Abstract
The Epstein-Barr virus (EBV) transcriptional coactivator EBNALP specifically associates and colocalizes with Hsp72 in lymphoblastoid cell lines. We now find that overexpression of Hsp72 more than doubled EBNALP coactivation with EBNA2 of a transfected EBV LMP1 promoter in B lymphoblasts, did not affect EBNA2 or EBNALP protein levels, and strongly up-regulated EBNA2 and EBNALP coactivation of LMP1 protein expression from the endogenous EBV genome in latency I infected Akata cells. The Hsp72 ATP, protein binding, and the C-terminal regulatory domains were required for full activity. An EBNALP deletion mutant, EBNALPd45, which does not associate with Hsp72, coactivated with EBNA2, but was not affected by Hsp72 overexpression, despite Hsp72 up-regulation of wild-type EBNALP coactivation with EBNA2 at all levels of EBNALP expression, indicating the importance of Hsp72 association with EBNALP for Hsp72 up-regulation of coactivation. Of importance, a 90% RNAi knockdown of Hsp72 reduced EBNALP coactivation with EBNA2 of transfected EBV LMP1 and Cp promoters by approximately 50%. Overexpression of the Hsp72 C-terminal interacting and regulatory protein, CHIP, strongly down-regulated EBNALP coactivation, independently of CHIP ubiquitin ligase activity. CHIP effects were Hsp72 dependent, indicating a background downmodulating role for CHIP in Hsp72 augmentation of EBNA2 and EBNALP coactivation. Based on these and other cited data, we favor a model in which Hsp72 chaperones EBNALP shuttling of repressors from EBNA2-enhanced promoters.
Introduction
Epstein-Barr virus (EBV) causes lymphoproliferative diseases in immune-deficient individuals, African Burkitt lymphoma (BL), Hodgkin disease (HD), and nasopharyngeal carcinoma (for review see Kieff1 and Rickinson and Kieff2 ). EBV infection of B lymphocytes results in expression of EBV nuclear antigen proteins (EBNAs), latent infection membrane proteins (LMPs), noncoding small RNAs, Bam A rightwards transcripts, and microRNAs3 and continuous lymphoblast cell proliferation. EBNALP, EBNA2, EBNA3A, EBNA3C, and LMP1 are critical for conversion of primary B lymphocytes to lymphoblast cell lines (LCLs). EBNA2 and EBNALP are the first EBV genes expressed after B-lymphocyte infection, are encoded by alternative splice products of the same pre-mRNA, and coordinately activate transcription from cell and virus promoters.1
EBNA2 activates specific cell and virus promoters and EBNALP potentiates EBNA2 effects.4–13 EBNA2 interacts with specific enhancer elements through cell DNA-binding proteins, including RBP-Jκ, PU.1, and AUF1.14–17 The EBNA2 C-terminal acidic activation domain (E2AD) recruits cell proteins important for basal and activated transcription.8,18–21 A second EBNA2 activation domain, E2AD2, also contributes to promoter up-regulation.22,24 EBNALP is comprised of 4 tandem 66 amino acid repeats and 45 unique C-terminal amino acids. The EBNALP repeats interact at a very low level with EBNA2 and coactivate transcription, whereas the EBNALP C-terminal 45 amino acids regulate coactivation and EBNA2 association.10,23,25 Deletion of only 10 or 34 of the C-terminal 45 amino acids stabilizes EBNALP association with EBNA2 and abrogates coactivation, whereas deletion of all 45 amino acids restores substantial coactivation and destabilizes association with EBNA2.10,23–25 EBNALP coactivation with EBNA2 is primarily dependent on the E2AD and secondarily dependent on the E2AD2.24 EBNALP is highly phosphorylated and serine phosphorylations are critical for EBNALP coactivation with EBNA2.6,26,27 Hsp72, HA95, DNA-PKcs, PKA, Hax-1, ERR1, and Sp100 associate or interact with EBNALP.4,28–33 Hax-1 associates with the EBNALP repeat domain,28 whereas ERR1 associates with EBNALP C-terminal 45 amino acid regulatory domain.30 Sp100 is implicated in EBNALP coactivation,4 whereas HA95 and PKA are implicated in negative regulation of EBNA2- and EBNALP-mediated transcription.31 The role of Hsp72 in EBNALP coactivation has not been evaluated and is the purpose of the experiments reported here.
Hsp72 is prominent in EBNALP immune precipitates,29,32,33 colocalizes with EBNALP in ND10 bodies,34 and further colocalizes with EBNALP to nucleoli when cells are grown at high concentration or after heat shock.34 Hsp72 associates with the EBNALP C-terminal 45–amino acid regulatory domain,32 which is critical for efficient B-lymphocyte conversion to LCLs.35 Hsp72 is important in posttranslational protein folding and in refolding following heat stress.36–38 Hsp72 has ATP or ADP and polypeptide-binding activities that are mediated by the Hsp72 N- and C-terminal domains, respectively (Figure 1A).39,40 Polypeptide binding stimulates ATPase activity, indicating that the 2 domains function coordinately.41 Hsp72 can also regulate intrinsic protein activities. For example, Hsp72 binding inhibits c-Jun N-terminal kinase and apoptosis signal regulating kinase-1.42,43
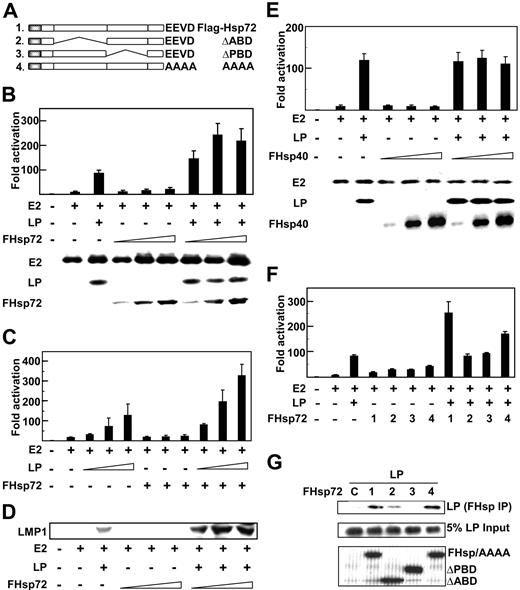
