Abstract
The antitumor potency of the mTOR inhibitor rapamycin (sirolimus) is the subject of intense investigations. Primary effusion lymphoma (PEL) appears as an AIDS-defining lymphoma and like Kaposi sarcoma has been linked to Kaposi sarcoma–associated herpesvirus (KSHV). We find that (1) rapamycin is efficacious against PEL in culture and in a murine xenograft model; (2) mTOR, its activator Akt, and its target p70S6 kinase are phosphorylated in PEL; (3) rapamycin inhibits mTOR signaling as determined by S6 phosphorylation; (4) KSHV transcription is unaffected; (5) inhibition of IL-10 signaling correlates with drug sensitivity; and (6) addition of exogenous IL-10 or IL-6 can reverse the rapamycin growth arrest. This validates sirolimus as a new treatment option for PEL.
Introduction
Organ transplant recipients are at an increased risk of Kaposi sarcoma (KS) and certain viral lymphomas because iatrogenic T-cell immunosuppression activates the causative agent of KS the Kaposi sarcoma–associated herpesvirus (KSHV/HHV-8).1–6 KSHV was originally identified in the context of HIV-1–induced T-cell depletion7 and is, by an overwhelming body of evidence, associated with KS as well as the B-cell lymphoproliferative diseases multicentric Castleman disease and primary effusion lymphoma (PEL).8–10 KHSV normally resides latent in a yet to be defined B-lymphocyte compartment.11 Immunosuppression is thought to disturb host surveillance of this virus, leading to reactivation, increased systemic viral load,12–15 and infection of endothelial cells. KSHV-infected LYVE-1+ (lymphatic vessel hyaluronan receptor-1), CD34+ lymphatic endothelial cells constitute the proliferating tumor cells in a KS lesion.16–20 In 2005, Stallone et al21 reported that switching from the immunosuppressant drug cyclosporine A to the immunosuppressant drug rapamycin (sirolimus) cured cutaneous KS in a group of 15 kidney transplant recipients. Over a 3-month period all KS lesions disappeared, whereas graft function remained level. This study separated the immunosuppressive function from the anticancer effect of rapamycin in a clinical setting and prompted us to evaluate rapamycin for PEL.
The mammalian target of rapamycin (mTOR) executes essential functions of Akt, which is also called protein kinase B, with regard to cancer cell growth and proliferation (reviewed in Hay22 ). Akt is among the most frequently activated kinases in human cancer. Receptor-tyrosine kinases activate Akt through the generation of phosphoinositol-3,4,5 phosphate (PI3K), which leads to Akt phosphorylation at Thr 308 (through PI3K-dependent kinase 1) and Ser 473 (through PI3K-dependent kinase 2). Generation of PI3K is counteracted by the phosphatase and tensin homolog deleted on chromosome 10 (PTEN) tumor suppressor gene. Phosphorylation at both Thr 308 and Ser 473 is required for full activation of Akt. Akt phosphorylates and thereby inhibits tuberous sclerosis complex 2 (TSC2), which heterodimerizes with TSC1. The TSC1-TSC2 heterodimer has GTPase activity, which inhibits the small G protein Rheb. Rheb is required for mTOR activation. Hence, TSC1 and TSC2 are considered tumor suppressor genes. The mTOR activation is marked by phosphorylation at Ser 2441 and 2448 and association with Raptor, the regulatory-associated protein of mTOR. The mTOR-Raptor complex has kinase activity and executes the rapamycin-sensitive functions of mTOR, such as phosphorylation of the p70 subunit of S6 kinase (p70S6K) at Thr 389 and Ser 371, and phosphorylation of 4E-BP1 at Ser 65 and Thr 37/46. Phospho-p70S6K phosphorylates small ribosomal protein S6 at Ser 235/236, which in turn facilitates protein translation. Phospho–4E-BP1 releases eIF4E from the inactive 4E-BP1/eIF4E heterodimer, which also facilitates translation. Hence, rapamycin-inhibiting mTOR inhibits translation, which leads to G1 cell-cycle arrest (cytostasis).
Rapamycin binds to FK506-binding protein 12 and the rapamycin-FKB12 complex inhibits mTOR kinase activity (reviewed in Sawyers23 ). Rapamycin inhibits cell proliferation with IC50s of 0.2 to 2 μM. It can cause apoptosis, but in most instances causes G1 cell-cycle arrest by impeding translation of essential proteins. Rapamycin was established clinically since 1999 as a second-generation immunosuppressive agent in the context of organ transplantation, because it inhibits IL-2 translation and secretion and thus inhibits T-cell proliferation. In the context of organ transplantation the cell autonomous G1 arrest phenotype of rapamycin is augmented by inhibition of IL-2, which is a paracrine and autocrine growth factor for T cells. Recently, rapamycin has been explored as an anticancer agent of solid tumors outside the context of lymphocyte inhibition. For instance, the rapamycin analog CCI-779 attenuates tumor growth in PTEN+/− mice and PTEN−/− cancer cells,24,25 and the prostate intraepithelial neoplasia phenotype of Akt transgenic mice can be antagonized by another rapamycin analog RAD001.26 Furthermore, rapamycin blocks focus formation induced by oncogenenic alleles of PI3K or Akt but not Src, Ras, or Myc.27 These observations place mTOR downstream of and epistatic to PTEN and Akt.
Materials and methods
Cell culture
Cell lines were cultured in RPMI 1640 supplemented with 100 μg/mL streptomycin sulfate, 100 U/mL penicillin G (Invitrogen Life Technologies, Carlsbad, CA), 2 mM l-glutamine, 0.05 mM 2-mercaptoethanol, 0.075% sodium bicarbonate, 1 U/mL human IL-6 (Roche, Indianapolis, IN), and 10% FBS at 37°C in 5% CO2. To test the in vitro effects of rapamycin (Sigma, St Louis, MO) and cyclosporine A (Roche), 8 × 105 cells were suspended in the presence of indicated amounts of drug in 5 mL medium and followed for 4 days. The cell numbers and viability were determined by trypan blue stain in quadruplicate counts. The number of metabolically active cells was also assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT)–based colorimetric assay. Cells were incubated for 4 days with 50 nM rapamycin at a seeding density of 1.6 × 105 cells/mL. The amount of MTT, which is converted to formazine represents the number of viable cells. The degree of the conversion was determined by measuring the absorbance at a wavelength of 570 nm.
