Abstract
Human leukocyte antigen (HLA)–E belongs, with HLA-G and HLA-F, to the nonclassic major histocompatibility complex (MHC) class I (Ib) molecules, broadly defined by a limited polymorphism and a restricted pattern of cellular expression. In contrast to HLA-G, the expression and function of HLA-E and HLA-F in physiologic and pathologic processes remain poorly established. In the present study, we show that HLA-E protein expression in normal human nonlymphoid organs is mainly restricted to endothelial cells (ECs). HLA-E is also basally expressed by B and T lymphocytes, natural killer (NK) cells and by macrophages. We demonstrate that tumor necrosis factor α (TNFα), interleukin-1β (IL-1β), and interferon γ (IFNγ) up-regulate the cell-surface expression of HLA-E on ECs in vitro and induce the release of soluble HLA-E (sHLA-E). HLA-E up-regulation protects IFNγ-activated ECs from NK-mediated cell lysis, while sHLA-E protects bystander cells. Finally, sHLA-E is not detected in normal sera, and increased serum levels correlate with disease activity in patients with antineutrophil cytoplasmic antibody–associated systemic vasculitis. Thus, HLA-E expression and release of sHLA-E are features of EC activation and emphasize immunoregulatory functions of the endothelium. The present identification of soluble HLA-E molecules may have important implications in understanding the pathogenesis of immune-mediated vascular diseases and for the diagnosis and monitoring of patients.
Introduction
Human leukocyte antigen (HLA)–E belongs, with HLA-G and HLA-F, to the nonclassical major histocompatibility complex (MHC) class I (Ib) molecules, broadly defined by a limited polymorphism and a restricted pattern of cellular expression.1 HLA-G plays an important role in immune tolerance during pregnancy, and in the escape of tumors from immune control. By contrast, the expression and function of HLA-E and HLA-F in physiologic and pathologic processes remain poorly established. Among Ib molecules, HLA-E was initially characterized by a broad pattern of mRNA expression in different cell types.2 Nevertheless, the surface expression of HLA-E requires the availability of a set of highly conserved nonameric peptides derived from the leader sequence of various HLA class I molecules, including HLA-A, HLA-B, HLA-C, and HLA-G,3,4 suggesting that HLA-E cell-surface expression may be limited to some cell type and/or cell-activation processes.
HLA-E has been identified as a ligand of CD94/NKG2A and CD94/NKG2C receptors expressed on natural killer (NK) cells and a subset of T cells.5,6 The interaction of HLA-E with the inhibitory CD94/NKG2A receptor results in inhibition of NK cell– and cytotoxic T lymphocyte (CTL)–dependent lysis.5,7 HLA-E molecules not only provide a protective pathway but also play a role in the regulation of T-cell function. HLA-E complexed with peptides can interact with αβ and γδ T-cell receptors (TCRs) expressed on CD8+ T cells to trigger conventional CTL function.8-10 HLA-E forms a heterodimer with β2-microglobulin, which binds and presents peptides derived from self- or foreign proteins10 after infection,11 immunization, or transplantation.12 Finally, in vitro studies using human cells10 and the demonstration that Qa-1 (homologous to HLA-E in mice)–deficient mice13 have defects in immunoregulation mediated by CD8+ T cells also provide evidence of the involvement of HLA-E–restricted CD8+ suppressor cells in controlling the adaptive immune response to both foreign and self-antigens.14,15 Together, these findings suggest that HLA-E may have unique regulatory functions in both the innate and cognate immune responses. Nevertheless, unlike HLA-G, the expression and function of HLA-E in physiologic and pathologic processes remain poorly established. Indeed, although HLA-E transcripts have been detected in almost all cell types, only few in vitro experiments have examined protein and cell-surface expression16-19 and in vivo analysis of HLA-E expression and function still need to be explored.
The endothelium is a functional barrier between the vessel wall and the bloodstream. It exhibits a variety of important functions, including control of coagulation, fibrinolysis, vascular tone, growth, and immune responses.20 It is now clear that activation of the endothelium disturbs the physiologic protective regulatory balance; this is a critical factor in the progression of inflammatory and autoimmune diseases, as well as in atherosclerosis and in transplant rejection.21 Human vascular ECs basally display classes I and II MHC peptide complexes on their surface as well as some costimulatory molecules, enabling them to activate CD8+ and CD4+ T cells.22 For instance, the vascular endothelium of an allograft is a primary target for immune-mediated rejection and activation of resting CD8+ T cells to effector CTLs, occurring after recognition of MHC class I–expressing ECs that leads to EC damage and vascular injury.23 Endothelial graft cells may also elicit humoral, donor-specific, anti-MHC response following transplantation. We have shown that, beyond complement activation, antibody-mediated cross-linking of donor MHC class I24 and class II25 molecules on human ECs results in EC activation, implicating specific signaling pathways. However, whether other MHC-related molecules, including the nonclassical class I MHC molecules, could be expressed on ECs and trigger either humoral or cellular response, or both, has been proposed but remains to be studied. Here, we investigated HLA-E protein expression in vivo and explored both regulation and function of HLA-E molecules on vascular endothelium in vitro and in vivo.
Materials and methods
Tissue samples and sera
Comparative expression of HLA-E and HLA-G proteins was studied in various normal human tissues, including first-trimester placenta tissue, kidney, lymph node, spleen, salivary gland, urinary bladder, thyroid, endometrium, skin, stomach, and liver. For each tissue, at least 3 independent samples were immunostained and analyzed. Tissues were fixed in 10% formalin and routinely processed for paraffin embedding. Serum samples (n = 22) from 10 patients with antineutrophil cytoplasmic antibody (ANCA)–associated vasculitis (Wegener granulomatosis [WG] and microscopic polyangiitis [MPA]) were analyzed. Disease activity was scored using the validated Birmingham Vasculitis Scoring Index (BVAS). Serum samples were obtained in the active phase (at diagnosis or relapse) and in remission (6 to 9 months later). The study was performed according to the guidelines of the local ethics committee (Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale [CCPRB], CHU de Nantes, France). Sera from healthy blood donors (n = 9 individuals and a pool of 20 donors) were provided by Establissement Français du Sang (EFS) (Nantes, France) and used as controls. Human aortic endothelial cells (HAECs) were isolated from unused aortic pieces collected at the time of kidney transplantation and harvested according to good medical practice and stored in the DIVAT Biocollection (French Health Minister Project no. 02G55). C-reactive protein (CRP) levels were determined by turbidimetry. Quantification of the levels of soluble interleukin-2 receptor (sIL-2Rα), IL-8, thrombomodulin, and vascular endothelial growth factor (VEGF) was carried out using enzyme-linked immunosorbent assay (ELISA) kits (Quantikine assays; R&D Systems, Abington, United Kingdom) performed, in triplicates according to the manufacturer's instructions.
Anti–HLA-E–specific antibodies
The following anti–HLA-E–specific monoclonal antibodies (mAbs) were used: MEM-E/02, MEM-E/07, and MEM-E/08 in which HLA-E specificity was previously defined by flow cytometry on the Third International Conference on HLA-G (Paris, July 2003) and confirmed in independent studies.19,26 These antibodies were produced and provided by Pr. Vaclav Horejsi (Institute of Molecular Genetics, Praha, Czech Republic). MEM-E/02 recognizes the denaturated form of HLA-E molecules and was used for Western blotting and immunohistochemistry. MEM-E/07 and MEM-E/08 mAbs recognize the native surface HLA-E molecule, and were used for confocal microscopy, fluorescence-activated cell sorting (FACS) analysis and cytotoxicity assays. In contrast with MEM-E/03 and MEM-E/06, which exhibit the broadest cross-reactivities, MEM-E/02, MEM-E/07, and MEM-E/08 are specific anti–HLA-E mAbs that exhibit only minimal cross-reactivities with some classical MHC antigens. MEM-E/02 (IgG1) does not cross-react with HLA-A, HLA-B, HLA-C, or HLA-G. MEM-E/07 (IgG1) cross-reacts with some classical MHC class I molecules: HLA-B7 (strongly); HLA-B8 (moderately); and HLA-B27 and HLA-B44 (weakly). MEM-E/08 (IgG1) is remarkably specific for HLA-E; it only shows weak cross-reactivity with the following classical MHC class I molecules: HLA-A24, HLA-B7, HLA-B27, HLA-B51, HLA-B54, and HLA-C7. Mouse IgG1 mAb MEM-G/01 reacts with the denatured HLA-G heavy chain of all isoforms and was purchased from Serotec (Cergy Saint-Christophe, France).