Hsp72 augments EBNA2 and EBNALP coactivation. (A) Schematic depiction of Hsp72 and Hsp72 mutants. (B) Hsp72 overexpression augments EBNALP coactivation with EBNA2 of the LMP1 promoter (LMP1p). BJAB B lymphoblasts were transfected with 5 μg LMP1p-Luciferase reporter plasmid, 10 μg EBNA2 expression plasmid (E2 +), 10 μg EBNALP expression plasmid (LP +), 10, 30, or 50 μg Flag-tagged Hsp72 expression plasmid (FHsp72), 2 μg β-gal transfection control expression plasmid, and an appropriate amount of control pSG5 expression vector DNA so that cells were transfected with equal amounts of DNA. Luciferase and β-gal activity activities were measured 24 hours after transfection. Luciferase activities were corrected for β-gal activity. EBNA2, EBNALP, and FHsp72 expression were determined by Western blotting with PE2, JF186, and M2 (Flag epitope specific) antibodies. (C) To assess the effect of Hsp72 overexpression on EBNA2 and EBNALP coactivation with varying levels of EBNALP, BJAB cells were transfected with 5 μg LMP1p-Luciferase reporter plasmid, 10 μg EBNA2 expression plasmid (E2 +), 1, 3, or 5 μg EBNALP (LP +) expression plasmid, 30 μg FHsp72 expression plasmid, or vector DNA to balance plasmid input for each transfection and processed as in panel B. (D) EBV latency I infected Akata BL cells were transfected with 10 μg EBNA2, 10 μg EBNALP, 10, 30, or 50 μg FHsp72, and appropriate amounts of control expression vector. LMP1 expression was assayed by Western blot with S12 mouse monoclonal antibody at 48 hours after transfection. (E) The effect of Hsp40 overexpression on EBNA2 and EBNALP coactivation of the LMP1p was assessed using a protocol identical to that in panel B except for FHsp40 expression vector instead of Fhsp72. (F) The effect of Hsp72 or Hsp72 mutants on EBNA2 and EBNALP coactivation of the LMP1p was assessed using a protocol similar to that used in panel B with 30 μg FHsp72 or FHsp72 mutant expression plasmids. (G) The association of EBNALP with Hsp72 and Hsp72 mutants was assessed in BJAB cells after transfection with 10 μg EBNALP and 10 μg FHsp72 or FHsp72 mutant expression plasmids. Lysates were immune precipitated with M2 Flag epitope–specific beads and Western blotted with JF186 monoclonal antibody to EBNALP. EBNALP input (5%) is shown. Lane 1 (C) is a control IP with M2 antibody in the absence of Flag tagged Hsp72 expression. Error bars indicate one standard deviation from the mean for data from multiple experiments in this and in subsequent figures.
Hsp72 augments EBNA2 and EBNALP coactivation. (A) Schematic depiction of Hsp72 and Hsp72 mutants. (B) Hsp72 overexpression augments EBNALP coactivation with EBNA2 of the LMP1 promoter (LMP1p). BJAB B lymphoblasts were transfected with 5 μg LMP1p-Luciferase reporter plasmid, 10 μg EBNA2 expression plasmid (E2 +), 10 μg EBNALP expression plasmid (LP +), 10, 30, or 50 μg Flag-tagged Hsp72 expression plasmid (FHsp72), 2 μg β-gal transfection control expression plasmid, and an appropriate amount of control pSG5 expression vector DNA so that cells were transfected with equal amounts of DNA. Luciferase and β-gal activity activities were measured 24 hours after transfection. Luciferase activities were corrected for β-gal activity. EBNA2, EBNALP, and FHsp72 expression were determined by Western blotting with PE2, JF186, and M2 (Flag epitope specific) antibodies. (C) To assess the effect of Hsp72 overexpression on EBNA2 and EBNALP coactivation with varying levels of EBNALP, BJAB cells were transfected with 5 μg LMP1p-Luciferase reporter plasmid, 10 μg EBNA2 expression plasmid (E2 +), 1, 3, or 5 μg EBNALP (LP +) expression plasmid, 30 μg FHsp72 expression plasmid, or vector DNA to balance plasmid input for each transfection and processed as in panel B. (D) EBV latency I infected Akata BL cells were transfected with 10 μg EBNA2, 10 μg EBNALP, 10, 30, or 50 μg FHsp72, and appropriate amounts of control expression vector. LMP1 expression was assayed by Western blot with S12 mouse monoclonal antibody at 48 hours after transfection. (E) The effect of Hsp40 overexpression on EBNA2 and EBNALP coactivation of the LMP1p was assessed using a protocol identical to that in panel B except for FHsp40 expression vector instead of Fhsp72. (F) The effect of Hsp72 or Hsp72 mutants on EBNA2 and EBNALP coactivation of the LMP1p was assessed using a protocol similar to that used in panel B with 30 μg FHsp72 or FHsp72 mutant expression plasmids. (G) The association of EBNALP with Hsp72 and Hsp72 mutants was assessed in BJAB cells after transfection with 10 μg EBNALP and 10 μg FHsp72 or FHsp72 mutant expression plasmids. Lysates were immune precipitated with M2 Flag epitope–specific beads and Western blotted with JF186 monoclonal antibody to EBNALP. EBNALP input (5%) is shown. Lane 1 (C) is a control IP with M2 antibody in the absence of Flag tagged Hsp72 expression. Error bars indicate one standard deviation from the mean for data from multiple experiments in this and in subsequent figures.
The carboxy terminus of Hsp72-interacting protein, CHIP, is a multifunctional cochaperone ubiquitin ligase that can ubiquitinate proteins bound to Hsp72; most ubiquitinated proteins are then degraded.44,45 In the absence of substrates bound to Hsp72, CHIP ubiquitinates Hsp72 and increases Hsp72 degradation.46 Through interaction with chaperones, CHIP also up-regulates heat shock factor 1 activity and Hsp72 transcription.46 The Hsp72 C-terminal EEVD residues interact with CHIP, Hsp40, or the Hsp72 ATP-binding site.40 Because of the key role of CHIP in Hsp72 affects, the experiments reported here also investigate some aspects of the role of CHIP in EBNALP coactivation with EBNA2.