Tumor formation
Cells were counted and washed once in ice-cold phosphate-buffered saline (PBS; Cellgro Mediatech, Herndon, VA), and indicated cell doses were diluted into 150 μL PBS and mixed with 150 μL growth factor–depleted Matrigel (BD Biosciences, Bedford, MA). Cells at the indicated doses were injected subcutaneously into the right flank or intraperitoneally into CB-17 severe combined immunodeficient (SCID) mice (Jackson Laboratory, Bar Harbor, ME) following our previously validated procedures.28 The mice were observed every 2 days for the presence of palpable tumors. Drug or vehicle was injected intraperitoneally at the indicated dosing schedule. Tumor diameters were determined by caliper measurements. Tumor volume was calculated as V = a × b × [MAX(a, b)]/2, where a and b are the long and short diameter of the tumor.29 The tumors were excised from the site of injection, weighed, and fixed in formalin (Fisher Diagnostics, Middletown, VA) or suspended in TRI reagent (Sigma-Aldrich, St Louis, MO) and processed for reverse transcription–polymerase chain reaction (RT-PCR).
RNA isolation and reverse transcription
Total RNA from suspension cells was isolated by using the Absolutely RNA Microprep kit (Stratagene, La Jolla, CA). Solid tumor pieces were suspended in 750 μL TRI Reagent (Sigma-Aldrich) and disrupted using an Ultra-Turrax T8 (IKA Labortechnik, Staufen, Germany). RNA was isolated according to the supplier's protocol, precipitated, and suspended in 50 μL DEPC-treated water at 56°C for 10 minutes. Subsequently, DNA was removed from the RNA isolation by using the DNA-free RNA kit (Zymo Research, Orange, CA). The RNA was reverse transcribed as described30 in a 20-μL reaction with 100 U Moloney murine leukemia virus reverse transcriptase (Life Technologies, Carlsbad, CA), 2 mM deoxynucleoside triphosphates, 2.5 mM MgCl2, 1 U RNAsin (all from Applied Biosystems, Foster City, CA), and 0.5 μg random hexanucleotide primers (Amersham Pharmacia Biotech, Piscataway, NJ). The RT reaction was sequentially incubated at 42°C for 45 minutes, 52°C for 30 minutes, and 70°C for 10 minutes. Finally, 0.5 μL RNAse H (Life Technologies) was added to the RT reaction, which was then incubated at 37°C for 30 minutes, heat-inactivated at 70°C for 10 minutes, and cDNA pools were stored at −80°C.
Real-time quantitative PCR
Quantitative real-time PCR primers and methods were previously described.30–32 Quantitative PCR (QPCR) was conducted on an Opticon2 Sequence Detector (Bio-Rad, Foster City, CA) using these cycling conditions: 2 minutes at 50°C, 10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 minute at 62°C. The cycle-threshold (CT) values were determined by automated analysis. The threshold was set to 3 times the standard deviation (SD) of the nontemplate control. CT values indicate the number of PCR cycles required to read a signal above threshold. Lower CT values correlate with higher target levels on a log2 scale. A CT of 40 indicates the undetectable levels. Dissociation curves were recorded after each run, and the amplified products were routinely analyzed by 2% agarose TAE gel electrophoresis.
Antibodies
Western blot assays were performed with antibodies to phospho-mTOR (ser2448), mTOR, phospho-Akt (ser473), Akt, TSC1, TSC2, phospho-TSC2 (not sure which one used), eIF4E, 4E-BP1, phospho-S6 (ser235/236), p70 S6 kinase (Cell Signaling, Beverly, MA), and β-actin (Sigma).
Immunoblotting and immunoprecipitation
Protein extracts were prepared using NP-40 lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40) supplemented with protease inhibitor cocktail (Sigma), 0.03% 2-mercaptoethanol, 10 μM phenylmethylsulfonyl fluoride, 30 mM β-glycerol phosphate, 50 mM NaF, and 1 mM sodium orthovanadate. Protein extract was separated on 10% SDS–polyacrylamide gel electrophoresis gels, except for analysis of mTOR (with 7.5%) and 4E-BP1 (with 12%). Proteins were transferred to Hybond P membranes (Amersham, Piscataway, NJ). Nonspecific bands were blocked for 1 hour at room temperature and then in appropriate primary antibody overnight at 4°C. After incubating with a horseradish peroxidase–linked secondary antibody, proteins were visualized by enhanced chemiluminescence (Amersham). For immunoprecipitation, BC-3 cells were seeded at 1.6 × 105 cells/mL. After 24 hours, the cells were treated with 50 nM rapamycin for the times indicated.7 Methyl-GTP-Sepharose 4B pull-down assay was performed as described earlier.33,34
Immunohistochemistry
Tumors were removed and fixed in 10% Neutral Buffered Formalin (Fisher Diagnostic) for 2 days, embedded in paraffin blocks, and processed by routine methods, and 7-μm sections were obtained. Sections were deparaffinized using Histochoice (Sigma-Aldrich) and dehydrated. Dewaxed sections were microwaved for 15 minutes in 1 mM EDTA (pH 8.0), cooled, and treated with 3% H2O2 (Sigma) in 10% methanol to inhibit endogenous peroxidase activity, blocked for 15 minutes at RT in solution-B (10% horse serum [Vector Laboratories, Burlingame, CA], 5% BSA, and 0.3% Triton X-100 in PBS) and incubated overnight at 4°C with the appropriate primary antibody: phospho-Akt (S473) polyclonal antibody (1:100) or phosphor-mTOR (S2448) polyconal antibody (1:100) or Phospho-P70 S6 kinase monoclonal antibody (1:200) from Cell Signaling. Solution B was used as negative control. Following washing, sections were stained, for 1 hour, with a goat anti–rabbit biotinylated horseradish peroxidase H-conjugated secondary antibody followed by Avidin DH (VECTASTAIN ABC kit; Vector Laboratories). Sections were washed in PBS and were incubated, for 5 minutes with Vector Nova Red substrate for peroxidase (Vector Laboratories). Slides were counterstained with hematoxylin (Sigma), dehydrated in 95% alcohol and absolute alcohol, cleared in xylene, and mounted in Permount (Sigma). Images were recorded using a LEICA DM LA histology microscope (Leica, Heidelberg, Germany) equipped with a 10×/0.25 numerical aperture (NA) or a 40×/0.75 NA N plan objective and a Leica DPC 480 camera, and were stored as .tif files with Mac OS X10.4. Image layout was done using Photoshop CS (Adobe, San Jose, CA).
Cell-cycle analysis
The cells were fixed in 80% ethanol in PBS. After pelleting, the fixed cells were resuspended in PBS containing of 20 μg/mL propidium iodide and 40 μg/mL RNAse A. Flow cytometric analysis was performed with a Becton Dickinson FACScan (BD Biosciences, San Jose, CA) and SUMMIT program (Dako, Fort Collins, CO).
