Abstract
Intercellular adhesion molecule 4 (ICAM-4) is a unique member of the ICAM family because of its specific expression on erythroid cells and ability to interact with several types of integrins expressed on blood and endothelial cells. The first reported receptors for ICAM-4 were CD11a/CD18 and CD11b/CD18. In contrast to these 2, the cellular ligands and the functional role of the third β2 integrin, CD11c/CD18, have not been well defined. Here, we show that ICAM-4 functions as a ligand for the monocyte/macrophage-specific CD11c/CD18. Deletion of the individual immunoglobulin domains of ICAM-4 demonstrated that both its domains contain binding sites for CD11c/CD18. Analysis of a panel of ICAM-4 point mutants identified residues that affected binding to the integrin. By molecular modeling the important residues were predicted to cluster in 2 distinct but spatially close regions of the first domain with an extension to the second domain spatially distant from the other residues. We also identified 2 peptides derived from sequences of ICAM-4 that are capable of modulating the binding to CD11c/CD18. CD11c/CD18 is expressed on macrophages in spleen and bone marrow. Inhibition of erythrophagocytosis by anti–ICAM-4 and anti-integrin antibodies suggests a role for these interactions in removal of senescent red cells.
Introduction
In general, the major roles of the red blood cells, such as the transport of oxygen and carbon dioxide throughout the body, have not been thought to require adhesion of these cells.1 However, recently it has been demonstrated that erythrocytes express on their surface proteins known to provide adhesive functions. Many of these adhesion proteins belong to the immunoglobulin superfamily of proteins. These molecules may participate in normal red-cell physiology by mediating cellular interactions during their life cycle as well as the pathology of human diseases. In bone marrow, erythroblasts surround central macrophages, forming erythroblastic islands where cell-adhesion events play critical roles in regulating erythropoiesis. A physiologic interaction between red cells and leukocytes, platelets, and endothelial cells can also occur during normal hemostatic conditions (clot formation), pathologic occlusive conditions, and inflammation. Finally, the interaction of senescent red cells with splenic macrophages is important for red-cell clearance.1-7
ICAM-4 is a glycoprotein expressed on red blood cells and erythroid precursor cells concurrently with glycophorin A and Rh glycoproteins.8-10 The protein was originally described as the LW blood group antigen (Landsteiner-Wiener), but elucidation of its primary structure revealed significant similarity to the intercellular adhesion molecules (ICAMs). It is a 42-kDa glycoprotein composed of 2 immunoglobulin-like domains, a transmembrane part, and a short cytoplasmic tail.11 The primary cellular counter-receptors for the ICAMs are the leukocyte-specific β2 integrins, which consist of 4 heterodimeric glycoproteins with specific α chains (CD11a, CD11b, CD11c, CD11d) and a common β2 chain (CD18).12-15 However, ICAM-4 is an unusual ICAM in that it has recently been found to interact with several types of integrins expressed on blood and endothelial cells. Like all other members of the ICAM family,16-20 ICAM-4 binds to the CD11a/CD18 (LFA-1, αLβ2) integrin expressed on leukocytes.21 ICAM-4 also interacts with the granulocyte/monocyte-enriched β2 integrin CD11b/CD18 (Mac-1, αMβ2),21 as do ICAM-1 and ICAM-2.22,23 In addition to β2 integrins, ICAM-4 has been shown to bind to αv integrins (αvβ1, αvβ3, and αvβ5) on nonhemopoietic cells, α4β1 on hemopoietic cells, and αIIbβ3 on platelets.24-27
Using mutational analysis, we have previously mapped the binding sites on ICAM-4 for the CD11a/CD18 and CD11b/CD18 integrins.28 The binding site for CD11a/CD18 was shown to be confined to the first Ig-like domain of ICAM-4, whereas both Ig domains are involved in CD11b/CD18 binding. Unlike the other ICAMs, ICAM-4 does not contain the conserved functionally important glutamate residue in the first domain, which is replaced by an arginine-52 residue. Mutation of arginine-52 back to glutamate did not affect CD11a/CD18 binding and even reduced the interaction with CD11b/CD18. Despite the lack of the conserved glutamate residue in ICAM-4, we have demonstrated that the CD11a and CD11b I domains contain an ICAM-4–binding region.29 These data suggest that the β2 integrin–binding motifs of ICAM-4 differ from those of other ICAMs.
We and others have previously suggested that ICAM-4 could mediate red-cell interactions with macrophages.4,21 These interactions are evidently important during the life span of red cells from erythropoiesis to erythrocyte senescence. CD11c/CD18 is the major β2 integrin on monocytes/macrophages30 ; therefore, we thought that it is important to study its binding characteristics. Whereas many ligands have been described for the CD11b/CD18 integrin,23,31-34 much less is known about CD11c/CD18. In this report we show that ICAM-4 directly binds to the I domain of CD11c/CD18 and mediates erythrophagocytosis. Furthermore, using ICAM-4 mutants as well as synthetic peptides of ICAM-4, we have identified critical sites on ICAM-4 required for its interaction with CD11c/CD18.
Materials and methods
Antibodies
The monoclonal antibodies (mAbs) used in this study were as follows. The 7E4 and 2E7 mAbs react with the β2 chain,35 TS1/22 (ATCC, Rockville, MD) recognize the α chain of CD11a/CD18, and MEM170 is specific for the I domain of CD11b/CD18.36 The anti-CD11c mAbs used included CBRp150/4G1 and 3.937,38 which were provided by Dr T. Springer (Harvard Medical School, Boston, MA) and Dr N. Hogg (Cancer Research UK, London, United Kingdom), respectively, Bly6 (BD PharMingen, San Diego, CA), BU15 (Serotec, Oxford, United Kingdom), and BL4H4 (Monosan, Hameenlinna, Finland). The mAbs against the first domain of ICAM-4 (BS46 and BS56) were from Dr H. Sonneborn (Biotest, Dreieich, Germany).28,39 The mAbs 4C8 and 1A1 against ICAM-4 were from Dr J.-P. Cartron (Institut National de Transfusion Sanguine, Paris, France). A mouse IgG1-negative control was purchased from Chemicon (Boronia, Australia) and a human IgG1 used as a control was from Sigma (St Louis, MO). FITC-conjugated rabbit anti–mouse F(ab′)2 (Dakopatts a/s, Copenhagen, Denmark) was used for the flow cytometry studies (fluorescence-activated cell sorting; FACS).
Purification of CD18 integrins and soluble recombinant Fc proteins
CD11a/CD18, CD11b/CD18, and CD11c/CD18 integrins were purified from human blood as described previously (Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article).37,40 cDNA clones encoding the extracellular domains (residues 1-208) of native or mutagenized ICAM-4 and deletion mutants of ICAM-4 containing domain 1 (residues 1-101) or domain 2 (residues 102-208) in the pIg vector were used to produce soluble Fc-fusion proteins in COS-1 cells.28 The wild-type and Ile314Gly mutant CD11c I domains were expressed and purified as described.41
Cells and cell lines
Approval was obtained from the Division of Biochemistry, University of Helsinki institutional review board for these studies. Informed consent was obtained in accordance with the Declaration of Helsinki.