Materials and methods
Expression plasmids
Plasmids pSG5-Flag EBNALP, d10, d34, and d45 express N-terminally Flag epitope–tagged wild-type (WT) or mutant EBNALPs.10 An Hsp72 expression plasmid was a gift of Dr Michael Sherman of Boston University. Flag-tagged full-length Hsp72 was generated by polymerase chain reaction (PCR) using forward 5′GAAGGAATTAAGCGGCCGCATGGCCAAAGCCGCGGCGATC3′ and reverse 5′CTAGGATCCTTACTAATCCACCTCCTCAATG3′ primers, which have an underlined NotI site upstream of the Hsp72 start codon and a BamHI site downstream of the stop codon. The resulting PCR products were subcloned into NotI and BamHI sites of pSG5-Flag. The EEVD/AAAA variant47 was generated by PCR-based mutagenesis kit (Stratagene, La Jolla, CA). Plasmid pSG5-Flag-Hsp72 ΔABD or ΔPBD has been described previously.43 Briefly, the ATP-binding domain (ABD), amino acid residues 120 to 428, were deleted by BglII digestion to release the BglII-BglII fragment (ABD), whereas the peptide-binding domain (PBD), amino acid residues 436 to 618, were deleted by XmaI digestion to release the XmaI-XmaI fragment (PBD). A CHIP expression vector was a gift from Dr Hamid Band of Harvard Medical School. WT CHIP, ΔTPR, and ΔU expression vectors were generated by PCR. K34A and H260Q, were generated using a QuickChange site-directed mutagenesis kit (Stratagene) using primers and mutagenesis protocols described by the manufacturer. The resulting PCR products were subcloned into pSG5-Flag after appropriate digestion. The hsp72 siRNA sequence 5′ACCTCATCGAC TTCTACACGTCCATCTCAAGAGGATGGACGTGTAGAAGTCGATTT3′ was subcloned into the BbsI site of psiRNAneo as suggested by the manufacturer (InvivoGen, San Diego, CA). The nucleotide sequence was selected using siRNA Wizard program (www.sirnawizard.com) provided by InvivoGen and siRNA expression vectors were verified by DNA sequencing. Hsp40 expression vector was a gift from Dr Richard Morimoto, Northwestern University. The flag tagged hsp40 were generated by PCR using appropriate primers and subcloned into XhoI and BamHI sites of pSG5-Flag expression vector.
Cell lines and transfections
BJAB and Akata are EBV-negative and positive Burkitt lymphoma (BL) cell lines, respectively.48,49 BL cell lines were cultured in RPMI 1640 medium (GIBCO/BRL, Grand Island, NY) supplemented with 10% FCS. For transfections, 1 to 1.5 × 107 cells were electroporated with the indicated amounts of expression and control plasmids using BioRad gene pulser (Hercules, CA). LMP1 promoter (−512/+72 × 2)-Luciferase reporter (LMP-Luc) or 8 copies of Cp-Luciferase reporter (Cp-Luc) activation by EBNA2 and coactivation by EBNALP were assayed by Luciferase activity. Briefly, 5 μg LMP1-Luc or Cp-Luc and 2 μg control β-gal expression plasmids were cotransfected with 10 μg EBNA2 and EBNALP expression vectors. Indicated amounts of WT or mutant Hsp72, CHIP, or control pSG5 expression plasmids were also cotransfected in indicated experiments to balance the total transfected DNA amounts. The Hsp72 knockdown cell lines were generated by electroporation of 10 μg siRNA expression plasmid into 107 BJAB cells and selection with 500 μg/mL G418 after linearization by ClaI.
Immunoprecipitation and immunoblot assays
Immune precipitation and immunoblot assays were done 24 hours after electroporation of 10 μg expression vector into 1 to 1.5 × 107 BJAB cells. Collected cells were lysed in buffer containing 50 mM Tris-HCl, 150 mM NaCl, 0.5 mM PMSF, 1 μg/mL aprotinin and leupeptin, and 1% NP40. Lysates were mixed at 4°C for 1 hour with primary monoclonal antibody; M2 or protein A Sepharose beads were added to the mixture and collected after 30 minutes at 4°C. Beads were washed with lysis buffer and analyzed by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) and immunoblot with indicated antibodies. For direct immunoblot analysis, cells were lysed in SDS sample buffer, subjected to SDS-PAGE, immunoblotted with indicated antibodies, including JF186 for EBNALP,50 PE2 for EBNA2,51 M2 (Sigma, St Louis, MO), Hsp90 (Upstate, Lake Placid, NY),β-tubulin (Upstate), and rabbit polyclonal Hsp72 (Upstate), and visualized by an enhanced chemiluminescence protocol (Perkin Elmer, Shelton, CT).
Results
Hsp72 up-regulates EBNALP coactivation with EBNA2
Since Hsp72 is significantly associated with EBNALP in LCLs and binds to the C-terminal 34 amino acids of EBNALP,29,32–34 we investigated whether Hsp72 affects EBNA2 and EBNALP coactivation. Overexpression of Hsp72 in non–EBV-infected BJAB BL lymphoblasts, in which EBNA2 or EBNALP was also expressed, augmented EBNA2 and EBNALP coactivation of Luciferase expression from an LMP1 promoter and Luciferase reporter plasmid (Figure 1B,C,F and data not shown). EBNA2 up-regulated the LMP1 promoter and Luciferase reporter 9- to 15-fold, and EBNALP coactivated EBNA2 to 90- to 115-fold total activation. While EBNA2 expression with increasing Hsp72 overexpression resulted in 15- to 20-fold activation, close to that of EBNA2 alone, Hsp72 overexpression with EBNA2 and EBNALP expression resulted in 245- to 300-fold activation, much greater than the EBNA2 and EBNALP 95- to 115-fold coactivation (Figure 1B,C,F). EBNALP and EBNA2 expression levels were largely unaffected by Hsp72 expression (Figure 1B and data not shown). Thus, Hsp72 overexpression augments EBNA2 and EBNALP coactivation of the LMP1 promoter without substantially affecting EBNA2 or EBNALP levels.
