Abstract
The antithrombin A384S mutation has a relatively high frequency in the British population but has not been identified in other populations. This variant has been associated with cases of thrombotic disease, but its clinical relevance in venous thrombosis remained unclear. We have conducted a secondary analysis of the prevalence of the mutation in a large case-control study, including 1018 consecutive Spanish patients with venous thromboembolism. In addition, we evaluated its functional consequences in 20 carriers (4 homozygous). This mutation, even in the homozygous state, did not affect anti-Xa activity or antigen levels, and it only slightly impaired anti-IIa activity. Thus, routine clinical methods cannot detect this anomaly, and, accordingly, this alteration could have been underestimated. We identified this mutation in 0.2% of Spanish controls. Among patients, this variant represented the first cause of antithrombin anomalies. Indeed, 1.7% patients carried the A384S mutation, but 0.6% had any other antithrombin deficiency. The mutated allele was associated with an increased risk of venous thrombosis with an adjusted OR of 9.75 (95% CI, 2.2-42.5). This is the first study supporting that antithrombin A384S mutation is a prevalent genetic risk factor for venous thrombosis and is the most frequent cause of antithrombin deficiency in white populations.
Introduction
Thrombosis is the leading cause of death in western societies, with an annual incidence of 1:1000 persons in the United States and Western Europe.1 Because of the high incidence of mortality and morbidity, thrombotic disorders consume a significant portion of the national health care expenditure.2–5 The negative impact of thrombosis could be significantly reduced if at-risk persons were identified and managed prior to clinical manifestation.6
Considerable progress has been made in determining the molecular bases of inherited thrombophilia in the past 40 years. Since Egeberg's description of antithrombin deficiency in 1965, a number of monogenic autosomal dominant disorders have been well established as risk factors for venous thromboembolic disease. At the beginning of the 1980s, deficiencies of other anticoagulant proteins, such as protein C and protein S, were also linked to venous thrombosis. All these disorders are rare, but they strongly increase the risk of venous thrombosis (10- to 20-fold). They follow a dominant inherited pattern with variable penetrance, suggesting that venous thrombosis might be a monogenic disease. However, all these deficiencies together accounted only for 4% to 8% of consecutive patients with venous thrombosis and 16% to 20% of families with thrombophilia. In 1994, the discovery of the factor V (FV) Leiden and, 2 years later, the identification of the genetic variation affecting the prothrombin gene (G20210A), dramatically changed the molecular view of venous thrombosis. These polymorphisms are relatively frequent in the normal population (2%-10%), but they mildly increase the risk of venous thrombosis (3- to 5-fold). Moreover, many symptomatic patients simultaneously carry more than one genetic risk factor, supporting that thrombophilia represents a polygenic rather than a monogenic clinical phenotype. In addition, genetic factors interact with environmental risk factors, clearly reflecting that thrombophilia is a complex and multifactorial trait.7 However, there are still up to 60% of consecutive cases with venous thrombosis and 25% of families with thrombophilia with no recognized genetic risk factors. Unfortunately, despite considerable efforts during 2 decades evaluating hundreds of polymorphisms affecting hemostatic-related proteins, no further genetic risk factors for venous thrombosis have been clearly identified.8
The first mutation in the antithrombin gene was characterized in 1984.9 Since then, more than 127 different mutations have been identified, which mostly occur in the heterozygous state.10 All these data, together with animal models, support the key role of antithrombin in hemostasis, as the most important endogenous anticoagulant, and the pathologic consequences of its deficiency. The heterozygous state is a rare defect with an estimated prevalence in the healthy general populations between 1/1400 and 16/9669 (0.07%-0.16%).11 Deficiency of this potent anticoagulant in thrombophilic patients is estimated between 1% and 8% and lower in consecutive patients with venous thrombosis.12 As for any rare mutation, most of the identified mutations affecting the antithrombin gene appear as a single entry in the database.10 Surprisingly, an antithrombin variant, A384S (antithrombin Cambridge II), had a high frequency in a study of blood donors in western Scotland (1/630),13 but it had not been studied in other populations. This variant has been associated with cases of thrombotic diseases, but its clinical relevance in venous thrombosis remains unclear. The aim of this study was to evaluate the prevalence of this mutation, which determines a mild deficiency of the most important physiologic anticoagulant, in a Spanish population and its role in venous thrombosis in a large cohort of patients and controls.
