In the United States, 59 000 people were diagnosed with non-Hodgkin lymphoma during 2006, and 19 000 died of the disease. Translational molecular research is a high priority; however, routine measurement of predictive gene expression levels has been difficult until now.
In this issue of Blood, Sakhinia and colleagues describe a promising research method to translate advances in the molecular characterization of lymphomas into clinical practice. The authors describe a reverse transcriptase–polymerase chain reaction (RT-PCR) methodology to quantify expression levels of “signature” genes previously highlighted in microarray-based discovery studies. Their methodology is flexible, and it can be applied successfully to frozen samples archived as part of standard clinical care.
The sensitivity of the method is notable. As shown in Figures 1-4 of their report, RT-PCR expression values appear to have a dynamic range of around 128-fold between the lower and upper limits that typically contain 99% of the data (the “whiskers” in the box plots). This broad dynamic range should help to accurately classify tumors into distinct groups based on precisely measured expression levels, a prerequisite for optimally informative association studies.
After controlling for the probability of statistical false discovery, RT-PCR was able to identify significant differences in expression levels of signature genes between follicular lymphoma (FL), diffuse large B-cell lymphoma (DLBCL), and reactive lymph nodes (LNs); differences between FL and DLBCL combined versus LNs; and differences between FL and DLBCL, thereby replicating and going beyond earlier studies.
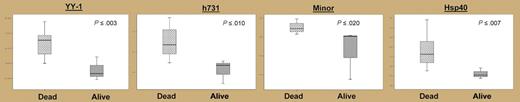
Of more importance from a clinical perspective, the investigators were able to begin to correlate sensitive RT-PCR expression levels with outcomes in the clinic (see figure), albeit with considerable uncertainty due to inherent limitations in sample size. As shown in their Kaplan-Meier survival curves (Figure 6), survival after FL and DLBCL is characterized by an extremely rapid decrease in the survival curve during the first few years following diagnosis, followed by a lower, but still substantial, mortality rate over the long term. In exploratory analysis, the authors found that expression levels in the lowest one fourth of observed values for the YY.1 gene appear to be associated with a comparatively favorable prognosis for both FL and DLBCL (see figure). Larger studies will be needed for replication. Is the survival advantage of low YY.1 levels due to a blunting of the early hazard, or to a lower failure rate over the long term, or both? Even larger studies will be needed to dissect such temporal associations, but these might provide important clues about disease heterogeneity.
Expression levels (log2) of genes with statistically significant differences in survival after DLBCL. See the complete figure in the article beginning on page3922.
Expression levels (log2) of genes with statistically significant differences in survival after DLBCL. See the complete figure in the article beginning on page3922.
Whatever specific genes emerge from such studies, it is important to realize that survival curves for lymphoma as observed reflect the natural history filtered through the efficacy of treatments in use during the calendar period of diagnosis. Therefore, it may be very fruitful to bring the RT-PCR technology developed by Sakhinia et al into the domain of clinical trials. Expression analysis of archived samples from completed trials might help to identify genes that are prognostic for FL and DLBCL, or even better, predictive of response to therapy.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ▪