To determine whether Hsp72 augmentation of EBNA2 and EBNALP coactivation is dependent on the EBNALP expression level and coactivation effect, Hsp72 effects were assayed at various levels of EBNALP expression. EBNA2 activated the LMP1 promoter 15-fold. EBNALP expressed from 1, 3, or 5 μg expression vector coactivated EBNA2 to 35-, 75-, or 130-fold and Hsp72 overexpression further augmented Luciferase activation to 85-, 195-, or 310-fold, indicating that Hsp72 overexpression more than doubles reporter activity at all EBNALP expression and coactivation levels (Figure 1C). EBNALP coactivation levels off and sometimes decreases following transfection with 5 to 10 μg EBNALP expression vector, as is evident from the 95-fold coactivating effect of 10 μg EBNALP expression vector in Figure 1B versus the 130-fold effect of 5 μg expression vector in Figure 1C. Despite EBNALP coactivation with 10 μg expression vector to 95-fold in Figure 1B, Hsp72 overexpression increased the 95-fold EBNALP affect to 240-fold (multiple similar experiments not shown). Thus, Hsp72 overexpression more than doubled EBNA2 and EBNALP coactivation over a broad range of EBNA2 and EBNALP coactivation.
The effect of Hsp72 overexpression was also evaluated in the context of EBNA2 and EBNALP coactivation of LMP1 expression from the endogenous EBV genome in EBV latency I infected Akata BL cells (Figure 1D).7,23 EBNA2 and EBNALP are not expressed in latency I infected Akata cells, and both EBNALP and EBNA2 expression are required to substantially induce LMP1 expression.7,23,31 As expected, EBNA2 alone had minimal effect, whereas EBNA2 and EBNALP coactivated LMP1 expression (Figure 1D).23 EBNALP and EBNA2 expression in latency I infected Akata cells does not result in higher EBNALP or EBNA2 levels than are evident with expression of either protein alone, consistent with the notion that EBNA promoters are not turned on by EBNA2 and EBNALP expression.23 Hsp72 overexpression with EBNA2 and EBNALP expression in latency I infected Akata cells substantially increased LMP1 expression above levels achieved with EBNA2 and EBNALP (Figure 1D). Thus, Hsp72 overexpression up-regulates EBNA2 and EBNALP coactivation of transiently transfected and endogenous LMP1 promoters.
To evaluate the specificity of the Hsp72 effect, the potential effect of Hsp40 overexpression on LMP1 promoter coactivation by EBNA2 and EBNALP was assessed (Figure 1E). Increasing Hsp40 expression did not affect EBNA2 or EBNALP protein expression and did not affect EBNA2 activation or EBNA2 and EBNALP coactivation. These data indicate that Hsp72 specifically augments EBNA2 and EBNALP coactivation.
To assess the role of Hsp72 ATP-binding (ABD), peptide-binding (PBD), and C-terminal EEVD domains in Hsp72 augmentation of EBNA2 and EBNALP coactivation, the effects of Hsp72 were compared with those of Hsp72 ΔABD, ΔPBD, or AAAA mutants (Figure 1A,F). Hsp72 and Hsp72 ΔABD, ΔPBD, and AAAA were expressed at similar levels (Figure 1G and data not shown). However, Hsp72 ΔABD and ΔPBD did not up-regulate EBNA2 activation or EBNA2 and EBNALP coactivation, Hsp72 AAAA was intermediate in phenotype and up-regulated EBNA2 and EBNALP coactivation from 90- to 170-fold, and Hsp72 up-regulated EBNA2 and EBNALP coactivation from 90-fold to 250-fold (Figure 1F). These data indicate that the Hsp72 ABD and PBD are critical for Hsp72 augmentation of EBNA2 and EBNALP coactivation and that the C-terminal EEVD domain is also important.
Hsp72 association with EBNALP is required for augmentation of EBNALP and EBNA2 coactivation
To evaluate whether EBNALP associates with the Hsp72 PBD, as expected for a client protein, Flag-tagged Hsp72 or Hsp72 mutant proteins were expressed in BJAB cells along with EBNALP. Hsp72 and Hsp72 mutant proteins were equally expressed and did not affect EBNA2 or EBNALP expression levels (Figure 1G). EBNALP coimmune precipitated equally with Hsp72 and with Hsp72 AAAA, but failed to precipitate with Hsp72 ΔPBD and was intermediate in precipitating with Hsp72 ΔABD (Figure 1G). Thus, EBNALP association with Hsp72 is dependent on the Hsp72 PBD and to a lesser extent on the ABD, but is independent of the C-terminal EEVD, correlating with the more prominent effects of the PBD and ABD deletions on Hsp72 augmentation (Figure 1F).
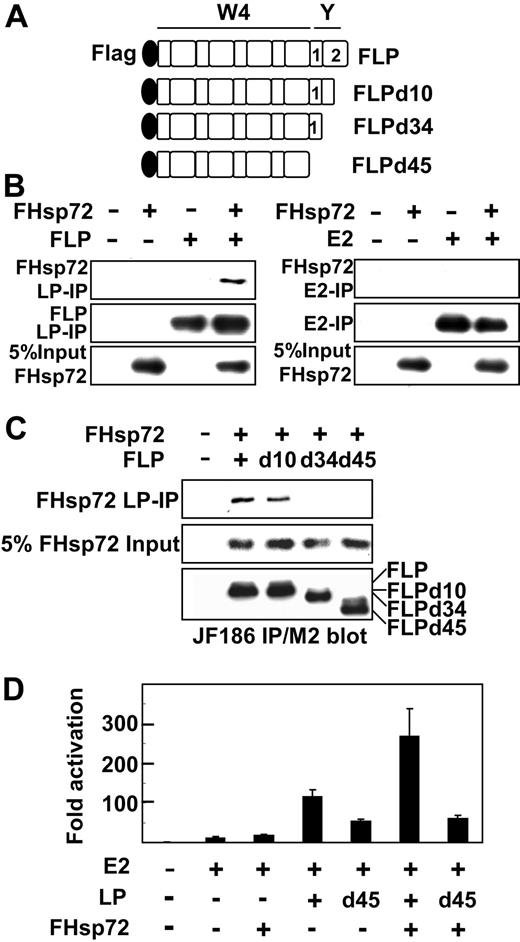
The EBNALP C-terminal 34 amino acids (Figure 2A) affect EBNALP association and coactivation with EBNA2. Deletion of the most C-terminal 10 amino acids results in EBNALP stable association with EBNA2 and inability to coactivate with EBNA2, whereas deletion of the C-terminal 45 amino acids restores unstable association and substantial coactivation.10,24,25 To further assess whether Hsp72 association with the EBNALP C-terminal 34 amino acids32 is important in the up-regulatory effects of Hsp72 on EBNA2 and EBNALP coactivation, the effects of deletion of the EBNALP C-terminal 34 amino acids on Hsp72 association and augmentation of EBNA2 and EBNALP coactivation were assessed. As expected, EBNALP associated with Hsp72, whereas EBNA2 did not associate with Hsp72 (Figure 2B). Furthermore, EBNALP deleted for the C-terminal 10 amino acids associated with Hsp72 in BJAB cells, whereas EBNALP deleted for the C-terminal 34 or 45 amino acids did not associate with Hsp72 (Figure 2C). Also as expected, EBNALP deleted for C-terminal 10 or 34 amino acids was substantially deficient in coactivation with EBNA2, whereas EBNALP deleted for C-terminal 45 amino acids (EBNALPd45) coactivated with approximately 50% of EBNALP effects (Figure 2D and data not shown).10 These data indicate that Hsp72 association is not required for EBNALPd45 partial coactivation with EBNA2.