Blood samples from common LW and Rh phenotypes (ICAM-4–positive red cells) were obtained from healthy volunteers using heparin as an anticoagulant.
The wild-type and L929 cells expressing ICAM-1, ICAM-2, or ICAM-4 have been previously described.29 All the transfectants were grown in IMDM medium supplemented with 1 mg/mL G418, 10% FBS, 100 U/mL penicillin, and 100 μmg/mL streptomycin. The CD11c/CD18-transfected L929 cell line was provided by Dr Y. van Kooyk (Vrije Universiteit Medical Center, Amsterdam, The Netherlands) and purified by sorting with a FACStar Sorter (Becton Dickinson, Immunocytometry Systems, San Jose, CA), and the G418-resistant cell populations were analyzed with a Becton Dickinson (Immunocytometry Systems) FACScan flow cytometer. For FACS analysis macrophages were pretreated with 3% BSA and 20% rabbit serum, and the stainings were performed in the presence of 1% BSA, 3% rabbit serum. The COS-1 (ATCC, Manassas, VA) cells were grown in DMEM.
Adhesion assays
For cell-adhesion assays of transfected and wild-type L929 cells 2 mM MnCl2 was added to the buffers. The biotinylated peptides were immobilized to streptavidin plates (Immobilizer Streptavidin Plate; Nunc, Kamstrup, Denmark) in 25 mM Tris, pH 8.0, 150 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 2 mM MnCl2 for 1 hour at room temperature or by overnight incubation at 4°C.
For the binding study with or without divalent cations, the adhesion assays were performed with buffers containing 5 mM EDTA, 5 mM EGTA and 2 mM MgCl2, 2 mM MgCl2 and 2 mM CaCl2, or 2 mM MnCl2.
Solid-phase ELISA assay
Ninety-six–well plates (Greiner, Solingen, Germany) were coated at 4°C with indicated amounts of CD11c/CD18 in assay buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 2 mM MnCl2). After blocking nonspecific sites with 1% HSA for 1 hour at room temperature (RT) the wells were washed 3 times with assay buffer. The recombinant ICAM-Fc proteins (25 μmg/well) were then added to the wells and incubated for 2 hours at RT. The wells were washed prior to the addition of a peroxidase-conjugated anti–human Fc mAb (1:1000 dilution; Amersham Biosciences, Freiburg, Germany) to the wells. After a 1-hour incubation at 37°C, the plates were washed, and the bound proteins were detected with 100 μmL/well 0.5 mg/mL o-phenylenediamine dihydrochloride added for 10 minutes, stopped by the addition of 50 μmL 12.5% H2SO4, and read in an enzyme-linked immunoabsorbent assay (ELISA) reader. For inhibition experiments, the recombinant soluble Fc fusion proteins or protein-coated wells were pretreated with different mAbs (50 μmg/mL).
Synthetic peptides
Peptides synthesized as spots on derivatized cellulose membranes.
The protein sequence of the extracellular part of ICAM-4 was synthesized as 56 membrane-bound peptides 15 amino acids long (SPOTs) with a 3–amino acid overlap using Abimed Auto-Spot robot ASP 222 (with F-moc chemistry; Abimed, Langensfeld, Germany).
Soluble peptides.
ICAM-4 sequence-derived peptides were synthesized by solid-phase synthesis using F-moc chemistry in an Applied Biosystems (Weiterstadt, Germany) model 433A automatic peptide synthesizer and purified by reverse-phase chromatography. The sequences of the peptides were confirmed by MALDI-TOF mass analysis.
The following peptides were synthesized: a 13–amino acid peptide derived from the Ig-like domain 1 of ICAM-4 (residues 44-56 of ICAM-4 = P-D1; sequence, PQPQNSSLRTPLR), a 13–amino acid peptide derived from the Ig-like domain 2 of ICAM-4 (residues 161-173 of ICAM-4 = tP-D2; sequence, VTLTYEFAAGPRD). Biotinylated versions of the same peptides were also synthesized to enable possible detection and immobilization. For the inhibition studies a control peptide (Pcontr1) of 10 residues with the sequence of ELSGRLPWLY was used, as well as a biotinylated control peptide derived from the cytoplasmic part of ICAM-5 (Pcontr2; sequence, GGGKKGEY).
Peptide-binding assays
For interaction studies, the PepSpot membrane containing the ICAM-4–derived peptides was blocked o/n at RT with TBST (Tris-buffered saline, pH 8.0, 0.05% Tween 20) containing 2.5% dried milk, 1.5% BSA, and 5% sucrose. The membrane was washed 3 times with Tris-buffered saline and incubated for 1.5 hours at 37°C with purified CD11c/CD18 integrin at a concentration of 5 μmg/mL in blocking buffer diluted 3:1 with TBST in the presence of 1 mM CaCl2, 1 mM MgCl2, and 1 mM MnCl2. After washes the bound integrin was visualized on X-ray film according to the protocol of enhanced chemiluminescence (ECL) Western blotting (Amersham Biosciences) using the CD11c/CD18 mAb CBRp150/4G1 (1 μmg/mL) and peroxidase-conjugated rabbit antimouse antibody (1:5000 dilution).
Erythrophagocytosis assay
Human buffy coat cells separated by Ficoll-Hypaque centrifugation were placed in 12-well culture dishes (Greiner) at 3.5 million cells/well. The adhered monocytes were differentiated to macrophages as described.42 The phagocytosis experiments were adapted from the method of Bratosin et al.43 First, the macrophage Fc receptors were blocked with heat-inactivated bovine serum for 10 minutes. Monoclonal anti–ICAM-4 or anti-integrin antibodies (40 μmg/mL) were incubated with macrophages for 20 minutes before adding 15 × 106 PKH-26–labeled (cell linker kit; Sigma) red cells. After 2 hours of incubation at 37°C, macrophages were washed 3 times with RPMI 1640 medium. Noninternalized red cells were lysed with hypotonic buffer, and macrophages were detached with lidocaine solution. Cells were fixed with 70% ethanol and analyzed by flow cytometry. The results were presented as relative (%) phagocytosis derived from mean values of the data. Experiments were repeated 3 times in duplicates.