Cell culture
Human umbilical vein ECs (HUVECs), HAECs and vascular smooth muscle cells (SMCs) were isolated and cultured as described.24,25 ECs were HLA typed and selected to avoid non–HLA-E–specific cross-reactivity with the anti–HLA-E mAbs listed earlier in “Materials and methods.” The following EC types were used: HUVEC no. Gd (HLA-A02, HLA-A26, HLA-B08, and HLA-B515 ); HAEC no. 8186 (HLA-A01, HLA-A02, HLA-B51,5 and HLA-B525 ); HAEC no. 9007 (HLA-01,0 HLA-02, HLA-B35, and HLA-B39); HAEC no. 9054 (HLA-A2, HLA-A26, HLA-B51,5 and HLA-B6215 ); HAEC no. 10238 (HLA-A24,9 HLA-A33,19 HLA-B14, and HLA-B35); HAEC no. 11202 (HLA-A02, HLA-A24, HLA-B18, and HLA-B51); HAEC no. 11636 (HLA-A02, HLA-A29,19 HLA-B44,12 and HLA-B35); and HAEC no. 14756 (HLA-A01, HLA-A30,19 HLA-B18, and HLA-B57). ECs were cultured in EC growth medium (ECBM) supplemented with 10% FCS, 0.004 mL ECG supplement (ECGS)/heparin, 0.1 ng/mL hEGF, 1 ng/mL human basic firbroblast growth factor (hbFGF), 1 μg/mL hydrocortisone, 50 μg/mL gentamicin, and 50 ng/mL amphotericin B (C-22010; PromoCell, Heidelberg, Germany). For activation, confluent EC monolayers were incubated with recombinant human TNFα (100 U/mL; kindly provided by Prof P. Neuman, BASF, Ludwigshafen, Germany), IFNγ (100 U/mL; Imukin, Boehringer Ingelheim, Germany), IL-1β (5 ng/mL; R&D Systems) for the indicated period of time in ECBM supplemented with 2% FCS. Culture supernatants were collected at the indicated times after activation and kept frozen. When needed, supernatants were concentrated (10 ×) using Microcon YM-3 (Millipore, Bedford, MA). PBMCs from random healthy volunteers were purified by Ficoll/Hypaque density centrifugation. NK cells were purified (> 95% of CD3−CD56+ and/or CD16+, as assessed by FACS) by negative selection using NK Cell Isolation kits according to the manufacturer's recommendations (Miltenyi Biotec, Paris, France).
Immunohistochemistry
Paraffin sections (4 μm thick) were mounted on pretreated slides, deparaffinized with toluene, rehydrated through a graded series of ethanol, and rinsed in distilled water. Tissue sections were then subjected to epitope retrieval in a microwave oven using citrate buffer (pH 6.0). Tissue sections were stained using a 2-step visualization system based on a peroxidase-conjugated dextran backbone (Dako Envision+ System; Dako, Trappes, France). The following mAbs were used: anti–human HLA-E (MEM-E/02) and anti–human HLA-G (MEM-G/01). Sections were incubated with primary antibody at room temperature (RT) for 30 min and then incubated with the HRP-conjugated secondary antibody for 30 minutes at RT. Immunostaining was visualized using the 3,3-diaminobenzidine/H202 substrate, and tissues were counterstained with haematoxylin.
For immunofluorescence, ECs were grown to confluence on glass coverslips. Cultures were washed with PBS and fixed for 20 minutes in PBS containing 4% paraformaldehyde. Cells were incubated overnight (ON) at 4°C with blocking buffer (5% BSA in PBS) and then incubated with an anti–HLA class I (W6/32) or anti–HLA-E (MEM-E/07) mAbs (10 μg/mL) in blocking buffer with or without Triton X-100 for 1 hours. Slides were rewashed and incubated with FITC-conjugated goat antimouse antibodies (5 μg/mL; Jackson Laboratories, West Grove, PA) for 1 hour. Endoplasmic reticulum (ER) and Golgi staining were performed using rhodamine-B-hexyl ester (2.5 μg/mL; Molecular Probes, Eugene, OR) and anti-Golgi mAbs (5 μg/mL; anti–golgin-97, clone CDF4; Molecular Probes), respectively. Anti-Golgi mAbs were revealed using TRITC-conjugated goat antimouse antibodies (5 μg/mL; Jackson Laboratories). Nuclear staining was performed using To-Pro-3 (1:1000 dilution; Molecular Probes). Slides were washed in PBS, dried, and mounted with ProLong antifade reagent (Molecular Probes). Fluorescence microscopy was performed with a Leica DM-IRBE laser-scanning confocal microscope (Leica, Heerbrugg, Switzerland) using a 63 × 1.4 oil p-Aplo lens (Leica) and analyzed using Leica TCS NT software.
RNA analysis and reverse transcriptase–PCR
RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) and analyzed by competitive polymerase chain reaction (PCR) as described previously.27 Briefly, total RNA (2 μg) was reverse transcribed with oligo (dT), treated with RNase H, and made up to 50 μL. cDNAs were diluted 1:2 for competitions. Competitor templates were initially diluted 1:10,6 followed by 4 serial dilutions of 1:3 each. The primer sequences were: HLA-A (334 bp) sense: 5′-CTACCCTGCGGAGATCA-3′, antisense: 5′-GCTCCCTCCTTTTCTATCTG-3′; HLA-B (255 bp) sense: CTACCCTGCGGAGATCA, antisense: ACAGCCAGGCCAGCAACA; HLA-E (257 bp) sense: 5′-CTACCCTGCGGAGATCA-3′, antisense: 5′-AGAGAACCAGGCCAGCAAT-3′; and HPRT (78 bp) sense: 5′-GGACAGGACTGAACGTCTTGC-3′, antisense: 5′-TTGAGCACACAGAGGGCTACA-3′. PCR products were sequenced by Genosys (Sigma-Aldrich, Saint Quentin Fallavier, France). Internal standards were obtained by mutagene PCR amplifications to generate mutated fragment by the deletion of 5 nucleotides as previously described.28 PCR products were separated on a 4% acrylamide gel and analyzed by capillary electrophoresis on an ABI PRISM 310 DNA Sequencer (PE Applied Biosystems, Foster City, CA) using GeneScan Analysis software (Applied Biosystems).
Semi-quantitative PCR for HLA-E and β-actin was carried out for 20 and 18 cycles, respectively, as follows: 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, with a final extension at 72°C for 3 minutes. PCR primers were: 5′-CCACCATGGTAGATGGAACCC-3′, and antisense: 5′-GCTTTACAAGCTGTCAGACTC-3′ for HLA-E; and sense: 5′-AATCTGGCACCACACCTTCTACA-3′, and antisense: 5′-CGACGTAGCACAGCTTCTCCTTA-3′ for β-actin. PCR products were separated on 1.5% agarose gels in the presence of ethidium bromide.