Cytokine measurements
Levels of IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12(p40), IL-12(p70), IL-13, IL-15, IP-10, Eotaxin, IFNγ, GM-CSF, MCP-1, MIP-1α, RANTES, and TNFα in culture supernatants were determined by a bead-based immunoassay (Beadlyte Human 22-plex Cytokine Detection System; Upstate, Charlottesville, VA). Lyophilized cytokine standards and centrifuged samples were diluted in assay buffer (PBS, pH 7.4, 1% BSA, 0.05% Tween 20, 0.05% sodium azide), and 50μL was added to wells of a filter plate (96-well MultiScreen plates; Millipore, Bedford, MA) preequilibrated with assay buffer. Samples were incubated with 25 μL of antibody-coupled beads at 4°C for 16 hours on a plate shaker (300 rpm). Anticytokine-conjugated beads were vortexed and sonicated prior to dilution to reduce clumping. After 3 washes with assay buffer, 100 μL of diluted secondary/detection antibody mixture was added to wells and incubated at room temperature on a microplate shaker for 90 minutes followed by 30 minutes without shaking. After the addition of 25 μL streptavidin-PE, samples were incubated at room temperature for 30 minutes on a plate shaker and for 5 minutes after the addition of 25 μL stop solution. Unbound reporter was filtered through wells, and beads were resuspended in 125 μL assay buffer prior to analysis on a Luminex100 Total system running v1.7 software (Luminex, Austin, TX). Liquid handling, including reagent additions and washes, was performed using a Biomek 2000 Laboratory Automation Workstation with 96-filtration system (Beckman-Coulter, Fullerton, CA) that was programmed using BioWorks 3.0 software. Cytokine concentrations were calculated using STarStation v2.0 (Applied Cytometry Systems, Sacramento, CA) software using standard curves derived from recombinant cytokine standards.
IL-10 and IL-6 levels were independently determined using commercial enzyme-linked immunoabsorbent assay (ELISA) assay according to the manufacturer's recommendations (eBioscience, San Diego CA).
Statistical analysis
Calculations were performed using Excel (Microsoft, Redmond, WA) and SPSS v11.0 (SPSS Science, Chicago, IL).
Results
Rapamycin inhibits PEL growth in culture
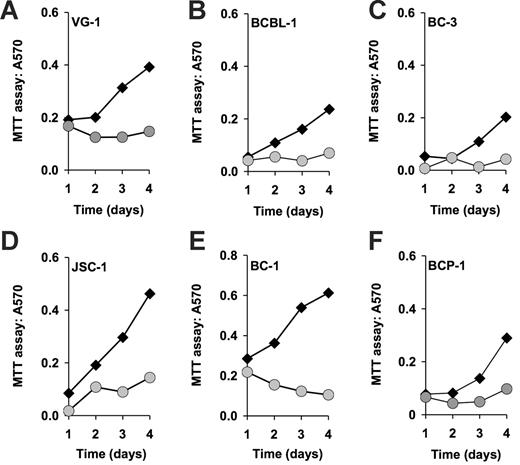
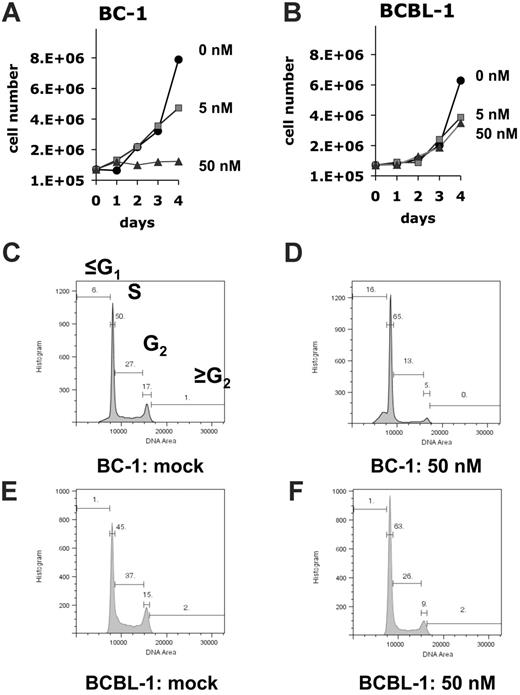
To determine the efficacy of rapamycin against PEL, we subjected a panel of established PEL cell lines to different doses of rapamycin. Cells (8 × 105) were seeded in 5 mL medium containing the indicated concentrations of rapamycin, and cell counts were determined every 24 hours over a period of 5 days. Neither fresh medium nor additional drug was added during the observation period. Using the MTT assay, we found that 6 independently derived PEL cell lines (BC-1, BC-3, JSC-1, BCBL-1, BCP-1, and VG-1) were inhibited by rapamycin at 50-nM concentration (Figure 1). Because the MTT assay measures mitochondrial activity, which is still substantial in viable, but growth-arrested cells, its dynamic range is limited. Therefore, we conducted dose-response experiments by traditional cell counting (Figure 2A-B). This showed that by comparison to BC-1 cells the BCBL-1 cell line was partly resistant to rapamycin, because increasing the dose from 5 to 50nM rapamycin did not result in more-efficient inhibition. This phenotype was also evident by cell-cycle analysis. Although BC-1 cells exposed to 50 nM rapamycin for 4 days stopped cycling as evidenced by a drop from 44% to 18% in the ≥ G1 (S + G2) DNA content cell fraction (Figure 2C-D), exposure of BCBL-1 cells to rapamycin resulted in only a drop from 52% to 34% (Figure 2E-F). With the exception of BCBL-1 all other PEL lines showed a similar cell-cycle arrest. The 2 most sensitive cell lines, BC-1 and BC-3, exhibited cell death phenotype as evidenced by a significant increase in their sub-G1 population (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The dichotomy in rapamycin IC50s for the different cell lines is not unexpected as it was previously encountered in the NCI 60 cell line screen.35 Although a general molecular mechanism underlying this phenotype has not been defined, we show here that BCBL-1's relative resistance to rapamycin correlates with a failure of drug to diminish IL-10 production.
Rapamycin-induced inhibition of cell proliferation as measured by MTT assay. Shown in each panel is the absorption at 570 nm in absence (black diamonds) or presence (gray circles) of 50 nM rapamycin on the vertical axis, and time in days after drug addition on the horizontal axis. Also shown is the percentage of inhibition at 96 hours after addition.
Rapamycin-induced inhibition of cell proliferation as measured by MTT assay. Shown in each panel is the absorption at 570 nm in absence (black diamonds) or presence (gray circles) of 50 nM rapamycin on the vertical axis, and time in days after drug addition on the horizontal axis. Also shown is the percentage of inhibition at 96 hours after addition.
Response to rapamycin in cell culture. (A) BC-1 or (B) BCBL-1 cells were seeded in medium containing the indicated concentrations of rapamycin. Plotted is the cell number on the vertical axis and time after seeding on the horizontal axis. This data represents 1 of 2 biologic replicates. (C-F) Cell-cycle analysis using propidium iodide stain at 4 days after exposure to 50 nM rapamycin or mock. In each panel plotted on the horizontal axis is the relative DNA content and on the vertical axis the number of cells. Also shown is the percentage of cells with ≤ G1, G1, S, G2 and ≥ G2 DNA content for each of the indicated cell lines and conditions.
