fold and 5.9-fold over the control, respectively; butyrate enhanced the promoter activity by only 2.5-fold in the luciferase assay. These results are consistent with a lack of correlation between HDAC inhibition and γ-globin transcription as previously observed.25 Unexpectedly, the most active inducer of the γ-globin gene promoter in both the reporter cell line and the primary erythroid cell cultures, compound RB7, exhibited a specific, dose-dependent enhancement of HDAC3 activity, rather than inhibition. Whether there is any functional significance to the stimulatory effect of RB7 on HDAC3 activity in vitro is under investigation.
To examine the local effects of these γ-globin gene–inducing SCFAD compounds on globin gene promoters, we initially investigated transcription factor and cofactor occupancy at integrated human β- and γ-globin promoters in the GM979 cell line, following exposure to the new SCFADs. Chromatin IP revealed no change in occupancy of HDAC1 and HDAC2 at the β-or γ-globin gene promoters in response to RB7 or AB treatment. However, HDAC3 was selectively displaced from the γ-globin promoter following RB7 and AB treatment of GM979 and primary human erythroid cells, thereby suggesting a role for the local association of HDAC3 in silencing of the γ-globin gene.
Consistent with previous reports,36,38,39 we found that HDAC3 and its corepressor NCoR exist as a complex in GM979 cells prior to treatment with the SCFAD. Twenty-four hours after treatment with SCFADs, we observed both the displacement of HDAC3 and NCoR from the γ-globin gene promoter and a reduction in the physical association between these proteins. It is not yet clear whether the dissociation follows the displacement of the complex or whether the test compound interferes with HDAC3-NCoR protein interaction, secondarily resulting in the displacement of both components of the complex from the promoter. Concurrent with the loss of the HDAC-NCoR complex, there was a significant increase in the acetylation status of histones H3 and H4 at the γ-globin gene promoter, indicating development of a more accessible chromatin state that likely facilitates access to the transcriptional machinery. The loss of the HDAC3/NCoR repressor complex from the γ-globin gene promoter was associated temporally with recruitment of RNA polymerase II to the promoter. No significant changes in local histone acetylation were observed at the β-globin gene promoter in response to SCFAD treatment, and no induction of β-globin gene expression was observed.
We further demonstrated that the same pattern of molecular events occurs at the β/γ-globin gene locus in cord blood erythroid progenitors when γ-globin gene expression is induced by AB or RB7. Although these primary cells at baseline expressed a higher relative proportion of γ-globin promoter-driven gene expression compared with GM979 cells, both compounds induced γ-globin gene expression over levels in control cells, displaced HDAC3 from the γ-globin gene promoter, and simultaneously induced recruitment of RNA polymerase II to the γ-globin gene promoter in the primary erythroid cells.
Additional evidence supporting a pivotal role of HDAC3 in SCFAD-mediated reactivation of the γ-globin promoter was obtained from RNA knockdown experiments. We demonstrate that significant depletion of total cellular HDAC3 protein is sufficient to increase transcription by the γ-globin gene promoter, but not the β-globin promoter. Silencing of HDAC3 in these cells reproduced the induction of the γ-globin gene promoter, although to a slightly lesser extent than did RB7 treatment. This less robust effect may be explained on the basis of incomplete knockdown of the HDAC3 protein. The approximate 20% of residual HDAC3 protein after siRNA treatment may be sufficient to prevent a more robust induction of this promoter.
Our findings suggest that the unexpected deacetylase-enhancing activity of RB7 (observed in in vitro assays) appears to be independent of its ability to displace HDAC3 from the γ-globin gene promoter, as another short-chain fatty acid, butyrate, had opposite effects on HDAC3 activity, yet both compounds resulted in displacement of HDAC3 from the γ-globin gene promoter. Although the in vitro enhancement of HDAC3 activity by RB7 is not yet correlated with its primary effect on γ-globin expression and HDAC3 displacement, this finding provides further evidence that generalized HDAC-inhibitory activity is not required for γ-globin induction by SCFADs.24-26
While SCFAD treatment results in reorganization of proteins at the γ-globin gene promoter with the displacement of the silencing complex HDAC3/NCoR, recruitment of RNA polymerase II, and a concurrent increase in the acetylation status of histones H3 and H4 at the promoter, it is not yet clear whether additional factors are recruited at the promoter to permit gene activation following these epigenetic changes. Further investigation is under way to identify potential factors that may participate in the recruitment of HDAC3/NCoR to the γ-globin gene promoter in the silenced state. Our results clearly establish HDAC3 as a critical molecule mediating the response of γ-globin gene expression to chemical inducers of HbF, and further demonstrate that suppression of HDAC3 is sufficient to induce the γ-globin gene. Collectively, these findings suggest that HDAC3 occupancy at the γ-globin promoter may play a similarly central role in the developmental silencing of the fetal globin genes, and this potential mechanism requires further investigation. In summary, these studies identify molecular events in association with fetal globin gene activation by SCFADs and provide a new molecular target for development of more potent therapeutics for the treatment of β-globin gene disorders.
Prepublished online as Blood First Edition Paper, July 18, 2006; DOI 10.1182/blood-2005-12-010934.
Supported by NIH grants DK-52962 and HL-78276 (S.P.P.), Department of Defense grant DAMD17-03-1-0213 (D.V.F.), NIH grants HL-52243 and HL-73442 (C.H.L.), and the National Cancer Institute (CA101992) (D.V.F.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.