Modeling of ICAM-4
The ICAM-411 (SWISS-PROT accession no. Q14773) model was built on the structure of ICAM-244 (Protein Data Base code 1ZXQ). Twenty models were made using the modeling program MODELLER (A. Sali, Department of Pharmaceutical Sciences, University of California–San Francisco).45 Each model was analyzed first at the overall structural level at which the models with the highest objective functions were discarded. The objective function describes the degree of fit of the model to the input structural data used in its construction, derived by the program MODELLER.45 Then the orientation of the amino acids in the models were analyzed one amino acid at a time, giving higher values to those amino acids that have been shown to be involved in ligand binding.28 In the final model most of the orientations of the amino acids were shared with the template structure. The objective function was near the average of the objective functions of all models. The detailed structural analysis of the ICAM-4 models was made using the Bodil Molecular Modeling Environment (M. S. Johnson, Structural Bioinformatics, Åbo Akademi University, Åbo, Finland).46 The ICAM-4 model was also validated with PROCHECK (J. M. Thornton, European Bioinformatics Institute, Cambridge, United Kingdom).47 Figure 7A and 7B were made using the program MOLSCRIPT48 (P. E. Kraulis, Stockholm University, Sweden) and Raster3D (D. J. Bacon and E. A. Merritt, Department of Biochemistry, University of Washington, Seattle).49,50
Results
Red cells and ICAM transfectants adhere to purified CD11c/CD18 integrin
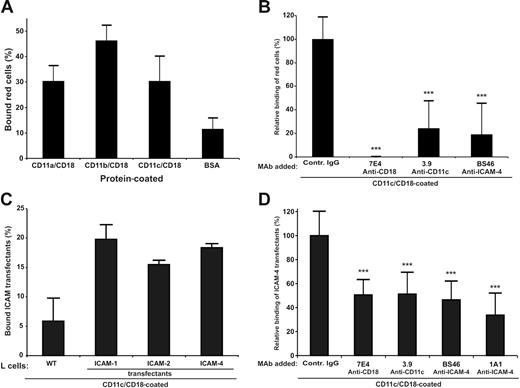
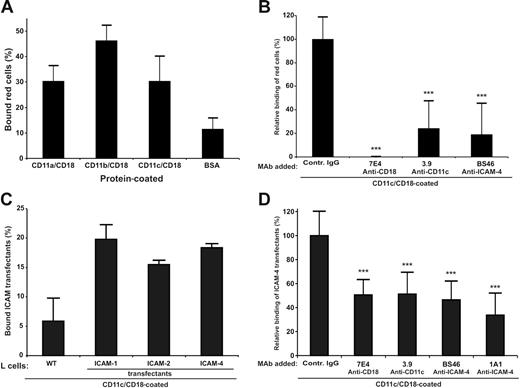
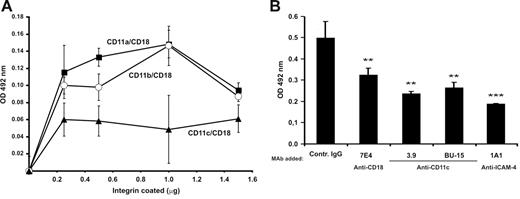
In SDS–polyacrylamide gel electrophoresis (PAGE) analysis of the purified CD11a/CD18, CD11b/CD18, CD11c/CD18, and the wild-type and mutant CD11c I domain preparations no major impurities were observed (Figure S1). Both the purified integrins and the recombinant I domains were tested for correct folding by ELISA using several anti-integrin antibodies. Red cells readily bound to coated CD11c/CD18 as well as to the other 2 CD18 integrins (Figure 1A). The binding to CD11c/CD18 was efficiently inhibited by monoclonal antibodies to ICAM-4 and CD11c/CD18 (Figure 1B). However, the adhesion was not completely blocked by the CD11c and ICAM-4 mAbs. There may exist other red-cell receptors for CD11c/CD18, and the purified immobilized CD11c/CD18 may not be recognized by the 3.9 antibody with equal efficiency as cellular integrins. Using purified CD11c/CD18, we also studied the adhesion of ICAM transfectants29 to coated CD11c/CD18 (Figure 1C). Their ICAM expression was ascertained by FACS analysis (Table 1). All the ICAM transfectants adhered to the coated integrin with similar efficiency (approximately 20% of the total added cells). Because the expression levels of ICAM-4 and ICAM-2 in L-cell transfectants were about 70% lower than ICAM-1 transfectants,29 these results suggest that ICAM-4 might be an even more potent ligand for CD11c/CD18 than ICAM-1. To a certain extent (10%-20%) the binding efficiencies to ICAMs varied between different preparations of CD11/CD18 integrins. However, the same preparations were used in all experiments reported here. The mAbs to CD11c/CD18 and ICAM-4 clearly inhibited the binding of ICAM-4 L cells to coated CD11c/CD18 integrin (Figure 1D). The ICAM-4 transfectants also bound to purified CD11c I domains, and the level of binding was similar both in the wild-type and the active Ile314Gly mutants41 (not shown).
Adhesion of red cells and ICAM-4 transfectants to CD11c/CD18 is mediated by ICAM-4. Adhesion assays were as described in “Materials and methods.” (A) The binding of erythrocytes to 1 μmg coated β2 integrins; (B) the inhibitory effect of anti–ICAM-4 (BS46), anti-CD18 (7E4), and anti-CD11c (3.9) monoclonal antibodies on the adhesion of red cells to purified CD11c/CD18. (C) Wells were coated with 1 μmg purified CD11c/CD18, and the adhesion of parental L cells and different ICAM L-cell transfectants was measured. (D) The effect of antibodies on binding of ICAM-4 L-cell transfectants to 1 μmg coated CD11c/CD18 was studied. The data in panels A and C are presented as a percentage of attached cells (amount of bound cells divided by input of cells). The results in panels B and D are expressed as a relative percentage of bound cells, where 100% is calculated from the total number of cells bound to the CD11c/CD18 in the absence of pretreatment with MAbs. The significance was determined by unpaired Student t test. Controls included unrelated mouse IgG antibody and wells with coated control protein (GPA) or without coated protein (BSA only). Background binding of cells to GPA or BSA was subtracted. The experiments were repeated 3 times with similar results. Data are expressed as mean ± SD, and statistical significances are shown, ***P< .005.
Adhesion of red cells and ICAM-4 transfectants to CD11c/CD18 is mediated by ICAM-4. Adhesion assays were as described in “Materials and methods.” (A) The binding of erythrocytes to 1 μmg coated β2 integrins; (B) the inhibitory effect of anti–ICAM-4 (BS46), anti-CD18 (7E4), and anti-CD11c (3.9) monoclonal antibodies on the adhesion of red cells to purified CD11c/CD18. (C) Wells were coated with 1 μmg purified CD11c/CD18, and the adhesion of parental L cells and different ICAM L-cell transfectants was measured. (D) The effect of antibodies on binding of ICAM-4 L-cell transfectants to 1 μmg coated CD11c/CD18 was studied. The data in panels A and C are presented as a percentage of attached cells (amount of bound cells divided by input of cells). The results in panels B and D are expressed as a relative percentage of bound cells, where 100% is calculated from the total number of cells bound to the CD11c/CD18 in the absence of pretreatment with MAbs. The significance was determined by unpaired Student t test. Controls included unrelated mouse IgG antibody and wells with coated control protein (GPA) or without coated protein (BSA only). Background binding of cells to GPA or BSA was subtracted. The experiments were repeated 3 times with similar results. Data are expressed as mean ± SD, and statistical significances are shown, ***P< .005.