Response to rapamycin in cell culture. (A) BC-1 or (B) BCBL-1 cells were seeded in medium containing the indicated concentrations of rapamycin. Plotted is the cell number on the vertical axis and time after seeding on the horizontal axis. This data represents 1 of 2 biologic replicates. (C-F) Cell-cycle analysis using propidium iodide stain at 4 days after exposure to 50 nM rapamycin or mock. In each panel plotted on the horizontal axis is the relative DNA content and on the vertical axis the number of cells. Also shown is the percentage of cells with ≤ G1, G1, S, G2 and ≥ G2 DNA content for each of the indicated cell lines and conditions.
Rapamycin delays PEL tumor progression in vivo
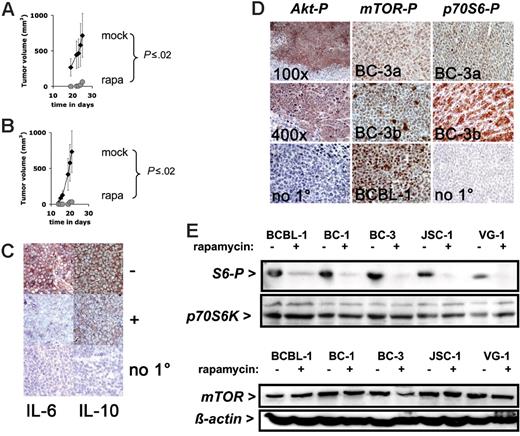
To investigate the effect of rapamycin on PEL growth in vivo, we used our PEL-SCID xenograft model.28 In this model PEL cells are injected subcutaneously in the presence of Matrigel, which allows us to quantify tumor growth noninvasively over time. We previously demonstrated that this model adequately reflects PEL tumorigenesis.28 First, to test whether rapamycin could prevent PEL tumor formation 1 × 106 BC-1 cells in Matrigel were injected subcutaneously into CB-17 SCID mice and treated continuously for the duration of the experiment with daily intraperitoneal injections (except on weekends) of 3 mg/kg per day (60 μg dissolved in 200 μL PBS) rapamycin or 200 μL vehicle (10% PEG 400/10% Tween 80/8% ethanol)36 over a period of 28 days. Rapamycin increased tumor-free time (P ≤ .005 by log-rank test) as well as slowed down the growth of tumors that did appear (Figure 3A) (P ≤ .025 by repeated measures ANOVA). This establishes that rapamycin is efficacious at preventing PEL tumor formation in vivo. In the same experimental design cyclosporine A did not have an antitumor effect (data not shown), which supports the notion that direct cytotoxicity rather than modulation of the host immune system causes rapamycin's antitumor phenotype in patients. Second, to test whether rapamycin could inhibit PEL tumor growth of established tumors 1 × 106 BC-1 cells in Matrigel were injected subcutaneously into CB-17 SCID mice, and tumors were allowed to form. Once the animals developed palpable tumors (∼ 2 mm diameter, day 14), the mice were treated continuously for the duration of the experiment. Tumor growth was significantly inhibited in rapamycin-treated animals, whereas mock-treated animals developed progressively growing tumors and had to be killed (P ≤ .005 by log-rank test) (Figure 3B). At termination, the difference in tumor size was significant to P ≤ .005 by Wilcox nonparametric test. This establishes that rapamycin is as efficacious against already established PEL tumors in vivo as it is at preventing tumor formation.
Response to rapamycin of experimental tumors. (A) Shown on the vertical axis is the tumor volume in mm3 and on the horizontal axis the time since inoculation with 106 BC-1 cells and simultaneous treatment with rapamycin (n = 5) or vehicle (n = 4). (B) Shown on the vertical axis is the tumor volume in mm3 and on the horizontal axis the time since inoculation with 106 BC-1 cells and treatment with rapamycin (n = 5) or vehicle (n = 4) animals after the establishment of tumors (day 14). Error bars indicate the SD for each group of animal. For rapamycin-treated animals the error bar is smaller than the symbol. (C) Expression of IL-6 in a mock-treated (−) BC-1 tumor (red color), which is reduced by rapamycin treatment (+). Expression of IL-10 in a mock-treated BC-1 tumor (−) (red color), which is reduced by rapamycin treatment (+). No staining in the absence of the specific primary antibody (no 1°). All sections are counterstained with hematoxilin (blue) and are at 400 × magnification. (D) Representative immunohistochemistry of mouse xenograft tumors using antibodies specific for phospho-Akt (Akt-P), phospho-mTOR (mTOR-P), and phospho-p70S6 kinase (p70S6-P). The phospho Akt staining was performed on BCBL-1 tumors at either 100 × or 400 × magnification. No 1° indicates the no primary antibody control reaction. The phospho-mTOR and phospho-p70S6K staining was performed on 2 different tumors (BC-3a and BC-3b) derived from the BC-3 cell line; No 1° indicates the no primary antibody control reaction. Pictures here were taken at 400 × magnification. (E) Western lot analysis of protein extracts for the indicated cell lines exposed to rapamycin or vehicle using anti–phospho-S6 and anti-p70S6 kinase antibody. (F) Western blot analysis of protein extracts for the indicated cell lines exposed to rapamycin or vehicle using anti-mTOR and antiactin antibody.
Response to rapamycin of experimental tumors. (A) Shown on the vertical axis is the tumor volume in mm3 and on the horizontal axis the time since inoculation with 106 BC-1 cells and simultaneous treatment with rapamycin (n = 5) or vehicle (n = 4). (B) Shown on the vertical axis is the tumor volume in mm3 and on the horizontal axis the time since inoculation with 106 BC-1 cells and treatment with rapamycin (n = 5) or vehicle (n = 4) animals after the establishment of tumors (day 14). Error bars indicate the SD for each group of animal. For rapamycin-treated animals the error bar is smaller than the symbol. (C) Expression of IL-6 in a mock-treated (−) BC-1 tumor (red color), which is reduced by rapamycin treatment (+). Expression of IL-10 in a mock-treated BC-1 tumor (−) (red color), which is reduced by rapamycin treatment (+). No staining in the absence of the specific primary antibody (no 1°). All sections are counterstained with hematoxilin (blue) and are at 400 × magnification. (D) Representative immunohistochemistry of mouse xenograft tumors using antibodies specific for phospho-Akt (Akt-P), phospho-mTOR (mTOR-P), and phospho-p70S6 kinase (p70S6-P). The phospho Akt staining was performed on BCBL-1 tumors at either 100 × or 400 × magnification. No 1° indicates the no primary antibody control reaction. The phospho-mTOR and phospho-p70S6K staining was performed on 2 different tumors (BC-3a and BC-3b) derived from the BC-3 cell line; No 1° indicates the no primary antibody control reaction. Pictures here were taken at 400 × magnification. (E) Western lot analysis of protein extracts for the indicated cell lines exposed to rapamycin or vehicle using anti–phospho-S6 and anti-p70S6 kinase antibody. (F) Western blot analysis of protein extracts for the indicated cell lines exposed to rapamycin or vehicle using anti-mTOR and antiactin antibody.