Materials and methods
Patients and controls
Our study included 1018 unrelated white patients with venous thromboembolism. The control group of our study included 1018 unrelated people without a history of vascular or thromboembolic disease. The features of these groups were previously described.14 Briefly, patients with a first objectively confirmed episode of venous thromboembolism before the age of 75 years who consecutively entered into the anticoagulation clinics from 4 Spanish hospitals in a time window between 2 and 4 years were enrolled. Objective diagnoses were done by clinical probability, D-dimer levels, compression ultrasonography, ventilation perfusion lung scan, and, when necessary, phlebography or pulmonary angiography. Patients with known malignant disorders were excluded. Controls were randomly and prospectively selected to match with the 1018 cases by age, sex, race, and geographic distribution.
Demographic parameters (age and sex) were recorded for all subjects. Relevant clinical data (family history of venous thrombosis, recurrence, type of thrombotic event, location of the thrombosis, and other thrombotic risk factors such as oral contraceptives, hormone replacement therapy) were recorded in patients. Functional protein S was assayed in automated coagulometers. Total and free protein S were assayed using enzyme-linked immunoabsorbent assay (ELISA) methods. Antithrombin (anti-Xa activity) and protein C were measured in automated coagulometers using chromogenic methods. Lupus anticoagulant was investigated by using the Exner method, and antiphospholipid antibodies were screened by ELISA methods that used cardiolipin or phosphatidylserine as antigen. The analysis of antibodies to β2-glycoprotein I was performed using ELISA methods.
All included subjects gave their informed consent to enter the study, which was approved by the ethics committees for each participating institution and performed in accordance with the Declaration of Helsinki, as amended in Edinburgh in 2000.
Genetic analysis
Blood samples were obtained by venipuncture collection into 1:10 volume of trisodium citrate (Vacutainer; Becton Dickinson, Meylon, France). Platelet-poor plasma fractions were obtained by centrifugation at 4°C for 20 minutes at 2200g (within 5 minutes after blood collection). Genomic DNA was purified by standard procedures. The exon 6 of the antithrombin gene was amplified by polymerase chain reaction (PCR) using the primers AT6F2, 5′-TGAGGAATTGCTGTGTCTGTG-3′, and AT6B, 5′-AGAGGTGCAAAGAATAAGAA-3′. The A384S mutation (antithrombin Cambridge II) was genotyped by PCR–allele-specific restriction assay (PCR-ASRA) using PvuII (New England Biolabs, Hitchin, England). Confirmation of genotypes was performed by sequencing. PCR products were purified from 1.5% agarose gels using Ultraclean Gel Spin (MoBio, Solana Beach, CA). The sequence reaction was performed with the ABI Prism Big Dye Terminator Cycle sequencing kit on an automated sequencer type 377 (Perkin-Elmer Applied Biosystems, Washington Chesire, United Kingdom), with forward and reverse primers used for amplification. FV Leiden, prothrombin G20210A, and protein Z-dependent protease inhibitor (ZPI) R67X polymorphisms were determined in patients and controls, as described.14,15
Measurement of plasma antithrombin activity and levels
Antithrombin activity was determined by chromogenic methods in citrated plasma from 20 carriers of the antithrombin Cambridge II mutation, including 4 in homozygous state, and 17 controls without this mutation. Anti-Xa assays were performed with heparin, bovine FXa, and S-2765 chromogenic substrate (Instrumentation Laboratory, Milan, Italy). Anti-IIa assays were performed with heparin, bovine thrombin, and chromogenic substrate (ethyl-malonyl-S-Pro-Arg-pNA.AcOH–CBS 61.50-; STA Antithrombin; Diagnostica Stago, Asnières, France). Both assays were performed with unfractionated heparin and low molecular weight heparin (Bemiparin) (both from Laboratorios Rovi S.A., Madrid, Spain).