Western blotting
When applicable, cells were treated for 12 to 72 hours with 100 U/mL TNFα, 100 U/mL IFNγ, and 5 ng/mL IL-1β. When applicable, cells were incubated with 2 μg/mL brefeldin A (BrfA) or 10 μM galardin as an MMP inhibitor (Sigma-Aldrich) for the last 6 hours. Alternatively, cells were treated for 1 hour with 2.5 μg/mL cyclohexamide (CHX) before stimulation. Cells (3 × 106) were washed in PBS and incubated in 300 μL lysis buffer (20 mM Tris-HCl [pH 7.4], 137 mM NaCl, 0.05% Triton X-100, and complete protease inhibitor cocktail from Sigma-Aldrich) for 15 minutes on ice. Equal amounts of protein (15 μg) or equal volumes of culture supernatants (20 μL) or sera (7 μL) were loaded under reducing conditions and resolved by 12% SDS-PAGE. Immunoblotting was performed on nitrocellulose membranes (Amersham-Pharmacia, Orsay, France) using the mAbs anti–HLA-E (MEM-E/02), anti–HLA-G (MEM-G/01), anti–MHC class I (W6/32 from American Tissue Culture Collection [ATCC], Manassas, VA), anti–β2-microglobulin (Exbio Praha, Czech Republic), and anti–VCAM-1 (R&D Systems). Anti-mouse IgG, HRP-linked Abs (Cell Signaling Technology, Beverly, MA) were used as secondary Abs in cheluminescent Western blot assays using the enhanced chemiluminescence (ECL) system (Amersham-Pharmacia). When applicable, blots were reprobed with mouse anti-GAPDH or antitubulin mAbs (Chemicon, Temecula, CA). For deglycosylation with endoglycosidase H (EndoH) and endoglycanase F (PNGase F) (both from Sigma-Aldrich), lysates of IFNγ-treated ECs were boiled in 0.5% SDS and 1% β-mercaptoethanol for 5 minutes. Afterward, the solution was adjusted to 1% Nonidet P-40, and either 50 mM sodium phosphate (pH 7.5) and 0.6 U PNGase F, or 50 mM sodium acetate (pH 6.5) with 15 mU EndoH (both with a protease inhibitor mixture). Digestion was performed overnight at 37°C followed by SDS-PAGE and immunoblotting, as described above.
Flow cytometry
Cells were harvested, washed twice with PBS containing 1% BSA and 0.1% NaN3, and then incubated on ice for 30 minutes with a saturating concentration of the primary antibody. After 3 washes, cells were incubated with an FITC-labeled goat anti–mouse F(ab′)2 IgG antibody (Jackson Laboratories) at 4°C for 30 minutes. Finally, cells were suspended in 1% paraformaldehyde in PBS. Negative controls were performed for each cell treatment by incubating the cells with isotype-matched control antibodies. The mouse mAbs used for this study were anti–HLA-A2/A28 (ATCC) and anti–HLA-E (MEM-E/07 or MEM-E/08). Fluorescence was measured on 10 000 cells/sample using a FACScalibur (Becton Dickinson, Mountain View, CA) and analyzed using CellQuestPro software (Becton Dickinson).
Cytotoxicity assays
ECs were cultured with or without 100 U/mL IFNγ for 3 days to generate optimal quantities of soluble HLA-E (sHLA-E) for analysis. Conditioned media were harvested and the presence of sHLA-E was assessed by Western blot analysis. Purified NK cells were preincubated for 20 minutes with antibodies, conditioned medium, or culture medium. Target cells (4 × 103/well) labeled with 51Cr were incubated with NK cells for 4 hours at various effector-target (E/T) ratios. Blocking experiments were performed by preincubating cells with saturating amounts of anti-NKG2A, anti-NKG2C, and anti-ILT2 mAbs (all from R&D Systems) or an isotype-matched irrelevant antibody for 10 minutes. Cytolytic activity was calculated using the following formula: % lysis = (experimental release − spontaneous release) × 100 / (maximum release − spontaneous release). The spontaneous release in all assays was less than 20% of the maximum release.
Statistical analysis
The data are expressed as the mean ± SD and were compared using the 1-way ANOVA test. Differences were considered significant at P values less than .05.
Results
HLA-E protein expression is restricted to leucocytes and ECs
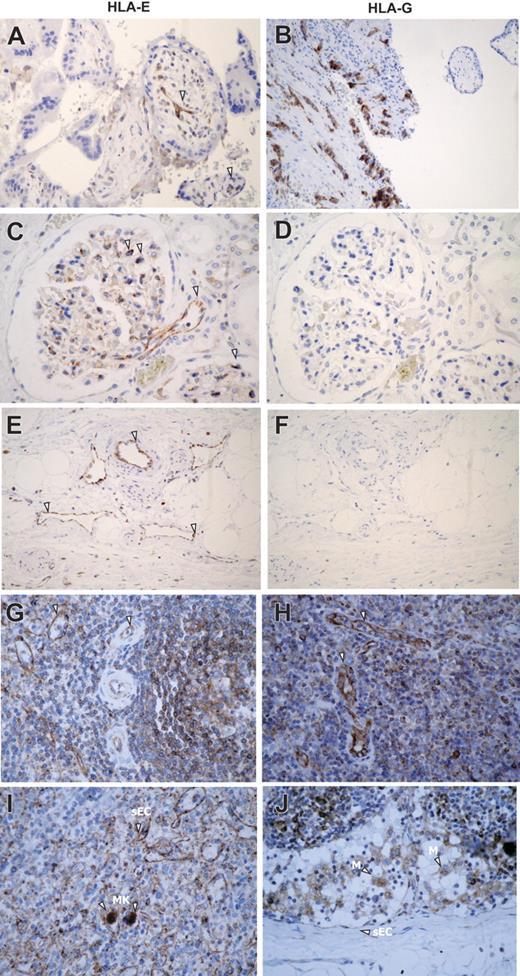
First, we examined HLA-E expression in human tissues by immunohistochemistry using mAbs specific for HLA-E or HLA-G as a control (Figure 1A). In placenta, comparative immunostaining indicated that HLA-E as well as HLA-G was expressed in extravillous trophoblast cells as previously reported.17,19 In all other normal, nonlymphoid, human tissues tested (kidney, skin, liver, salivary gland, urinary bladder, thyroid, stomach, and endometrium), HLA-E staining was restricted to ECs and was consistently observed on ECs from all types of vessels, including arteries, veins, capillaries, and lymphatics. As illustrated in Figure 1A, endothelial expression for HLA-E in kidney sections was present in vascular, capillary, and glomerular ECs. No staining was observed on mesenchymal, tubular, mesangial, or muscle cells, or adipocytes. In lymphoid organs such as spleen and lymph nodes (Figure 1B) endothelial expression for HLA-E was also observed on high endothelial venules (HEVs), concomitant with a strong expression in B and T lymphocytes as well as monocytes and macrophages (Figure 1B). Megakaryocytes but not erythrocytes also showed HLA-E immunostaining at a basal level. HLA-E distribution among mesenchymal, epithelial, and hematopoietic cells in various tissues is summarized Table S1, available on the Blood website (see the Supplemental Materials link at the top of the online article).
Immunoperoxidase staining showing HLA-E expression in human normal tissues. (A) Comparative staining for HLA-E (i, iii, v) and HLA-G (ii, iv, vi) in paraffin-embedded sections of human first-trimester trophoblast (i-ii) and kidney (iii-vi). (B) Comparative expression of HLA-E protein was studied in paraffin-embedded sections of human spleen (i-ii) and lymph node (iii-iv). Arrowheads indicate ECs, M, macrophages; MK, megakaryocytes; and sEC, sinusoidal ECs. Original magnification, ×400. Images were visualized under an Olympus BX50F4 microscope (Olympus Optical, Tokyo, Japan) equipped with 4×/0.13, 10×/0.30, 20×/0.50, and 40×/0.75 objective lenses and an Olympus Camedia C-4040Zoom camera; Olympus DP-Soft version 3.2 software was used to acquire and process images.