Soluble ICAM-4 binds to purified CD11c/CD18
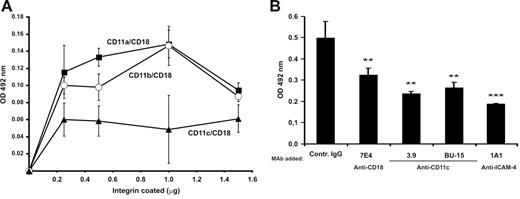
Further proof for the specific binding of ICAM-4 to the CD11c/CD18 integrin was obtained using a cell-free assay (Figure 2). By performing solid-phase ELISA assays with recombinant soluble ICAM-4Fc, we showed that ICAM-4 bound to coated purified CD11a/CD18, CD11b/CD18, and CD11c/CD18 in a dose-dependent fashion (Figure 2A). High amounts of coated integrin resulted in decreased binding possibly because of steric hindrance. Similar effects have been seen before.29 We also found that both CD11c/CD18-specific mAbs (3.9 and BU15) and anti-CD18 antibody 7E4 partially but significantly inhibited the binding, whereas the 1A1 (anti–ICAM-4) blocked more efficiently (Figure 2B).
Soluble ICAM-4Fc binds specifically to immobilized purified CD11c/CD18 in a solid-phase assay. (A) Dose-dependent binding of recombinant ICAM-4Fc to coated integrins. ICAM-4Fc + CD11a/CD18 (▪), ICAM-4Fc + CD11b/CD18 (○), ICAM-4Fc + CD11c/CD18 (▴). Background binding of control protein (human IgG1) was subtracted. (B) The effect of monoclonal antibodies on binding of soluble ICAM-4Fc to 0.4 μmg coated CD11c/CD18 in a solid-phase assay is shown. The ICAM-4Fc binding to CD11c/CD18 was examined in the presence of mAbs (50 μmg/mL) against the ICAM-4 (1A1), CD18 (7E4), and CD11c (3.9, BU15). Data shown are from 1 representative experiment of 3. Standard deviations and statistical significances are shown; ***P < .005, **P< .05. The significances were determined by unpaired Student t test.
Soluble ICAM-4Fc binds specifically to immobilized purified CD11c/CD18 in a solid-phase assay. (A) Dose-dependent binding of recombinant ICAM-4Fc to coated integrins. ICAM-4Fc + CD11a/CD18 (▪), ICAM-4Fc + CD11b/CD18 (○), ICAM-4Fc + CD11c/CD18 (▴). Background binding of control protein (human IgG1) was subtracted. (B) The effect of monoclonal antibodies on binding of soluble ICAM-4Fc to 0.4 μmg coated CD11c/CD18 in a solid-phase assay is shown. The ICAM-4Fc binding to CD11c/CD18 was examined in the presence of mAbs (50 μmg/mL) against the ICAM-4 (1A1), CD18 (7E4), and CD11c (3.9, BU15). Data shown are from 1 representative experiment of 3. Standard deviations and statistical significances are shown; ***P < .005, **P< .05. The significances were determined by unpaired Student t test.
Integrin transfectants adhere to ICAM-4
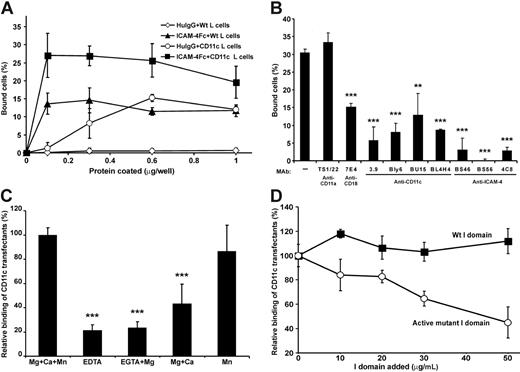
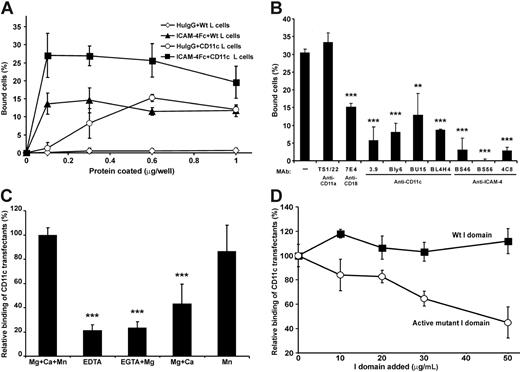
The surface expression of β2 integrins on CD11c/CD18 transfectants was studied by FACS (Table 1). As shown in Figure 3A the coated chimeric ICAM-4Fc protein supported the adhesion of CD11c/CD18 transfectants. Some background binding of untransfected L cells was observed, indicating additional interactions. To check the specificity of the interaction, we studied the effects of mAbs on the adhesion of transfectants to ICAM-4Fc (Figure 3B). Monoclonal antibodies against either the α or β chain inhibited the binding of CD11c/CD18 transfectants approximately by 50% to 85%, whereas the anti–ICAM-4 mAbs blocked the binding efficiently down to background levels. To obtain further evidence of specificity, we investigated the divalent cation requirements of the CD11c/CD18 transfectant binding to ICAM-4Fc (Figure 3C). In the absence of cations, the adhesion was efficiently but not totally abolished. As can be seen in Figure 3C, MgCl2 alone or in combination with CaCl2 was not sufficient to support the maximal binding of CD11c/CD18 L cells to ICAM-4Fc. Indeed, the presence of Mn2+ seems to be required for high-affinity binding of CD11c/CD18 transfectants to ICAM-4. Interestingly, the binding was efficiently inhibited by the soluble active mutant of CD11c I domain (Figure 3D).