Rapamycin molecular targets are present in PEL
Having established the efficacy of rapamycin against established and primary PEL cells, we characterized the molecular targets of rapamycin in PEL. In human tumors, activated (ie, fully phosphorylayted) Akt activates the mTOR pathway. Akt was previously shown to be activated in PEL.37 We used a phospho-Ser473–specific antibody to detect phospho-Akt in PEL xenograft tumors. Akt was uniformly phosphorylated in all tumor cells, except those in liquefied, necrotic areas (Figure 3D, left panels). No staining was observed without primary antibody. We measured phospho-mTOR using an antibody specific for phospho-Ser2448. All slides were stained in parallel and exposed to substrate for the same length of time. Phospho-mTOR was uniformly detectable in all cells in BCBL-1 cell xenograft tumors. It was uniformly detectable in all cells in BC-3 tumors, but different tumors exhibited different levels of staining (Figure 3D, middle panels). In one tumor individual cells exhibited different phospho-mTOR levels, suggesting that in addition to Akt other upstream signals, such as branched chain amino acid pools, influence mTOR activation. No staining was observed without primary antibody. The substrate of mTOR, p70S6K, was also phosphorylated in PEL tumors (Figure 3D, right panels). It was uniformly detectable in all cells in tumors, but even isogenic tumors exhibited different levels of staining, consistent with the notion that mTOR activity was not constitutive but subject to regulation in response to the cell cycle and nutrient stage. The variability in mTOR phosphoylation was more pronounced in the more demanding xenograft environment than in the uniform and nutrient-optimized tissue culture environment. There are many targets of mTOR and many are unknown. The presence of activated Akt and activated mTOR in PEL tumors is consistent with the efficacy of rapamycin and a continued need for mTOR signaling in PEL tumorigenesis.
Next, we tested whether rapamycin inhibited mTOR activity in PEL by Western blot analysis with phospho-S6 antibodies. Rapamycin inhibited ribosomal protein S6 phosphorylation in all cell lines tested, whereas levels of p70S6 kinase remained constant (Figure 3E). Rapamycin did not change the levels of mTOR (Figure 3F) nor did it change mTOR phosphorylation (S2448), TSC1, TSC2 protein levels, or TSC2 phosphorylation (S939) in treated PEL lines (Figure S1), because these molecules are upstream of the mTOR kinase. This outcome is consistent with the previously reported mechanism of action for rapamycin to inhibit mTOR kinase activity and phosphorylation of mTOR-Raptor–dependent regulators of protein translation.
Rapamycin does not affect KSHV transcription
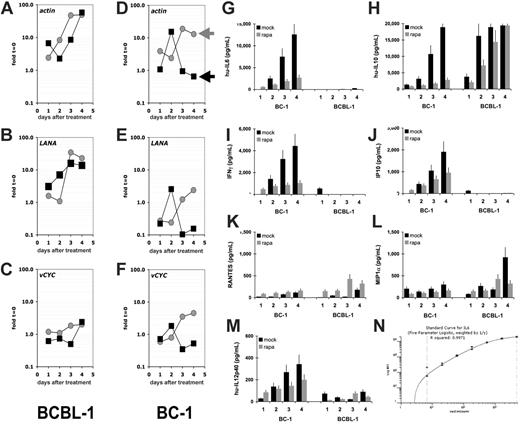
A defining feature of KS and PEL is the presence of KSHV (reviewed in Dittmer et al38 ). To date a possible effect of rapamycin on KSHV has not been explored. Viral latent genes are expressed in every tumor cell,39 and lytic mRNAs are found in a low, varying percentage of cells in culture. To investigate whether rapamycin had a differential effect on viral mRNA levels, we conducted microarray analysis using our established protocol.30,31 Rapamycin-sensitive BC-1 and rapamycin-resistant BCBL-1 cells were seeded at 1.6 × 105 cells/mL treated with rapamycin, and mRNA levels per 1 mL cells were determined by real-time QPCR (Figure 4). Relative to time zero rapamycin-treated (black squares) BC-1 cells showed a decline in actin mRNA as well as viral latent mRNAs, which is consistent with proliferation arrest, which also lowers the overall transcriptional activity. The level of actin mRNA and viral latent mRNAs increased over time in mock-treated cells (gray circles), because the cell number increased. There was no differential response between actin and viral mRNA levels, indicating that rapamcyin did not affect KSHV mRNA levels. In the rapamycin-resistant BCBL-1 cells actin mRNA levels increased regardless of drug treatment and so did viral latent mRNA levels, indicating that in rapamycin-resistant cells neither cellular nor viral mRNA levels were diminished. Next, we determined the relative mRNA levels for every mRNA in the viral genome (Figure S2). Because actin mRNA was dramatically altered by rapamycin, we did not normalize for cellular mRNA. At 24 hours after rapamycin addition, all viral mRNAs as well as actin were decreased in BC-1 cells. In contrast, they were unaltered in the rapamycin-resistant BCBL-1 cells, consistent with our observation that BCBL-1 cells are relatively resistant to rapamycin. We did not uncover a KSHV-specific effect of rapamycin or a rapamycin-inducible viral RNA.
Rapamycin does not affect KSHV transcription but cellular cytokine levels. Shown for each panel A to F on the vertical axis are fold changes relative to mRNA levels at t = 0 on a log10 scale and on the vertical axis days after addition to 50 nM rapamycin or vehicle control (mock). Rapamycin-treated cultures are represented by black squares, mock-treated cultures by gray circles. The results of rapamycin-resistant BCBL-1 cells are shown on the left column and of rapamycin-sensitive BC-1 cells on the right column. Individual panels refer to mRNA levels for the mRNAs actin, LANA, and vCYC. LANA refers to the latency-associated nuclear antigen (orf73) and vCYC to the viral cyclin homolog orf72. Shown for each panel are the concentrations in picogram per milliliter for the indicated cytokines in the vertical axis and time after addition of rapamycin (▪) or vehicle (⊡) on the horizontal axis for the cell lines BC-1 and BCBL-1: (G) hu-IL-6, (H) hu-IL-10, (I) IFN-γ, (J) IP-10, (K) RANTES, (L) MIP-1α, (M) IL12p40. These data represent 1 of 2 biologic replicates. Panel N shows a representative (IL-6) standard curve.
Rapamycin does not affect KSHV transcription but cellular cytokine levels. Shown for each panel A to F on the vertical axis are fold changes relative to mRNA levels at t = 0 on a log10 scale and on the vertical axis days after addition to 50 nM rapamycin or vehicle control (mock). Rapamycin-treated cultures are represented by black squares, mock-treated cultures by gray circles. The results of rapamycin-resistant BCBL-1 cells are shown on the left column and of rapamycin-sensitive BC-1 cells on the right column. Individual panels refer to mRNA levels for the mRNAs actin, LANA, and vCYC. LANA refers to the latency-associated nuclear antigen (orf73) and vCYC to the viral cyclin homolog orf72. Shown for each panel are the concentrations in picogram per milliliter for the indicated cytokines in the vertical axis and time after addition of rapamycin (▪) or vehicle (⊡) on the horizontal axis for the cell lines BC-1 and BCBL-1: (G) hu-IL-6, (H) hu-IL-10, (I) IFN-γ, (J) IP-10, (K) RANTES, (L) MIP-1α, (M) IL12p40. These data represent 1 of 2 biologic replicates. Panel N shows a representative (IL-6) standard curve.