Hsp72 association and coactivation with EBNALP or EBNALP C-terminal deletion mutants in BJAB cells. (A) Schematic depiction of N-terminally Flag-tagged EBNALP or EBNALP C-terminally deleted mutant proteins, FLP, FLPd10, FLPd34, and FLPd45. (B) EBNALP associates with Hsp72 and EBNA2 does not. BJAB cells were transfected with 10 μg FHsp72 expression vector and with 10 μg EBNA2 or EBNALP expression vector. Lysates were immune precipitated with PE2 (E2) or JF186 (LP) followed by Western blot analysis with M2, PE2, or JF186 antibody. Five percent of FHsp72 input is shown. (C) Hsp72 associates with FLP or FLPd10, but not with FLPd34 or FLPd45. BJAB cells were transfected with 10 μg FHsp72 expression vector or empty expression vector and with 10 μg FLP, FLPd10, FLPd34, FLPd45, or control expression vector. Lysates were immune precipitated with JF186 antibody to EBNALP followed by immune blot analysis with M2 or JF186 antibody. Five percent of FHsp72 input is shown. (D) Hsp72 augments EBNALP, but not EBNALPd45, coactivation with EBNA2. BJAB cells were transfected with 10 μg EBNA2, EBNALP, or EBNALPd45, and FHsp72 expression vector, 5 μg LMP1p-Luciferase Reporter, 2 μg β-gal control plasmid, and sufficient pSG5 vector to achieve equivalence in DNA input. Assays were done after 24 hours.
Hsp72 association and coactivation with EBNALP or EBNALP C-terminal deletion mutants in BJAB cells. (A) Schematic depiction of N-terminally Flag-tagged EBNALP or EBNALP C-terminally deleted mutant proteins, FLP, FLPd10, FLPd34, and FLPd45. (B) EBNALP associates with Hsp72 and EBNA2 does not. BJAB cells were transfected with 10 μg FHsp72 expression vector and with 10 μg EBNA2 or EBNALP expression vector. Lysates were immune precipitated with PE2 (E2) or JF186 (LP) followed by Western blot analysis with M2, PE2, or JF186 antibody. Five percent of FHsp72 input is shown. (C) Hsp72 associates with FLP or FLPd10, but not with FLPd34 or FLPd45. BJAB cells were transfected with 10 μg FHsp72 expression vector or empty expression vector and with 10 μg FLP, FLPd10, FLPd34, FLPd45, or control expression vector. Lysates were immune precipitated with JF186 antibody to EBNALP followed by immune blot analysis with M2 or JF186 antibody. Five percent of FHsp72 input is shown. (D) Hsp72 augments EBNALP, but not EBNALPd45, coactivation with EBNA2. BJAB cells were transfected with 10 μg EBNA2, EBNALP, or EBNALPd45, and FHsp72 expression vector, 5 μg LMP1p-Luciferase Reporter, 2 μg β-gal control plasmid, and sufficient pSG5 vector to achieve equivalence in DNA input. Assays were done after 24 hours.
Since EBNALPd45 does not associate with Hsp72 but still partially coactivated the LMP1 promoter with EBNA2, the importance of Hsp72 association with EBNALP in Hsp72 augmentation of EBNALP and EBNA2 coactivation could be assessed. Whereas and EBNA2 and EBNALP 110-fold coactivation was augmented by Hsp72 overexpression to 270-fold, EBNA2 and EBNALPd45 55-fold coactivation was not significantly affected by Hsp72 overexpression (Figure 2D and data not shown). These results indicate that Hsp72 association with EBNALP is necessary for Hsp72 overexpression augmentation of EBNALP and EBNA2 coactivation.
CHIP down-regulates EBNA2 and EBNALP coactivation
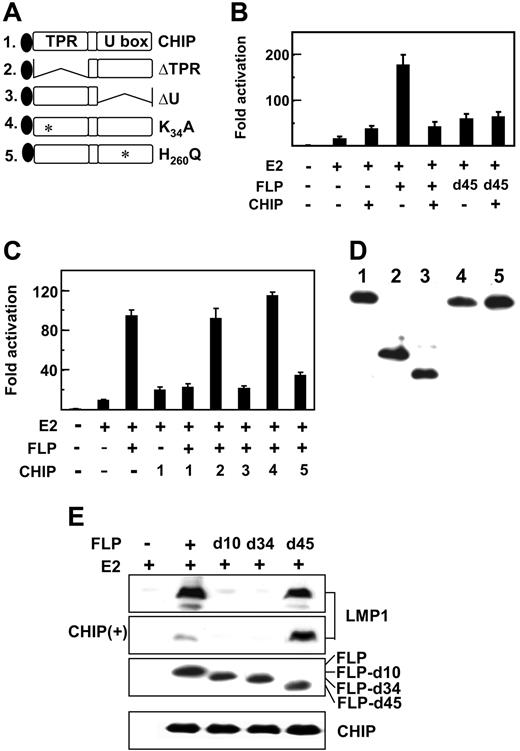
CHIP is a cochaperone E3 ubiquitin ligase that regulates Hsp72 transcription, stability, and chaperone effects through CHIP tetratrico peptide repeat (TPR) and U box E3 ligase domains (Figure 3A).44–46,52 To evaluate the potential role of CHIP in Hsp72 augmentation of EBNA2 and EBNALP coactivation of the LMP1 promoter, CHIP was overexpressed along with EBNA2 and EBNALP in BJAB cells cotransfected with an LMP1 promoter and Luciferase reporter plasmid. As in Figures 1-2, EBNA2 activated the LMP1 promoter 10- to 15-fold and EBNA2 and EBNALP coactivated to 95- to 170-fold (Figure 3B-C). Surprisingly, CHIP enhanced EBNA2 activation approximately 2.5-fold and substantially reduced EBNA2 and EBNALP coactivation to that observed with EBNA2 and CHIP (Figure 3B-C). In multiple experiments, CHIP expression up-regulated EBNA2 activation and inhibited EBNA2 and EBNALP coactivation.