CD11c/CD18 transfectants show specific cation-dependent binding to coated ICAM-4Fc. (A) The adhesion of parental L cells and CD11c/CD18 transfectants to ICAM-4Fc and human IgG proteins coated on plastic wells. CD11c/CD18 L cells + ICAM-4Fc (▪), parental L cells + ICAM-4Fc (▴), CD11c/CD18 L cells + human IgG (○), parental L cells + human IgG (◊). (B) The effects of anti–ICAM-4, anti-CD18, and anti-CD11c mAbs on adhesion of CD11c/CD18 transfectants to 0.5 μmg purified ICAM-4Fc fusion protein. Controls included wells with binding of wild-type L cells (not shown) and the effect of control antibody (anti-CD11a mAb). (C) The binding of transfectants to ICAM-4Fc in the absence or presence of divalent cations. (D) The binding of CD11c/CD18 transfectants to coated ICAM-4Fc in the presence of indicated concentrations of soluble wild-type CD11c I domain (▪) and the active mutant I domain (○). In panels A and B the results are shown as the percentage of input cells bound ± SD. In panels C and D the results are expressed as a relative percentage of bound cells, where 100% is given as the number of cells bound to the ICAM-4Fc in the presence of divalent cations (C) or in the absence of soluble I domain (D). Background binding of cells to HSA was subtracted. The experiments were repeated 3 to 5 times with similar results. Standard deviations and statistical significances are shown; ***P < .001, **P < .01.
CD11c/CD18 transfectants show specific cation-dependent binding to coated ICAM-4Fc. (A) The adhesion of parental L cells and CD11c/CD18 transfectants to ICAM-4Fc and human IgG proteins coated on plastic wells. CD11c/CD18 L cells + ICAM-4Fc (▪), parental L cells + ICAM-4Fc (▴), CD11c/CD18 L cells + human IgG (○), parental L cells + human IgG (◊). (B) The effects of anti–ICAM-4, anti-CD18, and anti-CD11c mAbs on adhesion of CD11c/CD18 transfectants to 0.5 μmg purified ICAM-4Fc fusion protein. Controls included wells with binding of wild-type L cells (not shown) and the effect of control antibody (anti-CD11a mAb). (C) The binding of transfectants to ICAM-4Fc in the absence or presence of divalent cations. (D) The binding of CD11c/CD18 transfectants to coated ICAM-4Fc in the presence of indicated concentrations of soluble wild-type CD11c I domain (▪) and the active mutant I domain (○). In panels A and B the results are shown as the percentage of input cells bound ± SD. In panels C and D the results are expressed as a relative percentage of bound cells, where 100% is given as the number of cells bound to the ICAM-4Fc in the presence of divalent cations (C) or in the absence of soluble I domain (D). Background binding of cells to HSA was subtracted. The experiments were repeated 3 to 5 times with similar results. Standard deviations and statistical significances are shown; ***P < .001, **P < .01.
CD11c/CD18 integrin binding sites on ICAM-4
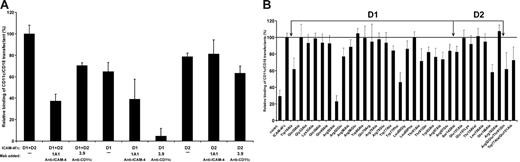
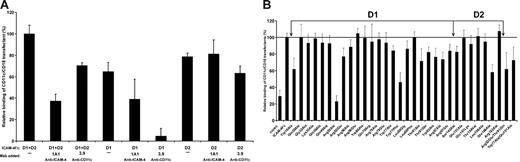
To establish which ICAM-4 Ig domains contain CD11c/CD18 binding sites, ICAM-4 deletion mutants lacking either domain D1 or D2 were tested in cell-adhesion assays (Figure 4A). Neither domain alone was sufficient for the maximal binding of CD11c/CD18 transfectants. However, both domain deletion mutants retained partial binding capacity. As expected, the anti–ICAM-4 mAb, 1A1, the epitopes of which reside within domain D1, affected only the binding to the wild-type ICAM-4Fc and the deletion mutant carrying domain D1. The I domain–specific anti-CD11c mAb, 3.9, inhibited CD11c/CD18 transfectant binding partially to wild-type ICAM-4 and only slightly to the mutant lacking D1. Instead, the adhesion to the mutant lacking domain D2 was almost totally abolished by 3.9.
Effects of domain deletion mutations and amino acid substitutions on ICAM-4 binding to CD11c/CD18 transfectants. Adhesion of CD11c/CD18-expressing L cells to plastic-coated native and mutated ICAM-4Fc (0.5 μmg/well). Results are shown as the percentage of CD11c/CD18 transfectant-cell binding relative to native ICAM-4Fc (100%). (A) The binding of CD11c/CD18 transfectants to ICAM-4 domain deletion mutants in the absence or the presence of mAbs 1A1 (anti–ICAM-4) or 3.9 (anti-CD11c). Controls included wells with binding of wild-type L cells and the effect of control antibody (not shown). Background binding of cells to HSA was subtracted. Panel B shows the adhesion of CD11c/CD18 transfectants to a panel of ICAM-4 mutants. The mean ± SD from 3 experiments is shown.
Effects of domain deletion mutations and amino acid substitutions on ICAM-4 binding to CD11c/CD18 transfectants. Adhesion of CD11c/CD18-expressing L cells to plastic-coated native and mutated ICAM-4Fc (0.5 μmg/well). Results are shown as the percentage of CD11c/CD18 transfectant-cell binding relative to native ICAM-4Fc (100%). (A) The binding of CD11c/CD18 transfectants to ICAM-4 domain deletion mutants in the absence or the presence of mAbs 1A1 (anti–ICAM-4) or 3.9 (anti-CD11c). Controls included wells with binding of wild-type L cells and the effect of control antibody (not shown). Background binding of cells to HSA was subtracted. Panel B shows the adhesion of CD11c/CD18 transfectants to a panel of ICAM-4 mutants. The mean ± SD from 3 experiments is shown.
To define the binding sites of CD11c/CD18 in ICAM-4 in more detail, we tested 32 ICAM-4Fc mutant proteins.28 The residues targeted for mutation were all expected to be surface exposed, being able to participate in molecular interactions with counter-receptors. The mutant proteins were tested for correct folding by ELISA with the anti–ICAM-4 mAb, 1A1. This mAb also reacted with the Trp19Ala mutant not recognized by the BS mAbs in our previous report, indicating partially correct folding.
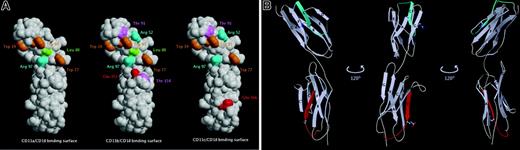
As shown in Figure 4B, 4 single point mutations caused a reduction in cell adhesion by 38% or greater. However, when the control binding (29%) is taken into account, the adhesion was reduced by 54% or greater. Three of these are located within domain D1 on the A strand at position Trp19, on the C strand at position Arg52, and in the E to F loop at position Trp77. One of these 4 mutations localized to D2, on the strand E at position Glu166. The Arg52Ala mutation almost completely eliminated binding. In addition to these 4 ICAM-4 mutants, the adhesion to mutants at positions Thr91, Trp93, and Arg 97 reduced adhesion by 25% to 30% (background not subtracted). When the Arg residues at positions 52 and 97 were mutated to opposite charge (Glu), the effect on cell adhesion was weaker than observed with substitution by an apolar amino acid (Ala), indicating that charge is needed at these positions. Furthermore, replacement of Trp at position 77 by Phe caused a severe reduction of CD11c/CD18 L-cell adhesion, whereas substitution by Ala had a minor effect. However, for the double mutants Arg52Glu/Thr91Gln and Trp77Ala/Glu151Ala a 40% and 30% reduction in cell adhesion was noted, respectively, confirming the importance of ICAM-4 residues Arg52, Thr91, and Trp77 in CD11c/CD18 binding. According to these data, of the 7 residues critical for ICAM-4 interaction with CD11c/CD18 integrin, 6 are spatially close, spanning the interface between the ABED and CFG faces of domain D1 with an extension of one important residue far from the others in the domain D2 (Figure 5A).