Rapamycin inhibits cellular cytokines IL-6, IL-10, and IFN-γ
PELs are dependent on autocrine growth factors. This has been shown in particular for the B-cell survival factors human IL-6 and IL-10.40–46 We used Luminex bead array technology to quantify cytokine secretion in response to rapamycin or mock treatment for the rapamycin-resistant cell line BCBL-1 and the rapamycin-susceptible cell line BC-1. The cytokines IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, eotaxin, IL-12p70, IL-12, IL-15, GM-CSF, IL-7, TNFα, MCP-1, and IL-8 were not secreted in significant amounts from either cell line (data not shown). IL-6, IFN-γ, IP-10, and IL-12p40 were secreted at high levels in BC-1 cells but not found in the medium of BCBL-1 cells, as previously shown for IL-644 (Figure 4). IL-6, IFN-γ, IP-10, and IL-12p40 secretion was inhibited by rapamycin in a time-dependent manner. RANTES and MIP-1α were secreted by both cell lines, but their levels were not affected by rapamycin. IL-10 is an essential survival factor for EBV-transformed B cells in posttransplantation lymphoproliferative disease (PTLD)47 as well as KSHV-transformed PEL, and was highly expressed in both BC-1 and BCBL-1, but only inhibited by drug in the rapamycin-sensitive BC-1 cell line. Hence, we conclude that suppression of IL-10 translation represents one molecular pathway that mediates the cytostatic effect of rapamycin against PEL.
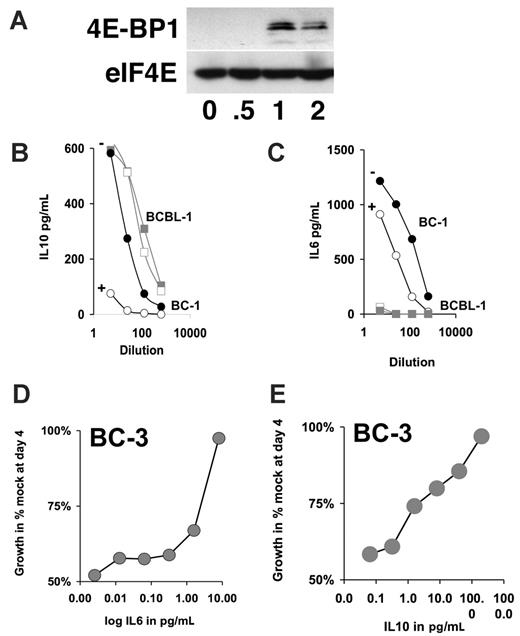
To independently confirm the Luminex analysis, we used ELISA to measure the amount of secreted IL-6 and IL-10. BC-1 or BCBL-1 cells were exposed to 50 nM rapamycin for 96 hours. BC-1 cells showed a dramatic reduction in IL-10 levels on rapamycin treatment, whereas BCBL-1 cells showed no significant changes in IL-10 production (Figure 5B). BCBL-1 cells did not secrete IL-6. By contrast BC-1 cells secret high levels of IL-6, and IL-6 secretion was reduced by rapamycin (Figure 5C). Consistent with rapamycin acting on a translational level neither IL-6 mRNA nor IL-10 mRNA levels were significantly altered by rapamycin treatment (Figure 6A). To independently confirm rapamycin's mechanism of action as a translational inhibitor we evaluated eIF4E/4E-BP1 complex formation by 7methyl-GTP Sepharose immune precipitation as previously described.34 Rapamycin caused eIF4E/4E-BP1 complex formation, which removes eIF4E from the translation initiation complex and inhibits translation independently and prior to the S6-dependent elongation (Figure 5A). This establishes a direct, rapamycin-sensitive signaling pathway from Akt to mTOR to eIF4E, S6-dependent translation to inhibition of IL-6 and IL-10 protein, without affecting mRNA levels in PEL.
IL-6 and IL-10 counteract the rapamycin-induced growth arrest. (A) Western blot analysis of 4E-BP1 and eIF4E after immunoprecipitation with 7methyl-GTP Sepharose. Samples are drawn at indicated times after exposure of BC-3 cells to 50 nM rapamycin. (B-C) Shown is the concentrations in picogram per milliliter for the indicated cytokine in the vertical axis and dilution of spent medium of 4 days after addition of rapamycin (open symbols) or vehicle (closed symbol) on the horizontal axis for the cell lines BC-1 (circles) and BCBL-1 (squares) as determined by ELISA. (D-E) Relative growth in percentage after 4-day culture in 50 nM rapamycin and the indicated concentrations of either IL-6 or IL-10 as determined by MTT assay.
IL-6 and IL-10 counteract the rapamycin-induced growth arrest. (A) Western blot analysis of 4E-BP1 and eIF4E after immunoprecipitation with 7methyl-GTP Sepharose. Samples are drawn at indicated times after exposure of BC-3 cells to 50 nM rapamycin. (B-C) Shown is the concentrations in picogram per milliliter for the indicated cytokine in the vertical axis and dilution of spent medium of 4 days after addition of rapamycin (open symbols) or vehicle (closed symbol) on the horizontal axis for the cell lines BC-1 (circles) and BCBL-1 (squares) as determined by ELISA. (D-E) Relative growth in percentage after 4-day culture in 50 nM rapamycin and the indicated concentrations of either IL-6 or IL-10 as determined by MTT assay.