Antigen levels were measured by immunodiffusion (Laurell Technologies, New Wales, PA).
Results are expressed as a percentage of a pool generated with plasma from 100 healthy controls.
Statistical analysis
Continuous variables were expressed as mean ± SD and as percentages for categorical variables. Comparison between groups was done using an unpaired, Student t test or a chi-square test, as appropriate. Univariate statistical analysis was performed by the chi-square test. The strength of the association of major risk factors and the polymorphism with the occurrence of disease was estimated using 2 × 2 tables and calculating the odds ratio (OR) with the EpiInfo software (Centre for Disease Control, Atlanta, GA) and the Cornfield method for the calculation of 95% confidence intervals (CIs). Multivariate analysis was performed using multiple logistic regression and included all the significant covariates and age in a single step (Enter method), with the SPSS statistical package for Windows 8.0 software (Chicago, IL). Differences with a 2-tailed P value less than .05 were considered as being statistically significant.
Results
Genetic identification
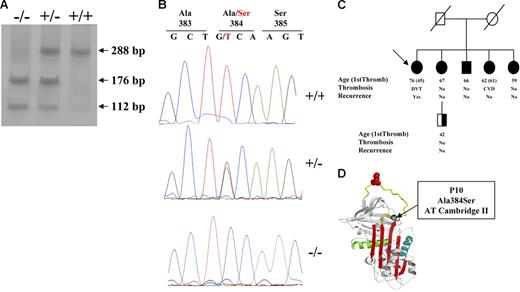
The G1246T mutation in the antithrombin gene (A384S; antithrombin Cambridge II) is clearly identified by PCR-ASRA (Figure 1A). Because antithrombin Cambridge I (G1246C; A384P)16 also gives the same restriction pattern, it is necessary to confirm the mutation by sequence analysis (Figure 1B). No subject was a carrier of the antithrombin Cambridge I.
Identification of antithrombin Cambridge II (A384S). (A) PCR-ASRA pattern. (B) Sequence analysis. (C) Pedigree of the family with 5 antithrombin Cambridge II homozygous subjects, with proband identified with an arrow. Type of thrombotic event, age, and recurrence are indicated (NA indicates not available). Filled symbols represent the genetic anomalies identified in this family. (D) Localization of the residue affected by the mutation in the reactive loop (A384S; P10).
Identification of antithrombin Cambridge II (A384S). (A) PCR-ASRA pattern. (B) Sequence analysis. (C) Pedigree of the family with 5 antithrombin Cambridge II homozygous subjects, with proband identified with an arrow. Type of thrombotic event, age, and recurrence are indicated (NA indicates not available). Filled symbols represent the genetic anomalies identified in this family. (D) Localization of the residue affected by the mutation in the reactive loop (A384S; P10).
Case-control study
Demographic, clinical, and genetic data of cases and controls are presented in Table 1. These features did not significantly differ in the samples from the 4 hospitals (data not shown).
The median age of patients at the time of the first thrombotic event was 45 years (interquartile range, 33-58 years), and the ratio of men to women was 1:1. Approximately two thirds of the patients had a single thrombotic episode (280 of 1018 had recurrent episodes of venous thrombosis), 218 patients reported risk factors such as oral contraceptive use, the presence of antiphospholipid antibodies, hormone replacement therapy, deficiency of antithrombin, protein C or protein S, and 177 patients (17.4%) reported a familial history of thrombosis. Antithrombin deficiency was diagnosed in 6 patients (0.6%). This diagnosis was performed by functional evaluation of anti-Xa activity. This method is routinely used in clinical practice, because it is the most reliable test to identify congenital antithrombin deficiency.17 The prevalence of the classic prothrombotic polymorphisms was similar to that described in other series. Thus, the FV Leiden was present in 145 patients (3 in homozygous state) (14.2%), and the prothrombin 20210A allele was identified in 101 patients (7 in homozygous state) (9.9%) (Table 1).