Immunoperoxidase staining showing HLA-E expression in human normal tissues. (A) Comparative staining for HLA-E (i, iii, v) and HLA-G (ii, iv, vi) in paraffin-embedded sections of human first-trimester trophoblast (i-ii) and kidney (iii-vi). (B) Comparative expression of HLA-E protein was studied in paraffin-embedded sections of human spleen (i-ii) and lymph node (iii-iv). Arrowheads indicate ECs, M, macrophages; MK, megakaryocytes; and sEC, sinusoidal ECs. Original magnification, ×400. Images were visualized under an Olympus BX50F4 microscope (Olympus Optical, Tokyo, Japan) equipped with 4×/0.13, 10×/0.30, 20×/0.50, and 40×/0.75 objective lenses and an Olympus Camedia C-4040Zoom camera; Olympus DP-Soft version 3.2 software was used to acquire and process images.
HLA-E expression at mRNA and protein levels was investigated in vitro using primary cultures of human vascular ECs. Because inflammation affects EC phenotype and functions,20,29 we studied the effects of the cytokines TNFα, IL-1β, and IFNγ on HLA-E expression and regulation. First, we quantified of HLA-A, HLA-B, and HLA-E mRNA levels in cultured ECs by competitive reverse transcription (RT)–PCR using locus-specific primers and a competitor template with an internal deletion as previously reported.27 Transcripts encoded by all 3 HLA loci were detected in resting ECs, demonstrating that ECs express HLA-E constitutively. ECs expressed similar levels of HLA-A and HLA-E but lower levels of HLA-B transcripts (Figure 2A; *P < .05). Both TNFα and IFNγ increased HLA-E transcript levels, peaking at 24 hours (Figure 2B).
HLA-E mRNA and protein expression in cultured human ECs. (A) Quantification and comparative analysis of mRNA steady state levels for HLA-A, HLA-B, and HLA-E in cultured ECs by competitive RT-PCR. Values are mean ± SD (n = 3). *P < .01 versus HLA-B. (B) Regulation of HLA-E mRNA in response to TNFα or IFNγ was assessed by semiquantitative RT-PCR. PCR amplifications for β-actin were used as controls. RNA 18S and 28S are shown below. (C) Confocal microscope images showing comparative cell-surface staining for HLA-A, HLA-B, HLA-C (i, ii) and HLA-E (ii, iv) on nonpermeabilized (i-ii) and permeabilized (iii-iv) vascular ECs. Nuclei were stained with To-pro-3 (red). (D) The colocalization of HLA-E (left panel; green) rhodamine-B-hexyl ester (for ER staining) or anti–golgin-97 (for Golgi staining, both middle panel; red) on permeabilized ECs. Merged images are shown on the right panel. Colocalization is shown in yellow. Original magnification, ×63. Scale bar equals 15 μm (applies to all figures).
HLA-E mRNA and protein expression in cultured human ECs. (A) Quantification and comparative analysis of mRNA steady state levels for HLA-A, HLA-B, and HLA-E in cultured ECs by competitive RT-PCR. Values are mean ± SD (n = 3). *P < .01 versus HLA-B. (B) Regulation of HLA-E mRNA in response to TNFα or IFNγ was assessed by semiquantitative RT-PCR. PCR amplifications for β-actin were used as controls. RNA 18S and 28S are shown below. (C) Confocal microscope images showing comparative cell-surface staining for HLA-A, HLA-B, HLA-C (i, ii) and HLA-E (ii, iv) on nonpermeabilized (i-ii) and permeabilized (iii-iv) vascular ECs. Nuclei were stained with To-pro-3 (red). (D) The colocalization of HLA-E (left panel; green) rhodamine-B-hexyl ester (for ER staining) or anti–golgin-97 (for Golgi staining, both middle panel; red) on permeabilized ECs. Merged images are shown on the right panel. Colocalization is shown in yellow. Original magnification, ×63. Scale bar equals 15 μm (applies to all figures).
Endothelial expression for HLA-E protein was investigated further on cultured human ECs. ECs were selected carefully according to their HLA type to avoid the reported cross-reactivity of the anti–HLA-E–specific mAbs with some classical MHC antigens, as described in “Materials and Methods.”
Immunofluorescence and confocal microscopy were used as a first attempt to examine surface and intracellular HLA-E expression in HAECs. Although weaker in intensity than staining for HLA-A, HLA-B, and HLA-C, HLA-E staining was found on nonpermeabilized cells (Figure 2C), confirming that HLA-E protein is localized at the outer surface of ECs. Intracellularly, HLA-E had a perinuclear distribution in permeabilized ECs (Figure 2C-D), where HLA-E fully colocalized with the ER, and partly with the Golgi apparatus (Figure 2D), suggesting that HLA-E molecules may be actively secreted.
Endothelial expression of HLA-E protein is up-regulated by proinflammatory cytokines
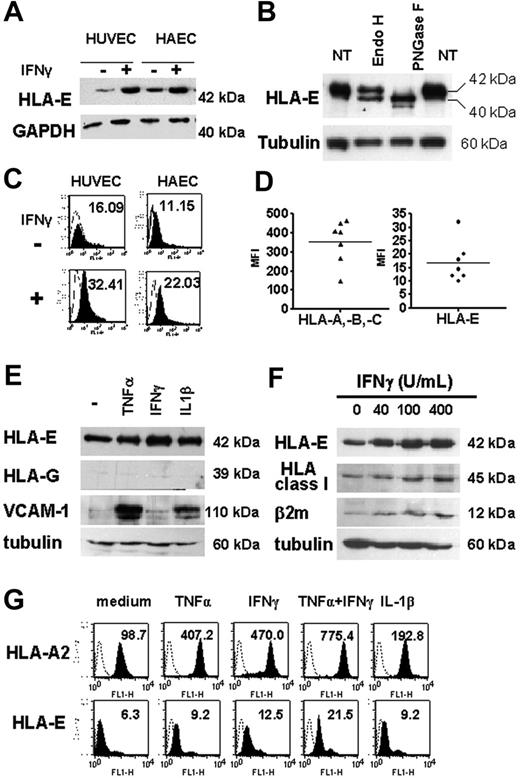
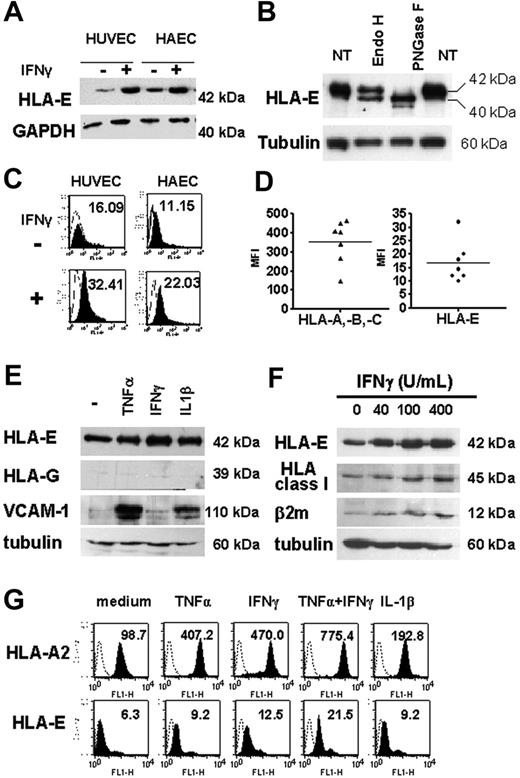
Western blot analysis, performed on total-cell lysates from HAECs and HUVECs and using MEM-E/02 as anti–HLA-E–specific mAbs, revealed a single band at 42 kDa, consistent with the molecular weight for HLA-E protein (Figure 3A), and distinct from classical HLA class I proteins detected at 45 kDa (Figure 3F). Cell lysates were treated with PNGase F or EndoH and analyzed by Western blot (Figure 3B). Our data indicates that EndoH partially converted the 42-kDa form of HLA-E to a single band of lower molecular weight (40 kDa), indicating that part of the cytoplasmic HLA-E protein was in an EndoH-sensitive form. However, approximately 50% of HLA-E was EndoH resistant, implying passage through the medial Golgi, a feature of mature MHC class I proteins. These HLA-E molecules likely represent the fraction found at the plasma membrane and is consistent with our results from confocal microscopy. In contrast, and as expected, most of HLA-E was PNGase sensitive. Indeed, PNGase F cleaves nearly all types of N-glycans and was used as a control for glycosylation. Using flow cytometry, we were able to detect HLA-E at the cell surface where basal expression of HLA-E protein was low but significant, and this was observed consistently in cultures from HAECs and HUVECs (Figure 3C). Comparative cell-surface expression for HLA-A, HLA-B, HLA-C, and HLA-E was assessed by flow cytometry using W6/32 and MEM-E/07 mAbs on 7 independent HAEC cultures (Figure 3D). Although lower than those of classical HLA class I (HLA-A, HLA-B, and HLA-C), basal expression of HLA-E protein was significant and was consistently observed on the membrane of the different cultures. Similar immunostaining was obtained with 2 different anti–HLA-E–specific mAbs (MEM-E/07 and MEM-E/08) both recognizing the native HLA-E molecule,26 although immunostaining with MEM-E/08 appeared weaker than with MEM-E/07 mAbs (data not shown). In response to IFNγ, both total and cell-bound HLA-E levels were increased and reached a maximal level at 48 hours after stimulation (data not shown).