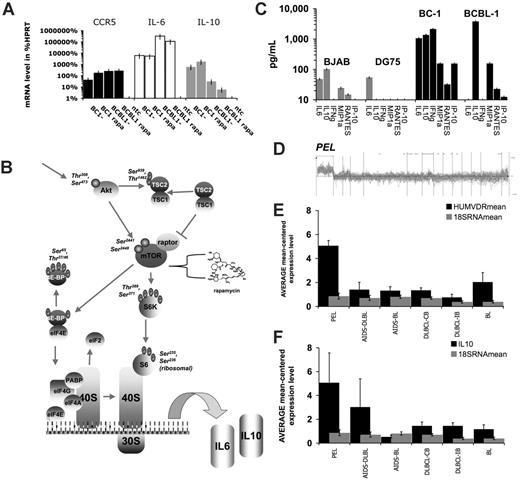
Cytokine mRNA levels predict PEL class membership. (A) Relative mRNA levels for CCR5, IL-6, and IL-10 as determined by real-time QPCR and expressed as the percentage of HPRT mRNA levels for BC-1 and BCBL-1 cells treated with rapamycin (rapa) or mock-treated; ntc indicates nontemplate control. (B) Summary of rapamycin signaling pathways in PEL (adapted from Science Slides 2005, VisiScience, Chapel Hill, NC). (C) Secreted cytokine levels in picogram per milliliter (log-scale) for virus-negative BJAB and DG75 and KSHV-positive BC-1 and BCBL-1 cell lines. Cytokines are listed on the horizontal axis. (D) Relative levels (vertical axis) of all mRNAs that can be used to predict class membership to PEL of a set of 101 lymphomas (horizontal axis). From this set the relative mRNA for vitamin D receptor (E) and IL-10 (F) levels were mean-centered across n = 101 Affymetrix datasets and averaged for PEL (n = 9), AIDS-associated Burkitt lymphoma (AIDS-BL) (n = 7), AIDS-associated diffuse large B-cell lymphoma (DLBCL) (n = 9), classic, non–AIDS-associated BL (n = 6), non–AIDS-associated DLBCL of immunoblast type (n = 8), non–AIDS-associated DLBCL of centroblast type (n = 23). Histograms show mean and standard deviation. Gray bars indicated 18S RNA levels as control.
Cytokine mRNA levels predict PEL class membership. (A) Relative mRNA levels for CCR5, IL-6, and IL-10 as determined by real-time QPCR and expressed as the percentage of HPRT mRNA levels for BC-1 and BCBL-1 cells treated with rapamycin (rapa) or mock-treated; ntc indicates nontemplate control. (B) Summary of rapamycin signaling pathways in PEL (adapted from Science Slides 2005, VisiScience, Chapel Hill, NC). (C) Secreted cytokine levels in picogram per milliliter (log-scale) for virus-negative BJAB and DG75 and KSHV-positive BC-1 and BCBL-1 cell lines. Cytokines are listed on the horizontal axis. (D) Relative levels (vertical axis) of all mRNAs that can be used to predict class membership to PEL of a set of 101 lymphomas (horizontal axis). From this set the relative mRNA for vitamin D receptor (E) and IL-10 (F) levels were mean-centered across n = 101 Affymetrix datasets and averaged for PEL (n = 9), AIDS-associated Burkitt lymphoma (AIDS-BL) (n = 7), AIDS-associated diffuse large B-cell lymphoma (DLBCL) (n = 9), classic, non–AIDS-associated BL (n = 6), non–AIDS-associated DLBCL of immunoblast type (n = 8), non–AIDS-associated DLBCL of centroblast type (n = 23). Histograms show mean and standard deviation. Gray bars indicated 18S RNA levels as control.
To determine whether PEL also expressed IL-6 and IL-10 in our xenograft model, we used immunohistochemistry. BC-1 xenograft cells expressed significant levels of IL-6 and IL-10 (Figure 3C). Rapamycin as noted in Figure 3A-B inhibited tumor growth. The residual tumors had barely detectable levels of IL-6 and IL-10 (Figure 3C). The no primary antibody control slides showed no staining (Figure 3C). This demonstrated that in vivo as well as in tissue culture IL-6 and IL-10 are expressed in PEL and that this expression is reduced by rapamycin coincident with the inhibition of proliferation.
IL-6 and IL-10 are proliferation and survival factors for PEL.40 To test whether the rapamycin-induced crossreduction in IL-6 and IL-10 secretion could be counteracted by providing IL-6 and IL-10 exogenously, we treated BC-3 cells with 50 nM rapamycin and added increasing amounts of either IL-6 or IL-10 (Figure 5D-E). Exogenous IL-6 or IL-10 was able to counteract the rapamycin-induced growth arrest in a dose-dependent manner. This establishes that the inhibition of IL-6 and IL-10 secretion is not only a surrogate marker for rapamycin action in PEL but, because of the autocrine feedback requirement for these 2 cytokines, constitutes a molecular mechanism that is targeted by rapamycin.
To evaluate whether sensitivity to rapamycin is a unique property of PEL, we evaluated the non-PEL cell lines DG-75 and BJAB. Neither cell line exhibited as high cytokine levels as PEL (Figure 6C) and neither cell line showed a significant, time, and rapamycin-dependent decrease in secreted cytokines (Table S2). We also extracted the rapamycin sensitivity data from the NCI 50 cancer cell lines screen.35 This screen included the leukemia cell lines CCRF-CEM, HL-60, K-562, MOLT-4, RPMI-8226, and SR as well as many solid-organ cell lines. The growth inhibitory concentration 50 (IC50) for the leukemia lines varied between 5 and 0.01 μM, suggesting that rapamycin sensitivity, reflecting a reliance on the Akt/mTOR pathway for oncogenesis, is not a property of all tumor cells per se, but maybe a hallmark of PEL and other KSHV-associated cancers.
To test the hypothesis that IL-10 or IL-6 expression was unique to PEL, Affymetrix data for n = 101 lymphomas were obtained from the study by Klein et al.48 We used ArrayMiner classmaker prediction algorithm to identify all genes that are capable of identifying PEL based on differential mRNA levels (Figure 6D). Among them we identified the vitamin D receptor, which had previously been identified as a PEL marker, using an unrelated tumor collection and microarray system.49 Vitamin D receptor protein levels also correlated with PEL class membership as did sensitivity to the vitamin D analog EB 1089 (seocalcitol), establishing vitamin D receptor mRNA and protein levels as the first PEL-specific marker that emerged in 2 independent transcriptional profiling studies and that is validated at the protein level. The classmaker prediction program also identified hu-IL10 mRNA levels as a PEL-specific marker. In contrast, IL-6 levels could not be used to establish PEL class membership, because many lymphomas exhibit elevated levels of IL-6 mRNA. Next, we mean centered the mRNA level across all 101 datasets and extracted the transcription data for PEL (n = 9 cases), AIDS-associated Burkitt lymphoma (BL) (n = 7 cases), AIDS-associated DLBCL of centroblast type (n = 9 cases), AIDS-associated DLBCL of immunoblast type (n = 4 cases), classic, non–AIDS-associated BL (n = 6), non–AIDS-associated DLBCL of immunoblast type (n = 8), non–AIDS-associated DLBCL of centroblast type (n = 23), and calculated mean and SD for vitamin D receptor, 18S RNA, and hu-IL10 (Figure 6E-F). Hu-IL10 levels as well as vitamin D receptor levels were significantly different among the tumor classes as determined by ANOVA, with P ≤ 2 × 10−6 and P ≤ 1 × 10−9, respectively. Expectedly, the levels of 18S RNA were not. Individual post-hoc comparison yielded significant differences (P ≤ .005 adjusted for multiple comparisons by Bonferroni correction) for IL-10 between PEL and all other lymphoma classes except AIDS-DLBCL of immunoblast type. No other lymphoma class exhibited significant differences in IL-10 levels in pairwise comparison. As expected vitamin D receptor levels were significantly different only in PEL and not any of the other lymphoma types. This establishes IL-10 as a PEL-specific cytokine and supports our contention that the sensitivity of PEL to rapamycin can, at least in part, be attributed to the inhibition of IL-10 expression.