Controls had a significant lower prevalence of these prothrombotic polymorphisms: 31 (3.0%) were heterozygous for the FV Leiden and 26 (2.6%) for the prothrombin 20210A allele (Table 1). Accordingly, these polymorphisms increased 5.29-fold (FV Leiden) and 4.20-fold (prothrombin 20210A) the risk of venous thrombosis (P < .001; 95% CI, 3.5-8.0 and P < .001; 95% CI, 2.7-6.7, respectively). In this case-control study, we recently identified a new prothrombotic nonsense polymorphism affecting the ZPI gene (ZPI R67X).14 Carriers of the ZPI 67X allele had a 3.40-fold risk of developing venous thrombosis (Table 1). The clinical features associated with these prothrombotic polymorphisms were described elsewhere.14
The antithrombin Cambridge II mutation was only identified in 2 controls (0.2%). Both controls were young women aged 31 and 42 years from distant regions of Spain (Barcelona and Murcia). In contrast, this mutation was present in 17 patients with venous thrombosis (1.7%). Thus, this mutation increased 8.63-fold the risk of venous thrombosis (P < .001; 95% CI, 1.9-54.2) (Table 1). In all cases, the mutation was identified in a heterozygous state.
The presence of this mutation did not significantly modify the age at the first thrombotic event (49 years in carriers and 45 years in noncarriers). Moreover, the antithrombin Cambridge II mutation did not increase the rate of recurrence (23.5% in carriers and 27.6% in noncarriers, P = .718) and was not associated with an increased familial history of venous thrombosis (11.8% in carriers and 17.5% in noncarriers, P = .544). Finally, the percentage of patients who displayed an additional risk factor was similar in carriers and noncarriers of this mutation (17.6% versus 21.5%, respectively, P = .703).
No patient carrying the antithrombin Cambridge II mutation had the prothrombin 20210A variant or the ZPI nonsense allele. However, 2 patients simultaneously displayed the FV Leiden and antithrombin Cambridge II mutation. Both were male patients who had their first episode of deep venous thrombosis at 46 and 59 years without additional risk factors, and they reported neither recurrence nor familial history of thrombosis. No control had such a combination.
Multivariate analysis, including age, prothrombotic polymorphisms (FV Leiden, PT G20210A, and ZPI R67X), and the antithrombin A384S mutation, confirmed that the antithrombin Cambridge II is an independent and strong risk factor for venous thrombosis (P = .002; OR = 9.75; 95% CI, 2.2-42.5) (Table 1).
In addition, we have analyzed the role of the antithrombin Cambridge II limited to persons with idiopathic first thrombosis, without additional genetic or acquired risk factors. Four hundred thirty-four cases fulfilled these features (median age, 52 years; 56.2% men). The subgroup of 952 controls selected for the analysis of cases with idiopathic venous thrombosis were chosen by excluding controls carrying the genetic risk factors (FV Leiden, PT 20210, and ZPI R67X) (median age, 47 years; 50.3% men). The antithrombin Cambridge II mutation was identified in 9 patients but 2 controls, supporting that this mutation is a genetic risk factor for idiopathic venous thrombosis (P < .001; OR, 10.06; 95% CI, 2.03-67.63).
Finally, we identified a family with 5 members carrying the antithrombin Cambridge II in homozygous state. A 7-year-old woman reported recurrent deep venous thrombosis, with the first episode at the age of 45 years. Moreover, one of her sisters also had ischemic cerebrovascular disease. However, 3 other homozygotes relatives did not report thrombotic episodes (Figure 1C).