Regulation of total and cell-surface HLA-E in cultured ECs upon inflammation. (A) HLA-E protein expression in untreated and IFNγ-activated ECs. Confluent monolayers of HAECs and HUVECs were incubated with culture medium or with 100 U/mL IFNγ for 48 hours. Cell lysates were immunoblotted using MEM-E/02 as anti–HLA-E mAbs. Immunoblots were reprobed with anti-GAPDH mAb to compare protein loading within samples. A representative immunoblot is shown. (B) Total-cell lysates from IFNγ-treated ECs were digested with EndoH or PNGaseF for 12 hours at 37°C, loaded onto a 10% SDS-PAGE, and examined by Western blotting with anti–HLA-E antibody. For controls, samples were incubated without the enzymes (NT). Immunoblots were reprobed with antitubulin mAbs. (C) FACS analysis showing cell-surface HLA-E expression (solid histograms) on HAECs and HUVECs either untreated or activated with IFNγ for 48 hours. Controls were performed by using an isotype-matched control antibody (empty histograms). Mean of fluorescence intensity are indicated. (D) Comparative analysis of HLA class Ia (HLA-A, HLA-B, and HLA-C) and HLA-E on the surface of 7 independently derived cultures of HAECs. Results are express as means of fluorescence intensity (MFI). Horizontal bars correspond to the means of value expressed as “mean of fluorescence intensity.” (E) Western blot analysis showing HLA-E, HLA-G, VCAM-1, and β2-microglobulin in cell lysates from HAECs treated for 48 hours with 100 U/mL TNFα, 2.5 ng/mL IL1β, or 100 U/mL IFNγ or culture medium alone (−). Immunoblots were reprobed with antitubulin mAbs to compare protein loading within samples. A representative immunoblot of 3 experiments is shown. (F) Western blot analysis showing HLA-E, classical HLA class I, VCAM-1, and β2-microglobulin in cell lysates from HAECs treated for 48 hours with 0 to 400 U/mL IFNγ. Immunoblots were reprobed with antitubulin mAbs to compare protein loading within samples. A representative immunoblot is shown. (G) Flow cytometry analysis comparing HLA-A2 and HLA-E expression at the cell surface after 48 hours of treatment with TNFα, IFNγ, IL-1β, TNFα plus IFNγ, or culture medium alone. MFIs are indicated.
Regulation of total and cell-surface HLA-E in cultured ECs upon inflammation. (A) HLA-E protein expression in untreated and IFNγ-activated ECs. Confluent monolayers of HAECs and HUVECs were incubated with culture medium or with 100 U/mL IFNγ for 48 hours. Cell lysates were immunoblotted using MEM-E/02 as anti–HLA-E mAbs. Immunoblots were reprobed with anti-GAPDH mAb to compare protein loading within samples. A representative immunoblot is shown. (B) Total-cell lysates from IFNγ-treated ECs were digested with EndoH or PNGaseF for 12 hours at 37°C, loaded onto a 10% SDS-PAGE, and examined by Western blotting with anti–HLA-E antibody. For controls, samples were incubated without the enzymes (NT). Immunoblots were reprobed with antitubulin mAbs. (C) FACS analysis showing cell-surface HLA-E expression (solid histograms) on HAECs and HUVECs either untreated or activated with IFNγ for 48 hours. Controls were performed by using an isotype-matched control antibody (empty histograms). Mean of fluorescence intensity are indicated. (D) Comparative analysis of HLA class Ia (HLA-A, HLA-B, and HLA-C) and HLA-E on the surface of 7 independently derived cultures of HAECs. Results are express as means of fluorescence intensity (MFI). Horizontal bars correspond to the means of value expressed as “mean of fluorescence intensity.” (E) Western blot analysis showing HLA-E, HLA-G, VCAM-1, and β2-microglobulin in cell lysates from HAECs treated for 48 hours with 100 U/mL TNFα, 2.5 ng/mL IL1β, or 100 U/mL IFNγ or culture medium alone (−). Immunoblots were reprobed with antitubulin mAbs to compare protein loading within samples. A representative immunoblot of 3 experiments is shown. (F) Western blot analysis showing HLA-E, classical HLA class I, VCAM-1, and β2-microglobulin in cell lysates from HAECs treated for 48 hours with 0 to 400 U/mL IFNγ. Immunoblots were reprobed with antitubulin mAbs to compare protein loading within samples. A representative immunoblot is shown. (G) Flow cytometry analysis comparing HLA-A2 and HLA-E expression at the cell surface after 48 hours of treatment with TNFα, IFNγ, IL-1β, TNFα plus IFNγ, or culture medium alone. MFIs are indicated.
Next, we compared endothelial expression for HLA-E after stimulation with TNFα, IL-1β, and IFNγ, alone or in combination. Immunoblotting showed an increase in HLA-E protein in ECs stimulated with these cytokines. Consistent with immunohistochemistry on human tissues, Figure 3E shows that no HLA-G expression was found in cultured ECs, even after treatment with cytokines; immunostaining for VCAM-1 and β2-microglobulin are shown as controls. FACS analysis confirmed that cytokines resulted in a significant increase in membrane-bound HLA-E compared with constitutive levels (Figure 3G). Combined treatment with TNFα and IFNγ (Figure 3G) produced further increases in HLA-E expression. Although moderate, IFNγ-induced up-regulation of HLA-E protein expression was dose dependent and paralleled those of classical class I HLA (Figures 3F-G). Indeed, these cytokines induced a 4- to 7-fold increase in HLA-A2 expression in ECs and up to a 3-fold increase in HLA-E over a 48-hour period in ECs.