CHIP coactivates with EBNA2 and inhibits EBNALP and EBNA2 coactivation. (A) Schematic diagram of Flag-tagged CHIP and CHIP deletion or point mutant proteins, CHIPΔTPR, CHIPΔU, CHIPK30A, and H260Q. (B-C) CHIP inhibits EBNALP, but not EBNALPd45, coactivation with EBNA2 in BJAB cells. BJAB cells were transfected with 10 μg E2, FLP, or FLPd45, or 30 μg CHIP or CHIP mutant expression vectors, 5 μg LMP1p-Luciferase reporter plasmid, 2 μg β-gal control expression plasmid, or vector DNA to balance the amount of transfected DNA. Assays were done after 24 hours. (D) Relative expression levels of CHIP or CHIP mutant proteins after expression for 24 hours in BJAB cells. Flag-tagged CHIP proteins were detected by immune blot with M2 antibody. (E) EBV latency I Akata BL cells were transfected with 10 μg E2 and 10 μg FLP, FLPd10, FLPd34, or FLPd45 expression vector and after 48 hours immune blotted for LMP1 expression. CHIP(+) indicates an immune blot from cells that were also transfected with 30 μg CHIP expression plasmid.
CHIP coactivates with EBNA2 and inhibits EBNALP and EBNA2 coactivation. (A) Schematic diagram of Flag-tagged CHIP and CHIP deletion or point mutant proteins, CHIPΔTPR, CHIPΔU, CHIPK30A, and H260Q. (B-C) CHIP inhibits EBNALP, but not EBNALPd45, coactivation with EBNA2 in BJAB cells. BJAB cells were transfected with 10 μg E2, FLP, or FLPd45, or 30 μg CHIP or CHIP mutant expression vectors, 5 μg LMP1p-Luciferase reporter plasmid, 2 μg β-gal control expression plasmid, or vector DNA to balance the amount of transfected DNA. Assays were done after 24 hours. (D) Relative expression levels of CHIP or CHIP mutant proteins after expression for 24 hours in BJAB cells. Flag-tagged CHIP proteins were detected by immune blot with M2 antibody. (E) EBV latency I Akata BL cells were transfected with 10 μg E2 and 10 μg FLP, FLPd10, FLPd34, or FLPd45 expression vector and after 48 hours immune blotted for LMP1 expression. CHIP(+) indicates an immune blot from cells that were also transfected with 30 μg CHIP expression plasmid.
To investigate whether CHIP inhibition of EBNA2 and EBNALP coactivation requires Hsp72 association with EBNALP, the effects of CHIP overexpression on EBNA2 and EBNALP or EBNALPd45 coactivation of the LMP1 promoter were compared in BJAB cells and in EBV latency I Akata BL cells (Figure 3B-C,E). In transiently transfected BJAB cells (Figure 3B-C) and in Akata cells (Figure 3E), CHIP overexpression inhibited EBNALP and EBNA2 coactivation and failed to affect EBNALPd45 coactivation (Figure 3E and to a lesser extent Figure 3B), indicative of CHIP down-regulation requiring Hsp72 association with EBNALP.
The role of CHIP functional domains in inhibiting EBNALP coactivation was evaluated by comparing CHIP and CHIP mutant effects (Figures 3A-D). CHIP ΔTPR and K30A are null deletion and point mutations in the tetratrico peptide repeat cochaperone domain, which mediates Hsp72 binding, whereas CHIP ΔU and H260Q are null deletion and point mutations in the ubiquitin ligase domain.47,53–57 CHIP mutants were similar to CHIP in expression level in BJAB cells (Figure 3D). Of interest, CHIP-ΔU and H260Q were similar to CHIP in strongly inhibiting EBNA2 and EBNALP coactivation, indicating that CHIP E3 ligase is dispensable for down-regulation (Figure 3C). In contrast, deletion or point mutation of the TPR cochaperone domain resulted in loss of CHIP down-regulating activity (Figure 3C). Thus, CHIP overexpression inhibition of EBNA2 and EBNALP coactivation requires HSP72 association with EBNALP but does not require CHIP ubiquitin ligase activity, consistent with the absence of an Hsp72 or CHIP effect on EBNA2 or EBNALP protein levels.
Hsp72 knockdown reduces EBNALP coactivation with EBNA2 and CHIP inhibition of EBNA2 and EBNALP coactivation
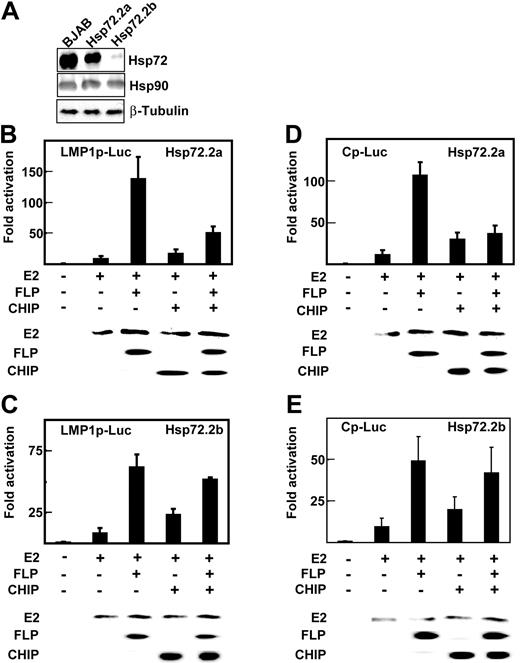
To further evaluate the role of Hsp72 in augmenting EBNA2 and EBNALP coactivation and in mediating CHIP inhibition of EBNA2 and EBNALP coactivation, Hsp72 knockdown BJAB BL lymphoblast clones were obtained by stably expressing an Hsp72 siRNA in BJAB cells. BJAB clone Hsp72.2a had Hsp72 reduced to 50% of wild-type BJAB levels, whereas clone Hsp72.2b had Hsp72 reduced to 10% of wild-type BJAB levels (Figure 4A). Hsp90 and β-tubulin expression were unaffected by Hsp72 siRNA expression (Figure 4A). Hsp72.2b BJAB cells grew more slowly than Hsp72.2a or BJAB cells (data not shown).