Antithrombin values
We have evaluated the functional consequences of this mutation in 37 subjects, 17 controls, and 20 carriers of the antithrombin A384S mutation (16 heterozygous and 4 homozygous). Both antigen and anti-Xa activity (in the presence of unfractionated heparin and low molecular weight heparin) of carriers and noncarriers are within the normal range. We did not observe significant differences in these parameters between carriers and noncarriers or between heterozygous and homozygous states (Table 2)Statistical differences were only achieved when comparing the anti-Xa activity in the presence of unfractionated heparin from noncarriers and homozygous (100.2% ± 4.3% versus 91.7% ± 10.5%, respectively; P = .016). However, the anti-IIa activity in the presence of unfractionated heparin was mild but significantly impaired by the Cambridge II mutation (100.0% ± 7.9%; range, 87.5%-118.0% for control versus 79.3% ± 7.8%; range, 66.9%-90.8% for carriers; P < .001) (Table 2). In addition, we observed a significant gene-dosage effect on this parameter. Thus, the anti-IIa activity of heterozygous carriers in the presence of unfractionated heparin was 81.8% ± 6.6% (range, 70.6%-90.8%), whereas homozygous subjects displayed 69.5% ± 2.4% (range, 66.9%-72.6%); P = .002 (Table 2). The functional consequences of the antithrombin Cambridge II mutation on anti-IIa activity were reduced when using low molecular weight heparin, but differences between carriers and noncarriers and between heterozygous and homozygous state still reached statistical significance (Table 2).
Discussion
A major challenge for biomedicine in the genomic era is to identify DNA modifications responsible for variation in complex diseases (eg, physiologic risk factors related to thrombotic disease). Identifying the loci that affect susceptibility of those complex phenotypes among the overall polymorphisms that exist in the human genome should bring about improved understanding of such diseases and provide new targets for clinical intervention. To date, most candidate gene studies have directly analyzed the association between disease status and a number of candidate SNPs that have known or predicted functional consequences.18 This design can easily determine whether a given variant confers significant risk of disease. For venous thrombosis, genetic risk factors have been identified in different candidate genes (antithrombin, protein C, protein S, FV, PT).
Herein, we present the results of a classic case-control association study in unrelated Spanish subjects designed to estimate the contribution of genetic and environmental factors to the risk of thrombotic disease. The results of this study show an adjusted OR of 9.75 of developing venous thromboembolism associated with the G1246T mutation in the antithrombin gene (A384S, antithrombin Cambridge II). The statistical power of our study is high because of the large sample size (92.5% and 79.5% with a confidence level of 95% and 99%, respectively). These results suggest that antithrombin Cambridge II itself is an independent risk factor for venous thromboembolism. These results, the wide CIs for the ORs in the case-control study and the identification of a family with 5 carriers of this mutation in homozygous state with moderate thrombotic clinic support that this variant lies between a mild prothrombotic polymorphism (such as FV Leiden) and a rare but severe mutation (such as an antithrombin type I deficiency).
Congenital antithrombin deficiency is an autosomal hereditary disorder with variable penetrance. Type I deficiencies usually associate with a severe risk for venous thrombosis. In contrast, overall type II deficiencies usually have milder thrombotic risk.19 Thus, type II deficiencies account for approximately 40% of patients with venous thrombosis and antithrombin deficiency, but 88% of identified antithrombin-deficient persons.13 Our study has important implications because it reveals another possible kind of antithrombin deficiency that is featured by no functional or antigenic deficiency, at least by using classic screening methods for antithrombin deficiency, which however have impaired functional activity under thrombotic situations. The missense substitution of A384S, discovered in 1991 and called antithrombin Cambridge II, is the result of a G1246T point mutation in exon 6 of the antithrombin gene.20 This mutation has been classically included in the group of type II deficiencies, affecting the P10 proximal hinge region of the reactive center loop of antithrombin (Figure 1D).10 However, the replacement of this residue by serine does not cause a complete loss of activity. Certainly, all available data from 41 carriers (from the study of Perry et al21 and this study) support that this mutation, even in the homozygous state, does not affect the anti-Xa activity (98.6% ± 15.4%). Moreover, 62 carriers, including 4 homozygous subjects (from the studies of Tait et al,13 Perry et al,21 and this study) have normal plasma levels of antithrombin antigen (99.5% ± 13.1%). Because these are the methods commonly used to identify antithrombin deficiency,17,22 we can understand why this mutation has not been identified in other populations, and, probably, its prevalence had been underestimated. Moreover, this variant only slightly impaired the anti-IIa activity (74.3% ± 8.5%) of 60 carriers (from the studies of Tait et al,13 Perry et al,21 and this study). Accordingly, plasma methods for detection of antithrombin deficiency might be complemented with the genetic test for G1246T point mutation in the antithrombin gene to avoid the underestimation and misdiagnosing antithrombin-deficient persons. This is particularly important, because antithrombin Cambridge II is the most frequent cause of deficiency of this serpin. Indeed, it is present in 0.2% of the normal population of at least 2 different white populations. Note that this percentage is similar to the antithrombin deficiency caused by the rest of the mutations identified so far. Moreover, this mutation was found in 1.7% of patients with venous thrombosis, more than twice the prevalence of all other antithrombin deficiencies in our study (0.6%). The relevance of this finding is not only restricted to patients with thrombosis but also to asymptomatic relatives and offspring that may carry the same genetic factor. At present, the most reasonable course of action is to carry out thromboprophylaxis under risk situations.23,24 Although the thrombotic risk associated with this mutation is milder than that associated with other antithrombin deficiencies, asymptomatic relatives carrying the G1246T mutation in the antithrombin gene could benefit from antithrombotic prophylaxis in risk situations.