Cytokine-activated ECs produce sHLA-E molecules
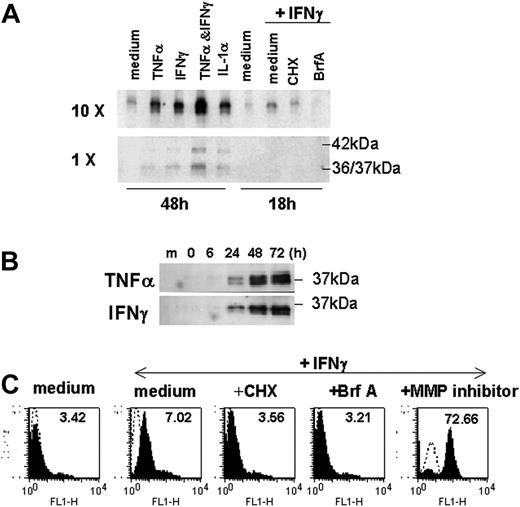
Next, we examined whether overexpression of HLA-E might be linked with the release of sHLA-E molecules, as described for other nonclassical class I MHC molecules, including HLA-G. Using Western blotting, we were able to detect the presence of sHLA-E in culture media from cytokine-activated ECs, whereas sHLA-E was below the detection threshold in conditioned medium from untreated ECs. Release of soluble HLA-E molecules was driven equally by TNFα, IFNγ, and IL-1β (Figure 4A). The combination of IFNγ and TNFα had an additive effect on its surface expression and release. sHLA-E was detected as a major band of 37 kDa (a doublet of 36 and 37 kDa), which may correspond to the metalloproteinase-dependent shedding of membrane-bound protein, but also as a minor band of 42 kDa corresponding to the full-length protein that may result from alternative gene splicing, as reported for other sHLA molecules.30 Time-course analysis revealed that sHLA-E protein was detectable by 12 to 24 hours after activation and was maximal at 72 hours after treatment with TNFα or IFNγ (Figure 4B). The kinetics of this process suggests that the cytokine-inducible release of sHLA-E likely occurs through new biosynthesis of the HLA-E protein. Our findings indicated that CHX, an inhibitor of protein synthesis, and BrfA, a specific inhibitor of exocytosis, prevented HLA-E expression and release by activated ECs (Figures 4A,C). In contrast, metalloproteinase inhibitor significantly increased expression of HLA-E at the membrane (Figure 4C) but abrogated release of sHLA-E, confirming that ECs, at least in part, generate sHLA-E by proteolytic shedding. Therefore, EC activation by inflammatory cytokines results in both increased membrane-bound HLA-E expression and release of sHLA-E.
Production of sHLA-E by cytokine-activated ECs. ECs were incubated with cytokines for 48 hours or cultured for 18 hours in the absence (medium) or in the presence of IFNγ after a preincubation with CHX for 1 hours or with BrfA or galardin as a metalloproteinase inhibitor (MP inhibitor) for the last 6 hours of culture. (A) Supernatants were collected; sHLA-E was then detected by Western blotting in normal (1 ×) or concentrated (10 ×) supernatants (20 μL/sample). One representative experiment of 5 performed (B) Time-course analysis by Western blotting of sHLA-E release by ECs treated with IFNγ. Data are from 1 representative experiment of 3 performed. (C) Cells were harvested for analysis of membrane-bound HLA-E by flow cytometry following immunostaining with MEM-E/07 mAbs (solid histograms) or an isotype-matched control antibody (histograms in dotted line). MFIs are indicated above. One representative experiment of 3 performed.
Production of sHLA-E by cytokine-activated ECs. ECs were incubated with cytokines for 48 hours or cultured for 18 hours in the absence (medium) or in the presence of IFNγ after a preincubation with CHX for 1 hours or with BrfA or galardin as a metalloproteinase inhibitor (MP inhibitor) for the last 6 hours of culture. (A) Supernatants were collected; sHLA-E was then detected by Western blotting in normal (1 ×) or concentrated (10 ×) supernatants (20 μL/sample). One representative experiment of 5 performed (B) Time-course analysis by Western blotting of sHLA-E release by ECs treated with IFNγ. Data are from 1 representative experiment of 3 performed. (C) Cells were harvested for analysis of membrane-bound HLA-E by flow cytometry following immunostaining with MEM-E/07 mAbs (solid histograms) or an isotype-matched control antibody (histograms in dotted line). MFIs are indicated above. One representative experiment of 3 performed.
As expected, flow cytometry analysis performed on freshly isolated PBMCs and cell lines (Jurkat, Raji, U937, and NKL) confirmed constitutive HLA-E expression on CD3+, CD4+, CD8+, CD19+, and CD14+ leukocyte subsets (Figure S1). When sHLA-E levels were measured, the presence of the sHLA-E was not detected under most conditions, except for purified NK cells and a NK cell line (NKL) activated by IL-2 (Figure S1). Together, these findings suggest that among HLA-E expressing cells, the production of sHLA-E is limited to a restricted set of activated cells, including endothelial and NK cells.
Membrane-bound and soluble endothelial HLA-E molecules protect cells from CD94/NKG2A-mediated cytolysis
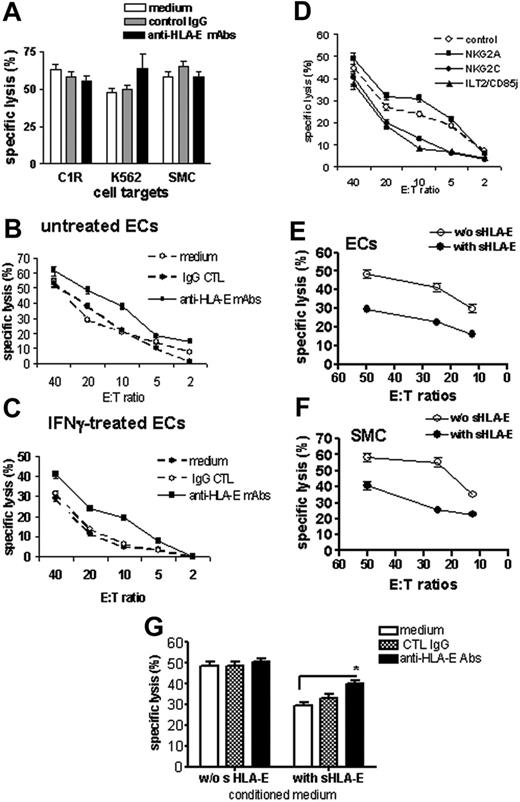
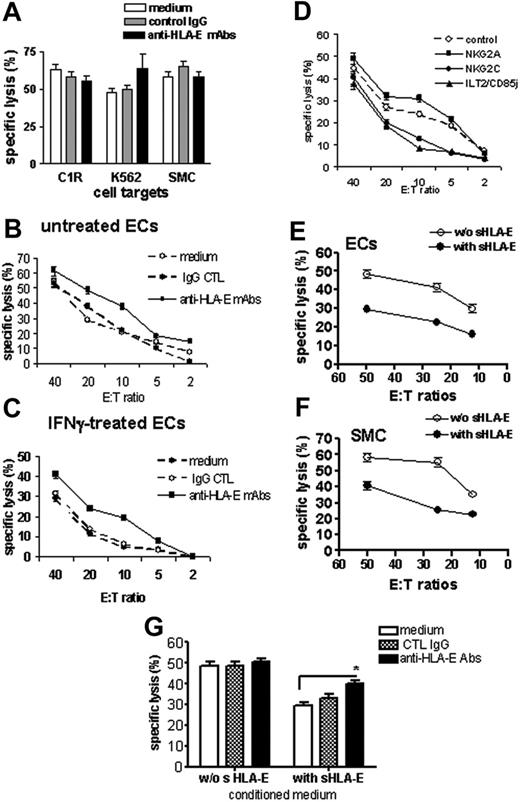
The functions of surface-bound and sHLA-E molecules, expressed and released by ECs, were evaluated in cell-mediated cytotoxicity assays. NK cells were purified from PBMCs and used as effector cells in cytotoxicity assays where target cells were the class I–deficient cell lines C1R and K562, and primary cultures of SMCs, which do not express HLA-E, or ECs (resting or activated for 48 hours with IFNγ). Experiments were performed in the presence or absence of a blocking anti–HLA-E mAbs (MEM-E/07). As shown in Figure 5A, cells that did not express HLA-E at the cell surface (C1R, K562, and SMCs) were efficiently lysed by NK cells with no effect of the anti–HLA-E–blocking mAb. Resting ECs were also efficiently killed by allogeneic NK cells, and blocking HLA-E slightly increased cell lysis (Figure 5B). This suggests that a basal level of surface HLA-E on quiescent ECs provides a moderate but significant degree of protection. EC activation with IFNγ decreased NK-mediated cytotoxicity dramatically compared with resting ECs, in the same experiment (Figure 5C). Blocking HLA-E on IFNγ-activated HAECs significantly restored cell lysis, suggesting that up-regulation of HLA-E at the cell surface upon IFNγ stimulation provides a protection against NK cytotoxicity. Remarkably, the selective blockade of the 2 potential HLA-E receptors NKG2A and NKG2C also confirmed HLA-E bioactivity (Figure 5D). Indeed, blocking NKG2A increased EC lysis, thus confirming our results that using anti–HLA-E mAbs while blocking the activating receptor NKG2C decreases EC cytotoxicity significantly. Interestingly, blocking ILT2, a receptor for HLA-G and HLA-F, also strongly reduced cytotoxicity. In an attempt to determine the biological activity of sHLA-E, we measured the cytotoxic activity of NK cells toward cells with no or low HLA-E expression (quiescent ECs and SMCs) in the presence of cell-conditioned medium with or without sHLA-E. These experiments indicate that cell lysis decreased significantly when sHLA-E was present (Figures 5E-F). Moreover, blocking sHLA-E with anti–HLA-E–specific mAbs partially restores NK-mediated EC lysis (Figure 5G). Consistent with a previous report,31 we found no down-regulation of CD94/NKG2A receptors on NK cells following coculture with ECs (data not shown).