Discussion
We interpret our data within the conceptual framework of the mTOR-signaling cascade (Figure 6), because Akt, mTOR, and p70S6 kinase are phosphorylated in PEL cells (Figure 3) and because rapamycin caused the expected phenotypes of cell proliferation arrest in tissue culture (Figures 1–2) and tumor cytostasis in vivo (Figure 3). This phenotype was caused by translational inhibition of IL-6 and IL-10 protein (Figure 4). Consistent with the predicted signaling pathways of mTOR, rapamycin treatment of PEL cells also caused eIF4E/4E-BP1 association (Figure 5A) that removes eIF4E from the translation initiation complex and inhibits translation independently and prior to the S6/40S/30S-dependent elongation step (Figure 6). Because rapamycin affects mTOR kinase function, neither mTOR, Akt, TSC1, TSC 2 levels nor phosphorylation was altered on treatment (data not shown). Recent reports show that Akt-mTOR signaling is more complex than predicted based on a linear signal transduction model. First, mTOR also functions in a nutrient sensor pathway independent of PI3K and Akt. This could explain the heterogeneity of phospho-mTOR and phospho-p70S6K in our experimental tumors that unlike cells grown in culture experience uneven nutrient and oxygen flow, depending on location within the tumor and distance to nearest blood vessels. Second, signaling through mTOR-Rictor is rapamycin insensitive as opposed to the canonical, rapamycin-sensitive mTOR-Raptor pathway. Very little is known about mTOR-Rictor targets, except mTOR-Rictor was recently shown to possess PDK2 activity and could thus activate Akt.50,51 The mTOR also feeds back to activate Akt by other means, for instance through p70S6K activating PI3K in response to insulin and IGF1.52 The importance of these signaling loops to the drug action of rapamycin in human cancer in general remains to be resolved.
The antiproliferative activity of rapamycin was first noted in the 1970s (reviewed in Sawyers23 ), and already at that time a dichotomy in rapamycin-sensitivity was noted. Although many cancer-derived cell lines undergo G1 arrest at 1-nM concentrations others require 1 μM rapamycin. This occurred despite the fact that 1 nM rapamycin blocked mTOR kinase activity (as measured by p70S6K phosphorylation) in cancer cells of both groups.25 This observation places the rapamycin-resistance phenotype downstream of p70S6K. Rapamycin-resistant cancers are either defective in G1 arrest checkpoint functions or normally essential mRNAs are no longer under subject to p70S6K. Most PEL cell lines tested here as well as primary PEL cells fell into the rapamycin-sensitive group. We did not find a correlation between rapamycin sensitivity and coinfection with Epstein-Barr virus (BC-1, JSC-1) or induction of a particular KSHV mRNA. BCBL-1 cells were unusual as they required 10-fold higher doses of rapamycin than the rest of the group to evidence growth retardation. In response to drug, BCBL-1 cells did not down-regulate IL-10 secretion unlike BC-1 cells (Figure 4), which suffices to explain the cell line difference in rapamycin responsiveness, although it does not preclude other mechanisms. For instance, BCBL-1 contains a mutated p53 allele, whereas BC-3, BC-1 cells do not (S.-H.S. and D.P.D., unpublished data, July 2006). We speculate that the p53 mutation in BCBL-1 may be the result of adaptation to liquid culture, because the same mutation was reported by Katano et al53 but not by Friborg et al.54 A mutant p53 would counteract the cell-cycle arrest phenotype in BCBL-1 cells epistatic to mTOR kinase inhibition.
Rapamycin is effective against EBV-associated lymphomas55 and EBV-associated PTLD. The latter is mediated by inhibiting IL-10,47 which suggests that EBV-transformed PTLD may exhibit similar cytokine dependencies as KSHV-transformed PEL. In contrast 2 uninfected B-cell lymphoma cell lines tested here did not exhibit an extensive cytokine secretion profile, nor was their cytokine profile significantly altered by rapamycin. In reanalyzing transcription profile data generated by Klein et al,48 we found that IL-10 could be used to identify PEL in a set of 100 lymphomas, whereas IL-6 mRNA levels were increased in lymphomas of many subtypes. This suggests that IL-10 may be a unique marker for postgerminal center lymphomas. Rapamycin also down-regulated IFN-γ, IP-10, and IL-12p40 in PEL. IP-10 has been shown to be a negative regulator of KSHV vGPCR-mediated signaling,56 which in turn is known to modulate Akt signaling.57–59 VGPCR-transformed cells are uniquely sensitive to rapamycin and rely on mTOR up-regulation to sustain proliferation and transformation of K1, another KSHV gene with established oncogenic potential also induces Akt signaling.60,61 This may provide another potential mechanism to explain the efficacy of rapamycin against KSHV-associated tumors.
The clinical implications of this study are exciting. Our data provide experimental evidence and a scientific rationale for using rapamycin (sirolimus) in the treatment of HIV-associated and non–HIV-associated PEL. To date, 2 patients with PEL have been treated with oral rapamycin and showed a response (D.H. and W.J.H., unpublished observation, October 2006). In KSHV-positive patients who received a transplant substituting rapamycin for cyclosporine A may protect against of PEL and other KSHV-associated lymphoproliferative diseases similar to the way it protected against KS.21 The strong dependence on autocrine and paracrine growth factors as well as the Akt pathway constitutes a defining feature of KSHV-associated malignancies regardless of cell origin (endothelial or lymphatic) (reviewed in Damania62 and Ablashi et al63 ). This renders KSHV-associated cancers an ideal target for rapamycin and related mTOR inhibitors.
Authorship
Contribution: S.-H.S. was the lead author and designed and conducted the experiments; D.R., L.W., M.R.S., F.D.F., D.D.P., and B.A.D. designed and conducted the experiments; D.H. designed the experiments and provided critical input and clinical expertise; W.J.H. provided critical input and clinical expertise; and D.P.D. was the corresponding author, designed the experiments, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dirk P. Dittmer, Department of Microbiology and Immunology and Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, CB# 7290, 804 Mary Ellen Jones, Chapel Hill, NC 27599-7290; e-mail: ddittmer@med.unc.edu.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank Donald Ganem, David Scadden, Giovanna Tosato, Richard Ambinder, and Ethel Cesarman for cell lines; Chelsey Hilscher and Lisa Rothlein for technical expertise.
This work was supported by a translational award from the Leukemia and Lymphoma Society, the AIDS-associated Malignancies Clinical Trials Consortium (AMC; CA700580), and NIH (grants CA109232, HL083469, CA098110, CA096500, CA112935, and AI057157). B.A.D. is a Leukemia and Lymphoma Society scholar.