Finally, an accurate diagnosis of antithrombin Cambridge II could have potential therapeutic relevance. The conservative A384S mutation at the proximal hinge (P10 position) of the reactive center loop of antithrombin slows the initial step of loop insertion because of steric effects and to the buckling of the strand that tip the balance from inhibitory to substrate in the presence of heparin.25 Thus, in the presence of full-length heparin the Cambridge II mutation displays a 3- and 7-fold increase in the stoichimetry of inhibition of FXa and thrombin, respectively.25 Our functional studies in blood samples from carriers confirm that the deleterious effect of this mutation is particularly evident with unfractionated heparin. Accordingly, further studies must be performed to clarify whether anticoagulant therapy of carriers with this mutation should avoid unfractionated heparin, as suggested elsewhere.25
Potential limitations might have influenced our findings. First, the relatively low prevalence of this mutation explains the very wide confidence intervals of our study. Moreover, although the size of our sample is large, the selection of patients and controls from different hospitals is a methodologic weakness of our study. In addition, there are slight variations in the anti-IIa activity observed in our study compared with that from previous studies which may be attributable to different factors, such as the concentration and type of unfractionated heparin, the chromogenic substrate, the source of FIIa, or slight methodologic variations. Therefore, confirmatory studies from other large case-control studies are critical to define relative risks, likelihood of recurrence, and prevalence of this mutation in other populations to encourage the inclusion of this mutation in routine thrombophilic studies to improve diagnosis and prevention strategies. In addition, it should be profitable to investigate the possible joint effects of the G1246T variant with other risk factors, such as FV Leiden, prothrombin G20210A, or ZPI R67X polymorphisms, and the effectiveness of anticoagulant treatment with heparins in carriers of this common mutation.
Authorship
Contribution: J.C., I.A., J.F., R.L., and V.V. designed the research; D.H.-E., R.G.-C., A.O., J.M., A.M., M.G., V.R., J.R.G.-P., J.M., E.P.-C, I.S., and M.G. analyzed the data; and J.C. and J.M.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Javier Corral, Centro Regional de Hemodonación, C/Ronda de Garay s/n; Murcia 30003, Spain; e-mail: javier.corral@carm.es
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Fundación Séneca (00583/PI/04, SAF2003-00840, and SAF2006-06212) (Ministerio de Ciencia y Tecnología [MCYT] and Fondo Europeo de Desarollo Regional [FEDER]), by Redes temáticas (RETICS) (Red Cardiovascular [RECAVA]) from Instituto de Salud Carlos III (ISCIII), and by Programa d'Estabilització d'Investigador de la Direcció d'Estrategia i Coordinació del Departament de Salut (Generalitat de Catalunya) (J.M.S.). A.O. has a Fondo de Investigaciones Sanitarias (FIS) predoctoral fellowship. D.H.-E. has an FIS postdoctoral fellowship.