Protective effect of cell-surface and sHLA-E molecules against CD94/NKG2A-dependent NK cell cytotoxicity. Cell surface expression of HLA-E mediates protection toward NK cell cytotoxicity. (A) Cytotoxicity assays were performed using target cells with no HLA-E expression at the cell surface, including the class I–deficient lymphoblastoid cell lines (C1R and K562) and primary cultures of SMCs. Target cells were preincubated with culture medium, control IgG (mouse IgG1; 10 μg/mL), or anti–HLA-E mAbs (10 μg/mL) for 20 minutes at RT. (B-C) Cytotoxicity assays were performed using ECs as target cells with a regulated HLA-E expression at the cell-surface ECs. ECs were untreated (B) or activated with 100 U/mL IFNγ for 48 hours (C). ECs were labeled with 51Cr and preincubated with culture medium, control IgG (mouse IgG1; 10 μg/mL), or anti–HLA-E mAb (10 μg/mL) for 20 minutes at RT before incubation with purified NK cells for 4 hours at 37°C. (D) Blocking experiments were performed after preincubation of NK cells with anti-NKG2A, anti-NKG2C, or anti-ILT2 receptor (10 μg/mL for each). Control was achieved using an isotype-matched control IgG. Soluble HLA-E provides protection toward NK cell cytotoxicity to cells with no or low HLA-E expression at the membrane. Resting ECs (E) and SMC (F) were preincubated with culture medium (without sHLA-E) or conditioned medium from IFNγ-treated HAECs (with sHLA-E) for 20 minutes at RT before incubation with freshly purified NK cells. (G) Resting ECs were preincubated with culture medium (without sHLA-E) or conditioned medium from IFNγ-treated HAECs (with sHLA-E) for 20 minutes at RT. ECs were then treated with culture medium, control IgG (mouse IgG1; 10μg/mL), or anti–HLA-E mAbs (10 μg/mL) for 20 minutes at RT before incubation with NK cells. For all these experiments, results, expressed as mean of specific lysis ± SD, are representative of at least 3 independent experiments. *P < .01 versus control.
Protective effect of cell-surface and sHLA-E molecules against CD94/NKG2A-dependent NK cell cytotoxicity. Cell surface expression of HLA-E mediates protection toward NK cell cytotoxicity. (A) Cytotoxicity assays were performed using target cells with no HLA-E expression at the cell surface, including the class I–deficient lymphoblastoid cell lines (C1R and K562) and primary cultures of SMCs. Target cells were preincubated with culture medium, control IgG (mouse IgG1; 10 μg/mL), or anti–HLA-E mAbs (10 μg/mL) for 20 minutes at RT. (B-C) Cytotoxicity assays were performed using ECs as target cells with a regulated HLA-E expression at the cell-surface ECs. ECs were untreated (B) or activated with 100 U/mL IFNγ for 48 hours (C). ECs were labeled with 51Cr and preincubated with culture medium, control IgG (mouse IgG1; 10 μg/mL), or anti–HLA-E mAb (10 μg/mL) for 20 minutes at RT before incubation with purified NK cells for 4 hours at 37°C. (D) Blocking experiments were performed after preincubation of NK cells with anti-NKG2A, anti-NKG2C, or anti-ILT2 receptor (10 μg/mL for each). Control was achieved using an isotype-matched control IgG. Soluble HLA-E provides protection toward NK cell cytotoxicity to cells with no or low HLA-E expression at the membrane. Resting ECs (E) and SMC (F) were preincubated with culture medium (without sHLA-E) or conditioned medium from IFNγ-treated HAECs (with sHLA-E) for 20 minutes at RT before incubation with freshly purified NK cells. (G) Resting ECs were preincubated with culture medium (without sHLA-E) or conditioned medium from IFNγ-treated HAECs (with sHLA-E) for 20 minutes at RT. ECs were then treated with culture medium, control IgG (mouse IgG1; 10μg/mL), or anti–HLA-E mAbs (10 μg/mL) for 20 minutes at RT before incubation with NK cells. For all these experiments, results, expressed as mean of specific lysis ± SD, are representative of at least 3 independent experiments. *P < .01 versus control.
Presence of soluble HLA-E in the patient's sera correlates with disease activity in patients with AASV
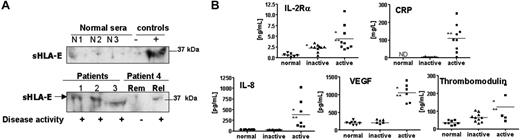
As a prerequisite to determine the clinical relevance of sHLA-E in physiologic or pathologic situations, we tested for sHLA-E in the sera of patients with ANCA-associated systemic vasculitis (AASV), including WG and MPA. This well-defined primary vasculitis subgroup is predominantly characterized by systemic inflammation of small vessels, and provides a relevant clinical model to investigate the relation between clinical inflammation and endothelial activation.32,33 Disease activity in 10 patients was assessed according to clinical features and levels of ANCA and CRP. Serum samples of patients (n = 10; 22 sera) with active (diagnosis or relapse) or inactive disease (remission) and of healthy volunteers (n = 9) were collected and assayed for sHLA-E. The presence of sHLA-E in sera, examined by immunoblot analysis of sera, revealed a band at 37 kDa, similar to that of sHLA-E released in cell-culture medium (Figure 6A). No significant amount of sHLA-E was detected in any of the healthy volunteers (Figure 6A; Table 1). In contrast, sera from 9 of 11 sera from patients with active disease (at diagnosis or relapse) showed the presence of sHLA-E, whereas sHLA-E was detectable in sera from only 3 of 11 patients in the remission phase (*P < .01; Table 1). The presence of sHLA-E was correlated with elevated levels of CRP and soluble IL-2Rβ (sIL-2Rβ), IL-8, VEGF, and thrombomodulin in the patients' sera as determined by enzyme-linked immunosorbent assays (ELISAs) (Figure 6B). In samples collected during the remission phase, we found significant elevations in sIL-2Rα levels relative to healthy volunteers (**P < .01). No evidence of sHLA-E was found in the sera from patients (n = 5) with sepsis and elevated CRP (data not shown). These data indicate that sHLA-E is released in significant amounts from cells in vivo in response to inflammatory processes affecting ECs, suggesting that shedding of sHLA-E may reflect vascular injury.