Hsp72 knockdown decreases EBNALP coactivation with EBNA2 and CHIP inhibition of EBNA2 and EBNALP coactivation. (A) Lysates of BJAB cells and BJAB cell clones 2a and 2b, which are stably transfected with psiRNA-Hsp72, were immune blotted with rabbit antibody to Hsp72 and mouse monoclonal antibodies to Hsp90 or β-tubulin. (B-E) Hsp72.2a BJAB cells that are 50% Hsp72 deficient or Hsp72.2b BJAB cells that are 90% Hsp72 deficient were transfected with 10 μg E2, 0 or 10 μg FLP, 30 μg Flag-tagged CHIP (CHIP) expression vector, and appropriate amounts of control expression vector, 2 μg β-gal control plasmid, and 5 μg LMP1p-Luciferase or Cp-Luciferase reporter plasmids. Cells were assayed for Luciferase activity after 24 hours. EBNA2, EBNALP, and CHIP expression were monitored by immune blot with specific antibodies.
Hsp72 knockdown decreases EBNALP coactivation with EBNA2 and CHIP inhibition of EBNA2 and EBNALP coactivation. (A) Lysates of BJAB cells and BJAB cell clones 2a and 2b, which are stably transfected with psiRNA-Hsp72, were immune blotted with rabbit antibody to Hsp72 and mouse monoclonal antibodies to Hsp90 or β-tubulin. (B-E) Hsp72.2a BJAB cells that are 50% Hsp72 deficient or Hsp72.2b BJAB cells that are 90% Hsp72 deficient were transfected with 10 μg E2, 0 or 10 μg FLP, 30 μg Flag-tagged CHIP (CHIP) expression vector, and appropriate amounts of control expression vector, 2 μg β-gal control plasmid, and 5 μg LMP1p-Luciferase or Cp-Luciferase reporter plasmids. Cells were assayed for Luciferase activity after 24 hours. EBNA2, EBNALP, and CHIP expression were monitored by immune blot with specific antibodies.
EBNA2 activated the LMP1 promoter 8- to 10-fold in the Hsp72.2a BJAB cells that have 50% Hsp72 expression (Figure 4B) and 9- to 12-fold in Hsp72.2b BJAB cells that have 10% Hsp72 expression (Figure 4C), as observed in BJAB cells (Figures 1,Figure 2-3). EBNA2 and EBNALP also coactivated the LMP1 promoter 125-fold in Hsp72.2a BJAB (Figure 4B), as observed in BJAB cells (Figures 1,Figure 2-3). However, EBNA2 and EBNALP coactivated only 63-fold in Hsp72.2b BJAB (Figure 4C). Similar differences were observed for the EBNA Cp promoter in Hsp72.2a and Hsp72.2b cells. EBNA2 and EBNALP coactivated the Cp promoter 110-fold in Hsp72.2a cells (Figure 4D) and only 50-fold in Hsp72.2b cells (Figure 4E). Control β-galactosidase and Luciferase and EBNA2 and EBNALP expression levels were similar in Hsp72.2a, Hsp72.2b, and BJAB cells (Figures 4B-D and data not shown). These data indicate that Hsp72 is required for normal up-regulation of EBNA2 and EBNALP coactivation of the LMP1 and Cp promoters.
CHIP overexpression increased EBNA2 activation from 8- to 12-fold to 20- to 25-fold in Hsp72.2a and Hsp72.2b cells (Figure 4B-4E) as was evident in BJAB cells (Figure 3B-C). In contrast, CHIP overexpression inhibited EBNA2 and EBNALP 125- and 110-fold coactivation of the LMP1 and Cp promoters in Hsp72.2a cells to 50- and 30-fold (Figure 4B,D) as in BJAB cells (Figure 3B-C), but failed to inhibit EBNA2 and EBNALP coactivation of the LMP1 and Cp promoters in HSP72.2b cells (Figure 4C,E). These data indicate that CHIP up-regulation of EBNA2 activation is Hsp72 independent, whereas CHIP down-regulation of EBNA2 and EBNALP coactivation is Hsp72 dependent.