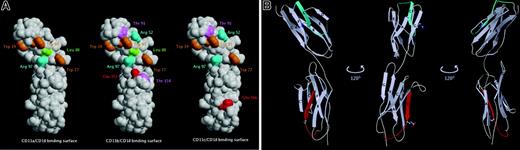
Critical residues of ICAM-4 involved in interaction with β2 integrins and location of ICAM-4 peptides that mediate adhesion to CD11c/CD18 integrin. (A) Three-dimensional representation of an ICAM-4 model constructed on the basis of ICAM-2 structure. Amino acids are designated by 3-letter codes and residue numbers. The solvent-accessible surfaces of ICAM-4 residues that when mutated cause a decrease in adhesion to CD11a/CD18 (left), CD11b/CD18 (middle), and CD11c/CD18 (right) are colored (Trp, orange; Arg, cyan; Leu, green; Thr, magenta; Glu, red). (B) A ribbon diagram of ICAM-4 displayed in 3 orientations 120 degrees to one another. The strands of the antiparallel β sheets are labeled by capital letters. The locations of the ICAM-4 peptides are colored: P-D1 (PQPQNSSLRTPLR), residues 44 to 56 (cyan); P-D2 (VTLTYEFAAGPRD), residues 161 to 173 (red). Side chains of the 2 peptide residues found to be involved in CD11c/CD18 binding by mutagenesis studies are shown as ball and stick.
Critical residues of ICAM-4 involved in interaction with β2 integrins and location of ICAM-4 peptides that mediate adhesion to CD11c/CD18 integrin. (A) Three-dimensional representation of an ICAM-4 model constructed on the basis of ICAM-2 structure. Amino acids are designated by 3-letter codes and residue numbers. The solvent-accessible surfaces of ICAM-4 residues that when mutated cause a decrease in adhesion to CD11a/CD18 (left), CD11b/CD18 (middle), and CD11c/CD18 (right) are colored (Trp, orange; Arg, cyan; Leu, green; Thr, magenta; Glu, red). (B) A ribbon diagram of ICAM-4 displayed in 3 orientations 120 degrees to one another. The strands of the antiparallel β sheets are labeled by capital letters. The locations of the ICAM-4 peptides are colored: P-D1 (PQPQNSSLRTPLR), residues 44 to 56 (cyan); P-D2 (VTLTYEFAAGPRD), residues 161 to 173 (red). Side chains of the 2 peptide residues found to be involved in CD11c/CD18 binding by mutagenesis studies are shown as ball and stick.
ICAM-4–derived peptides interact with CD11c/CD18 and inhibit cell adhesion
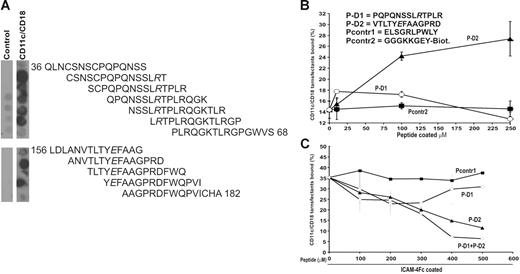
Using a PepSpot assay, we were able to identify 2 peptide regions derived from ICAM-4 which selectively bound to soluble purified CD11c/CD18 integrin. CD11c/CD18 reacted with 5 overlapping peptides spanning the ICAM-4 sequence of amino acids 39 to 65 located in domain D1 and with 3 peptides spanning the sequence of amino acids 159 to 179 located in domain D2 (Figure 6). Consistent with the mutation results the peptide region from domain D1 includes the important Arg52 (see Figure 4). Furthermore, the only critical amino acid mutation localized to D2 at position Glu166 was included in the selected peptide sequences from D2. According to these results, 2 peptides were chosen for further analysis in cell-adhesion assays: P-D1 (PQPQNSSLRTPLR) covering the C strand and parts of the loops at both ends of the C strand in the domain 1 and P-D2 (VTLTYEFAAGPRD) derived from the domain 2 and covering the E strand and part of the loop connecting the E strand to F strand (Figure 5B). Two available control peptides, Pcontr1 (ELSGRLPWLY) and Pcontr2 (biotinylated GGGKKGEY), were used in the cell-adhesion assays, but in the PepSpot assays the negative and flanking peptides also acted as controls.
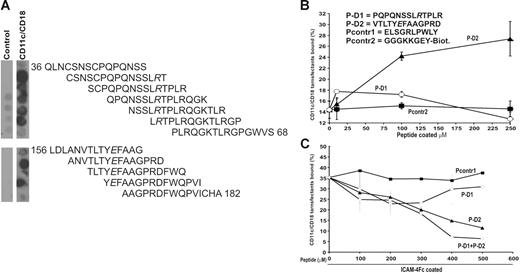
CD11c/CD18 binds selectively to peptides derived from the ICAM-4 sequence. (A) A total of 56 overlapping synthetic peptides 15 amino acid-long that corresponded to the protein sequence of the extracellular part of ICAM-4 were synthesized as immobilized spots on a cellulose membrane. The reactivities of the peptides with purified CD11c/CD18 integrin were tested. The CD11c/CD18 mAb CBRp150/4G and peroxidase-conjugated rabbit antimouse antibody were used to detect bound CD11c/CD18 integrin. The figure shows the reactivity of the overlapping peptides selected with the soluble CD11c/CD18 integrin. Negative control was carried out in the absence of the integrin. According to these results 2 peptides were chosen for solid-phase synthesis: P-D1 derived from the ICAM-4 Ig-like domain 1 and P-D2 derived from the domain 2. Both of the peptides included an amino acid shown to be involved in adhesion to CD11c/CD18 according to our mutational studies (see Figure 4). (B) The adhesion of the CD11c/CD18 transfectants to biotinylated versions of the selected ICAM-4 peptides and a control peptide (Pcontr2) captured by streptavidin microplates. Indicated amounts of the peptides were coated per well, and the cell adhesion was performed as described in “Materials and methods.” Sequences of the synthesized ICAM-4–derived peptides and the peptides used as control are listed in the figure. (C) The effects of the defined ICAM-4 peptides and the control peptide on adhesion of the CD11c/CD18 transfectants to coated ICAM-4Fc are shown. The cells were pretreated or not with the control peptide (Pcontr1), P-D1, P-D2 or the P-D1 and P-D2 peptides together at different final concentrations. A representative binding experiment with results expressed as the percentage of input cells bound are shown. Control peptides (▪), P-D2 (▴), P-D1 (○), P-D1 + P-D2 (◊).