Detection of soluble HLA-E in sera from patients with AASV and correlation with soluble markers of inflammation. (A) Representative immunoblots showing the detection of sHLA-E in sera from patients with AASV (“Materials and methods”). The top panel shows the absence of sHLA-E in the serum from 3 healthy volunteers compared with negative and positive controls. The bottom panel illustrates the presence of sHLA-E in 3 sera from patients (nos. 1-3) with active phase and in 2 sera from 1 patient (no. 4). Rem indicates remission; Rel, relapse). Results are representative of 5 independent experiments. (B) Quantitative analysis of soluble IL-2Rα, CRP, IL-8, VEGF, and thrombomodulin by ELISA assays. *P < .01 versus normal; **P < .01 versus inactive. ND indicates not determined. Horizontal bars correspond to the means of value for each group of sera.
Detection of soluble HLA-E in sera from patients with AASV and correlation with soluble markers of inflammation. (A) Representative immunoblots showing the detection of sHLA-E in sera from patients with AASV (“Materials and methods”). The top panel shows the absence of sHLA-E in the serum from 3 healthy volunteers compared with negative and positive controls. The bottom panel illustrates the presence of sHLA-E in 3 sera from patients (nos. 1-3) with active phase and in 2 sera from 1 patient (no. 4). Rem indicates remission; Rel, relapse). Results are representative of 5 independent experiments. (B) Quantitative analysis of soluble IL-2Rα, CRP, IL-8, VEGF, and thrombomodulin by ELISA assays. *P < .01 versus normal; **P < .01 versus inactive. ND indicates not determined. Horizontal bars correspond to the means of value for each group of sera.
Discussion
Here we found that in contrast to the ubiquitously expressed classical HLA class I molecules, expression of the nonclassical HLA-E was restricted to specific cells, including ECs and immune cells (B, T lymphocytes, NK cells, monocytes, and macrophages). The first conclusion of this study is that although HLA-E transcripts can always be detected,2 the HLA-E protein is not always expressed despite the availability of peptides linked to HLA class I expression in most cells. Therefore, this suggests that specific posttranscriptional regulatory mechanisms might be critical for HLA-E protein expression in endothelial and immune cells. These mechanisms remain to be elucidated. Interestingly, as reported here and in previous studies, and similar to HLA-G, HLA-E is also expressed in trophoblasts, as is HLA-G,16,17,19 and in some tumor cells.18,26,34,35 Here, the expression of HLA-G was checked both in vivo and in vitro and found to be negative in ECs even upon inflammation. Furthermore, we found no evidence for the binding of sHLA-E to an endothelial receptor, as recently reported for soluble HLA-G and the CD160 receptor on ECs36 (data not shown). Therefore, our findings provide another example of constitutive expression for nonclassical class Ib molecules in a selected set of cells and thereby raise the possibility that HLA-E drives specific function in immune and vascular homeostasis.
HLA-E is tightly up-regulated upon cytokine-induced activation at the EC surface. This up-regulation paralleled those of the classical HLA class I at the EC surface but remained moderate compared with those HLA class Ia. We also demonstrated a link between increased cell-surface expression and the release of sHLA-E in the extracellular compartment. Functionally, these experiments show the capability of sHLA-E to protect HLA-E–negative cells from NK cytolysis and provide the first evidence for a role of sHLA-E as an immunoregulatory molecule. Consistent with previous studies performed on HLA-E–transfected30 or HLA-G–transfected37 cells, our experiments using inhibitors of metalloproteinase suggest that HLA-E undergoes proteolysis after reaching the EC surface. Release of a secreted form of HLA-E resulting from alternative gene splicing is also supported by our data and is under further investigation. Soluble isoforms of classical or nonclassical class I HLA molecules have been shown to exhibit various functions. Soluble isoforms of HLA-G have been detected in biological fluids, such as amniotic fluid and serum, they inhibit NK cytolysis,37 and they induce apoptosis of activated T cells.38 Another soluble nonclassical MHC molecule, the MHC class I–related molecule MICA, acts as a competitive mimic that blocks recognition of membrane-bound ligand and as a suppressor that down-regulates NKG2D expression.39
We have recently shown in a concomitant study that IFNγ also up-regulates HLA-E expression and induces the release of sHLA-E from melanoma cells.35 We speculate that such abundant expression of membrane-associated and sHLA-E molecules in malignant tumor cells might promote tumor growth.
Furthermore, we found that sHLA-E release, as recently reported for HLA-G40 and MICA,41 may correlate with clinical outcome and is of potential relevance for the diagnosis and monitoring of patients. Indeed, we established the presence of sHLA-E in patients with active WG, a form of small-vessel vasculitis where endothelial activation in vessels parallels disease activity and systemic inflammation.33 Although little is known about the etiology of AASV, it is now becoming apparent that EC activation as well as immune mechanisms are central to the pathogenesis of these diseases.
Functionally, we found that enhanced HLA-E expression on IFNγ-activated ECs impaired CD94/NKG2A-dependent NK cytolysis, whereas sHLA-E molecules provided protection to HLA-E–negative target cells. To our knowledge, this is the first evidence for a biological activity of membrane-associated and sHLA-E molecules on normal nontransfected cells. This protective pathway could have substantial relevance by counteracting classical class I down-regulation on virally infected ECs (eg, cytomegalovirus [CMV]) and in the setting of vascularized allografts where other inhibitory NK receptors may not operate.42
Although not investigated in the present study, we speculate that HLA-E may also affect antigen presentation by the endothelium. Indeed, HLA-E displays features of antigen-presenting molecules whereas ECs display features of antigen-presenting cells (APCs).22,23,43 HLA-E recognition by human CTLs has been reported,10 and TCRαβ+ alloreactive CTLs specific for HLA-E have now been identified.12 Therefore, it can be hypothesized that HLA-E complexed with β2-microglobulin and peptide might also play a role in autologous and allogeneic peptide presentation by ECs to CD8+ T cells.
Self-peptide presentation by HLA-E is thought to be central in the generation of suppressive CD8+ T cells controlling the expansion of pathogenic autoreactive T cells.14,15 Because the HLA-E peptide complexes can interact with the CD94/NKG2 receptors expressed on NK or classical CD8+ T cells to regulate function either positively or negatively, HLA-E may not only serve as the target of CD8 suppression, but might also regulate the function of CD8+ suppressor T cells via the CD94-NKG2 receptors. Suppressor T cells are essential for the control of autoimmunity and are also involved in the control of the immune response to transplanted allografts.44 In some conditions, ECs also suppress CD8+ T-cell activation by professional APCs, resulting in a form of immune tolerance.45,46 Whether endothelial HLA-E contributes to T-cell suppression rather than to T-cell activation has yet to be elucidated.
To conclude, here we have established clearly that HLA-E expression and release of sHLA-E can be considered as markers of EC activation in inflammatory and autoimmune diseases. Given the putative impact of HLA-E bioactivity, we speculate that endothelial HLA-E may play a role in the pathogenesis of vascular diseases. Increased HLA-E and sHLA-E levels in response to inflammatory insults may serve as a protective response of EC to injury aimed at curbing NK-mediated lysis. Moreover, increased expression of HLA-E may either be deleterious through the recognition of this nonclassical HLA molecule by CTLs, mediating allograft rejection, or beneficial through the generation of suppressive CD8+ T cells, as shown in mice models with the Qa-1 molecule. Overall, this implies that endothelial HLA-E may regulate multiple functions and therefore underscores the fundamental role of ECs in immune regulation pathways.
Authorship
Contribution: S.C. designed the research and performed the experiments; A.M. performed immunohistochemistry analysis; M.H. provided sera from patients with AASV and analyzed the data; V.H. contributed new anti–HLA-E monoclonal antibodies; J.P.S. analyzed data; and B.C. designed and performed the experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Béatrice Charreau, INSERM U643, 30 bd J. Monnet, Nantes, F-44093 France; e-mail: Beatrice.Charreau@univ-nantes.fr.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
The authors wish to thank Jeanne Souchet, Estelle Drouet, and Bernadette Denis for technical assistance, and Annabelle Chauveau for excellent assistance with confocal microscopy.
This work was supported by a grant “Recherche et Greffe 2005” from l'Agence de Biomédecine and a grant from the Société de Néphrologie.