Discussion
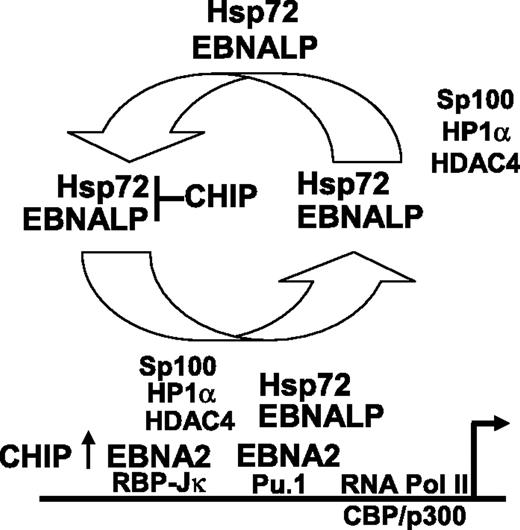
Hsp72 augmentation of EBNALP coactivation of transcription with EBNA2 is a surprising finding in these investigations. Hsp72 most commonly assists protein folding, associates with proteasomes, and binds CHIP, which ubiquitinates and targets misfolded proteins to degradation.58 Our data indicate that Hsp72 and CHIP do not alter EBNALP or EBNA2 protein levels. Instead, Hsp72 binding to the EBNALP C-terminal regulatory domain augments EBNALP and EBNA2 coactivation as is evident from the effect of forced high-level Hsp72 expression in increasing EBNA2 and EBNALP coactivation of a cotransfected LMP1 promoter and in increasing LMP1 expression from the endogenous EBV genome in EBV-infected Akata BL cells. Furthermore, a 90% knockdown of Hsp72 expression reduced EBNALP and EBNA2 coactivation from a cotransfected LMP1 or Cp EBNA promoter. Moreover, Hsp72 augmentation of EBNALP and EBNA2 coactivation of the LMP1 promoter is dependent on interaction with EBNALP and on the Hsp72 ABD, PBD, and C-terminal EEVD sequence. Hsp72 associates with the EBNALP C-terminal regulatory 34 amino acids,32 which are critical for full EBNALP coactivation and transient interaction with the EBNA2 acidic activation domain.10,23–25 Overall, current and previous data support a model in which Hsp72 chaperone interactions with the EBNALP C-terminal regulatory domain augment EBNALP transcriptional up-regulatory effects at EBNA2-enhanced promoters. Since EBNALP coactivation of EBNA2-mediated transcription appears to be due to EBNALP transport of Sp100 and HDAC4 associated repressor complexes from EBNA2-activated promoters,4,59 Hsp72 chaperone interaction with the EBNALP C-terminal regulatory domain is likely to be important for efficient EBNALP engagement or release of repressor complexes (Figure 5).
Proposed model of the role of Hsp72 and CHIP in EBNALP coactivation of EBNA2-mediated transcription. Hsp72 stably associates with EBNALP and chaperones EBNALP through transient interaction with EBNA2 and more stable interactions with Sp100, HDAC4, and HP1α repressor complexes.4,25,59 EBNALP shuttles Sp100, HDAC4, and HP1α from EBNA2-responsive promoters to other nuclear or cytoplasmic locations,4,59 and thereby coactivates with EBNA2 in enabling RNA polymerase II complex assembly and transcription. CHIP can downmodulate the Hsp72 effect on EBNALP coactivation with EBNA2, but also can have a positive effect on EBNA2 in the absence of EBNALP.
Proposed model of the role of Hsp72 and CHIP in EBNALP coactivation of EBNA2-mediated transcription. Hsp72 stably associates with EBNALP and chaperones EBNALP through transient interaction with EBNA2 and more stable interactions with Sp100, HDAC4, and HP1α repressor complexes.4,25,59 EBNALP shuttles Sp100, HDAC4, and HP1α from EBNA2-responsive promoters to other nuclear or cytoplasmic locations,4,59 and thereby coactivates with EBNA2 in enabling RNA polymerase II complex assembly and transcription. CHIP can downmodulate the Hsp72 effect on EBNALP coactivation with EBNA2, but also can have a positive effect on EBNA2 in the absence of EBNALP.
CHIP overexpression coactivated EBNA2 and strongly inhibited EBNALP coactivation with EBNA2 of LMP1 promoter–regulated Luciferase expression in transiently transfected BJAB and in latency I infected Akata BL cells. The effects of CHIP on EBNALP coactivation required the CHIP TPR protein interactive domain, but not the ubiquitin ligase domain, and did not affect EBNALP or EBNA2 protein levels. Furthermore, CHIP inhibition of EBNALP coactivation in latency I infected Akata cells was dependent on EBNALP residues necessary for Hsp72 binding. The affects of CHIP in EBV-uninfected BJAB BL cells were also Hsp72 dependent since CHIP strongly inhibited EBNALP coactivation with EBNA2 of LMP1p or Cp promoter–driven Luciferase reporters in BJAB cells, was slightly less inhibitory in BJAB cells that are 50% deficient in Hsp72 expression, and failed to inhibit LMP1p or Cp promoter driven Luciferase expression in BJAB cells that are 90% deficient in Hsp72 expression. Thus, CHIP inhibition of EBNALP coactivation with EBNA2 is Hsp72 dependent, independent of CHIP ubiquitin ligase activity, dependent on CHIP TPR activity, and opposite in effect to CHIP coactivation with EBNA2. These data are most consistent with a model in which CHIP overexpression mimics EBNALP in coactivating EBNA2, possibly through degradation of an EBNA2-associated repressor, but strongly inhibits EBNALP coactivation with EBNA2 by binding to the Hsp72 C-terminal regulatory domain and inhibiting Hsp72 ATPase cycle44,47 or protein-protein interactions necessary for chaperoning EBNALP removal of repressors from EBNA2-regulated promoters44,59 (Figure 5). Alternatively, Hsp72 binding may optimize EBNALP regulatory domain interaction with the EBNALP repeat domain that mediates increased transcription.10,23–25 The EBNALP repeat domain can be extensively complexed with the HA95 shuttling protein and must transiently interact with EBNA2 at promoter sites.10,24,25 Hsp72 could also recruit 19s or 20s proteasomal subunits to EBNA2 responsive promoters by binding BAG-1.60 Proteasomal subunits are implicated in transcriptional activation and repressor degradation.61,62 BAG-1 is known to associate with nuclear hormone receptors to regulate transcription.63,64
EBNALP is the principal EBV coactivator with EBNA2 in the first days of primary B-lymphocyte infection.65 EBNALP coactivation with EBNA2 is therefore likely to be most critical in the initial up-regulation of the EBV Cp and LMP1 promoters.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grant CA47006 from the National Cancer Institute of the USPHS and by NSC 93-2314-B-320-014, NSC 94-2311-B-320-002, and TCMRC94006 from NSC and TCU to C.-W.P.
National Institutes of Health
Authorship
Contribution: C.-W.P. designed research and performed research; B.Z. analyzed data and wrote the paper; H.-C.C., M.-L.C., and C.-Y.L. analyzed data; H.-Y.H. and S.-Z.L. provided the analytical tool; E.K. designed research, wrote the paper, and is the corresponding author.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Elliott Kieff, Program in Virology and Department of Medicine and Microbiology and Molecular Genetics, Channing Laboratory, Brigham and Women's Hospital and Harvard University, 181 Longwood Ave, Boston, MA 02115; e-mail: ekieff@rics.bwh.harvard.edu or pengcw@mail.tcu.edu.tw.