CD11c/CD18 binds selectively to peptides derived from the ICAM-4 sequence. (A) A total of 56 overlapping synthetic peptides 15 amino acid-long that corresponded to the protein sequence of the extracellular part of ICAM-4 were synthesized as immobilized spots on a cellulose membrane. The reactivities of the peptides with purified CD11c/CD18 integrin were tested. The CD11c/CD18 mAb CBRp150/4G and peroxidase-conjugated rabbit antimouse antibody were used to detect bound CD11c/CD18 integrin. The figure shows the reactivity of the overlapping peptides selected with the soluble CD11c/CD18 integrin. Negative control was carried out in the absence of the integrin. According to these results 2 peptides were chosen for solid-phase synthesis: P-D1 derived from the ICAM-4 Ig-like domain 1 and P-D2 derived from the domain 2. Both of the peptides included an amino acid shown to be involved in adhesion to CD11c/CD18 according to our mutational studies (see Figure 4). (B) The adhesion of the CD11c/CD18 transfectants to biotinylated versions of the selected ICAM-4 peptides and a control peptide (Pcontr2) captured by streptavidin microplates. Indicated amounts of the peptides were coated per well, and the cell adhesion was performed as described in “Materials and methods.” Sequences of the synthesized ICAM-4–derived peptides and the peptides used as control are listed in the figure. (C) The effects of the defined ICAM-4 peptides and the control peptide on adhesion of the CD11c/CD18 transfectants to coated ICAM-4Fc are shown. The cells were pretreated or not with the control peptide (Pcontr1), P-D1, P-D2 or the P-D1 and P-D2 peptides together at different final concentrations. A representative binding experiment with results expressed as the percentage of input cells bound are shown. Control peptides (▪), P-D2 (▴), P-D1 (○), P-D1 + P-D2 (◊).
The immobilized P-D2 peptide supported binding of CD11c/CD18 transfectants efficiently, whereas the P-D1 peptide supported adhesion only weakly. The binding profiles of the 2 peptides were clearly different as well (Figure 6B). Both peptides inhibited the interaction between ICAM-4 and CD11c/CD18. However, the P-D2 peptide was a significantly more effective blocker of the CD11c/CD18 L-cell binding. Figure 6C shows that the inhibition of CD11c/CD18 transfectant adhesion to ICAM-4Fc by the P-D2 peptide was concentration dependent, and 65% inhibition was obtained with a peptide concentration of 500 μmM. The P-D1 peptide was less active. The blocking activity was reduced with higher P-D1 amounts. When the peptides were tested together, there was an additional inhibition of binding.
Erythrophagocytosis is mediated by ICAM-4/β2 integrin interaction
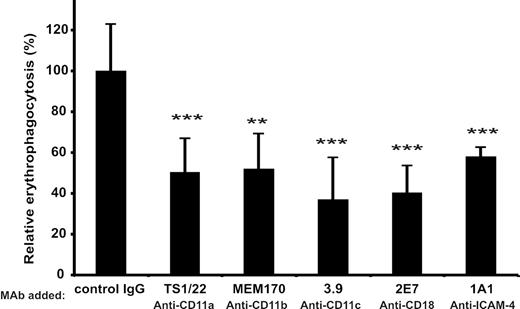
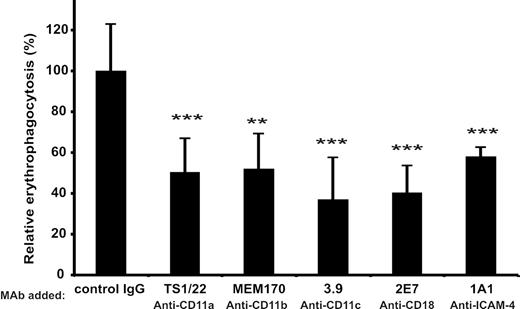
To find a possible physiologic role of the ICAM-4/β2 integrin interaction we studied erythrophagocytosis. The expression of β2 integrins on cultured, monocyte-derived macrophages was verified by FACS (Table 1). The uptake of PKH-26–labeled red cells by human macrophages was effectively inhibited by antibodies against ICAM-4 and β2 integrins. Figure 7 shows that these mAbs reduced the phagocytosis of red cells by 50% or more. Greater than 60% inhibition was obtained in the presence of the 3.9 CD11c mAb.
Phagocytosis of red cells by macrophages is inhibited by β2 integrin and ICAM-4 antibodies. (A) Effect of mAbs against the CD11a (TS1/22), CD11b (MEM170), CD11c (3.9), CD18 (2E7), and ICAM-4 (1A1) on erythrophagocytosis. The results are presented as relative phagocytosis derived from mean values of the data; ***P < .001, **P < .01.
Phagocytosis of red cells by macrophages is inhibited by β2 integrin and ICAM-4 antibodies. (A) Effect of mAbs against the CD11a (TS1/22), CD11b (MEM170), CD11c (3.9), CD18 (2E7), and ICAM-4 (1A1) on erythrophagocytosis. The results are presented as relative phagocytosis derived from mean values of the data; ***P < .001, **P < .01.
Discussion
ICAM-4 is remarkable because of its ability to bind several members of integrin subclasses. These include the αv integrins (αvβ1, αvβ3, and αvβ5), α4β1 and αIIbβ3,24-26 in addition to CD11a/CD18 and CD11b/CD18.21 The binding sites for all the ICAM-4 receptor integrins, except for CD11a/CD18, comprise residues on both Ig-like domains of ICAM-4. The regions of ICAM-4 identified as β2 and β3 integrin interaction sites appear clearly distinct, although they all share W77. The ICAM-4 amino acids predicted to be critical for binding to αvβ1 and αvβ5 are located in close proximity to those needed for β2 integrin interaction. However, R97 is the only residue that is shared between the 2 sites.25,26,28,51 The ability of ICAM-4 to interact selectively with several different integrin receptors suggests multiple functions for ICAM-4 in red-cell physiology and pathology.27
The CD11c/CD18 (αxβ2) integrin is most homologous to the CD11b/CD18 and CD11d/CD18 integrins with 60% to 66% amino acid identity, whereas it is only 37% identical to CD11a/CD18.52-54 CD11c/CD18 plays an important role in phagocytosis, chemotaxis, and regulation of the immune response.12,13 Little is known about the cellular ligands of CD11c/CD18, but the ligands appear to overlap in part with those of CD11b/CD18. In addition to several soluble and matrix ligands, the only reported cellular ligands for CD11c/CD18 are ICAM-137,55 and Thy-1.56 A recent study shows that the binding site of CD11c/CD18 on ICAM-1 resides on domain D4,57 and another report suggested that negatively charged residues in structurally decayed proteins serve as a pattern recognition motif for CD11c/CD18.58 Preliminary experiments indicated that CD11c/CD18 in THP-1 cells binds to purified ICAM-4, but we failed to see binding to red cells.21 This could be due to low affinity of CD11c/CD18, but now we have used an antibody, which allows purification in a functionally active form.37
In this study we have convincingly demonstrated that red-cell ICAM-4 binds to CD11c/CD18: (1) red cells and ICAM-4 L-cell transfectants adhered to immobilized purified CD11c/CD18; (2) the specificity was confirmed by the ability of antibodies against CD11c/CD18 and ICAM-4 to block the adhesion;, (3) with purified CD11c/CD18 and ICAM-4 a direct binding was observed; (4) adhesion of the CD11c/CD18 transfectants to recombinant-coated ICAM-4Fc was inhibited by soluble I domain, the antibodies against ICAM-4, and the α and β subunits of the CD11c/CD18; (5) ICAM-4 deletion mutants lacking either domain D1 or D2 showed lower binding to CD11c/CD18; (6) the panel of point mutants studied resulted in the identification of 7 residues of ICAM-4 important for binding to CD11c/CD18. ICAM-4 residues in domain D1 (Trp19, Arg52, Trp77, Thr91, Trp93, and Arg97) have previously been shown to be involved in binding to CD11b/CD18 as well.
Figure 5 shows a model of the external part of the ICAM-4 molecule, with critical amino acids marked defining distinct but overlapping β2 integrin binding footprints on ICAM-4. As expected, our model is consistent with the one published by Hermand et al28 because these both have the same template structure. Furthermore, the amino acids that were reported to reside on the surface of the protein51 are on the surface in our model as well. D1 residues showing the greatest specific effect on CD11c/CD18 binding, and which may be central in the binding site, are Arg52 at the bottom of strand C, Trp19 at the top of strand A, and Trp77 on the E to F loop. Trp19, Arg52, Thr91, and Trp93 lie approximately in the middle of domain D1.
Previous and current results show that both Ig-like domains of ICAM-4 are involved in binding to CD11b/CD18 and CD11c/CD18. However, the vital ICAM-4 residues in domain D2 mediating adhesion to the 2 integrins are different. Mutation of Glu166 at the bottom of strand E in D2 reduced binding to CD11c/CD18, whereas the recognition site for CD11b/CD18 is located in the C′ to E loop of D2 far from the CD11c/CD18 binding site. The model of ICAM-4 shows that the location of the Glu166 is spatially distant from the rest of the integrin-binding footprint. This could indicate that the integrin molecules interact with a large surface area of the ICAM-4 molecule. It is interesting to note that in all other ICAMs there is a glutamic acid residue in the first domains, which are needed for adhesion, but in ICAM-4 this is replaced by Arg52. Glu166 may substitute for this. The binding of the CD11b/CD18 and CD11c/CD18 integrins to additional sites present on domain D2 of ICAM-4 is consistent with the stronger binding to ICAM-4 observed for these 2 β2 integrins as compared with CD11a/CD18. The I domain–specific anti-CD11c mAb, 3.9, efficiently blocked the binding to the D2 deletion mutant but only slightly to the D1 deletion mutant. These data suggest that the I domain mainly binds to the D1 of ICAM-4, but they also indicate that multiple regions of the CD11c/CD18 could be involved in the interaction with ICAM-4.
Our results with synthetic peptides provided additional support for the role of ICAM-4 residues identified by site-directed mutagenesis. Using PepSpot analysis, we found 2 peptides derived from ICAM-4 reacting with CD11c/CD18. Both peptides included amino acids (Arg52 in D1 and Glu166 in D2) shown to be needed for CD11c/CD18 binding. The peptide binding and inhibition data are consistent with those of our mutational results and support the finding that both ICAM-4 domains are involved in binding to CD11c/CD18. The 2 peptides and the critical residues are exposed at the surface of the ICAM-4 molecule being available for interactions (Figure 5).
The role of ICAM-4 in red-cell physiology has remained poorly understood. CD11c/CD18 is expressed on macrophages30 (Table 1), and we report here that erythrophagocytosis was prevented by the inhibition of ICAM-4-β2 integrin binding, indicating that this interaction may be important in removal of erythrocytes from the circulation by splenic macrophages. Interestingly, in a recent report erythroblastic island formation was shown to decrease in ICAM-4–null mice, and the interactions between erythroblast ICAM-4 and macrophage αv integrin were reported to be critical for island integrity.59 Thus, ICAM-4 seems to be important for red-cell development and turnover. Furthermore, secreted isoforms of ICAM-4 may be important regulators of these molecular interactions.11,60
Although it has been commonly accepted that normal red cells are fairly nonadhesive during coagulation and thrombosis, increasing evidence shows that red cells are in fact able to form adhesive interactions during these physiologic processes. The ICAM-4/integrin interactions could be important during hemostasis where, in the developing thrombus, erythrocytes interact with activated neutrophils and monocytes. This possibility is supported by the fact that under low flow conditions adhesion of red cells to activated neutrophils involves interaction of ICAM-4 with CD11b/CD18.61 The finding that ICAM-4 binds to the platelet integrin αIIbβ3 indicates that this interaction may be involved in platelet-erythrocyte aggregate formation during coagulation.26 Interestingly, recent data indicate that during the pathogenesis of sickle red-cell vasoocclusion, adherent leukocytes bind not only to the inflamed endothelium but also to the erythrocytes.62 Furthermore, a recent report shows that activated ICAM-4 mediates binding of the sickle red cells to endothelial αvβ3 integrin.63 These findings indicate that drugs targeting ICAM-4-integrin interactions would be of great therapeutic value. Our results with synthetic peptides suggest that modulation of these interactions is possible.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: E.I., L.M.U., and A.T. did the experimental work and planned several experiments; E.I. wrote the first version of the manuscript; M.V. made the structural model; and C.G.G. planned much of the work and obtained most of the financial support.
Acknowledgments
We thank T.A. Springer for the CD11c I domain constructs, Cami Kantor-Aaltonen for synthetic peptides, and Jussi Hepojoki for PepSpot preparations. We also thank them for their generous help and valuable comments on peptides. We thank Leena Kuoppasalmi and Maria Aatonen for expert technical support and Yvonne Heinilä for secretarial assistance.
This work was supported by the University of Helsinki, the Academy of Finland, the Sigrid Jusélius Foundation, the Magnus Ehrnrooth Foundation, and the Finnish Cancer Society.