Abstract
The TEL-JAK2 gene fusion, which has been identified in human leukemia, encodes a chimeric protein endowed with constitutive tyrosine kinase activity. TEL-JAK2 transgenic expression in the mouse lymphoid lineage results in fatal and rapid T-cell leukemia/lymphoma. In the present report we show that T-cell leukemic cells from EμSRα-TEL-JAK2 transgenic mice present an aberrant CD8+ differentiation phenotype, as determined by the expression of stage-specific cell surface markers and lineage-specific genes. TEL-JAK2 transforms immature CD4−CD8− double-negative thymocytes, as demonstrated by the development of T-cell leukemia with full penetrance in a Rag2-deficient genetic background. This disease is similar to the bona fide TEL-JAK2 disease as assessed by phenotypic and gene profiling analyses. Pre-TCR signaling synergizes with TEL-JAK2 to transform immature thymocytes and initiate leukemogenesis as shown by (1) the delayed leukemia onset in Rag2-, CD3ϵ- and pTα-deficient mice, (2) the occurrence of recurrent chromosomal alterations in pre-TCR–deficient leukemia, and (3) the correction of delayed leukemia onset in Rag2-deficient TEL-JAK2 mice by an H-Y TCRαβ transgene that mimics pre-TCR signaling. Although not affecting leukemia incidence and mouse survival, TCRαβ expression was shown to facilitate leukemic cell expansion in secondary lymphoid organs.
Introduction
In the thymus, T cells develop from a common CD4−CD8− double-negative (DN) progenitor into 2 main lineages, αβ and γδ, which are defined by the selection of productive rearrangements in the respective T-cell receptor (TCR) loci (for a review, see Aifantis et al1 ). In mice, DN thymocytes are divided in 4 categories according to CD25 and CD44 expression: DN1 (CD25−CD44+), DN2 (CD25+CD44+), DN3 (CD25+CD44−), and DN4 (CD25−CD44−). Rag-mediated rearrangement of the TCRβ locus at the DN3 stage leads to cell surface expression of a functional TCRβ chain, which assembles with the surrogate pTα chain and CD3-signaling proteins to form the pre-TCR complex. Constitutive survival, proliferation, and differentiation signals emanating from the pre-TCR allow T cells to pass through the β-selection checkpoint and mature to the DN4, CD4−CD8+ immature single-positive (ISP) and CD4+CD8+ double-positive (DP) stages. The β-selected cells rearrange their TCRα locus, and only the minority of cells expressing a functional TCRαβ complex at the cell surface will either undergo negative selection and die or undergo positive selection and become mature CD4 or CD8 single-positive (SP) thymocytes
Chromosomal rearrangements or point mutations in oncogenes or tumor suppressor genes occur in hematopoietic stem cells (HSCs), uncommitted and committed lymphoid progenitors, or developing thymocytes, thus leading to T-cell leukemia. Chromosomal translocations involving the juxtaposition of proto-oncogenes to the promoter and enhancer sequences of TCR loci result in deregulated oncogene expression. Chromosomal translocations can also create fusion genes encoding fusion oncoproteins. Gene expression profiling analyses have identified coordinate gene expression variations associated with T-acute lymphoblastic leukemias (T-ALL) subgroups defined by the expression of particular oncogenes.2,3 These T-ALL gene expression profiles were linked to specific stages in normal thymocyte differentiation, thus supporting the notion that T-ALLs are arrested at particular developmental stages.2,3
The TEL/ETV6-JAK2 gene fusion, resulting from a t(9;12)(p24;p13) chromosomal translocation, was identified in T-ALL, B-ALL, and atypical chronic myeloid leukemia.4,5 The presence of the TEL Pointed domain results in TEL-JAK2 oligomerization, constitutive JAK2 tyrosine kinase activity, and as a result, potent hematopoietic cell-transforming activity.4,6–8 Transgenic EμSRα-TEL-JAK2 mice develop a highly penetrant and fatal clonal/oligoclonal T-cell leukemia/lymphoma, which invades lymphoid and visceral organs and is transplantable to syngeneic or immunodeficient mice and either T- or B-cell leukemia/lymphoma when combined with CD3ϵ deficiency.9 TEL-JAK2–transforming activity in T and myeloid cells was also demonstrated after retroviral-mediated gene transduction into HSC-enriched bone marrow cells followed by adoptive transfer into lethally irradiated syngeneic mice.7
The disease developed in EμSRα-TEL-JAK2 transgenic mice is clinicopathologically consistent with T-cell lymphoblastic leukemia/lymphoma.8 In the present report we show that T-cell leukemic cells from EμSRα-TEL-JAK2 transgenic mice present an aberrant differentiation phenotype. Using gene-targeted mice lacking TCRαβ or pre-TCR expression (or both), we found that none of these receptor complexes is essential for T-cell leukemia development in TEL-JAK2 transgenic mice and that DN thymocytes are in vivo targets for transformation by this fusion protein. However, though not being required, the pre-TCR complex cooperates with TEL-JAK2 to accelerate T-cell leukemia onset.
Materials and methods
Mice
The EμSRα-TEL-JAK2 transgenic mice (line 71)8 were bred with knock-out mouse strains on the C57BL/6 background. All mice were maintained under specific pathogen-free conditions in the animal facility of the Institut Curie (Orsay, France), in accordance with institutional guidelines approved by the Ministère de l'Agriculture et de la Forêt, Direction de la Santé et de la Protection Animale (France). The Tcra10 and Rag211 knock-out mice were obtained from CNRS-CDTA (Orléans, France). The Ptcra12 and Prkcq13 knock-out mice were kindly provided by Harald von Boehmer (Dana Farber Cancer Institute, Boston, MA) and Dan Littman (Skirball Institute of Biomolecular Medicine, New York, NY), respectively. The Marylin transgenic mice14 were kindly provided by Olivier Lantz (Institut Curie). The mice were humanely killed when terminally ill, due to either severe dyspnea caused by massive expansion of leukemic cells in the thymus, or extreme weakness caused by leukemic dissemination to vital organs such as bone marrow, lung, and liver. Statistical analyses and survival curves were calculated using Prism 4 (GraphPad, San Diego, CA).
Flow cytometry
Single-cell suspensions were prepared from lymphoid organs, stained with fluorochrome-labeled antibodies, and detected by a FACSCalibur cytometer (BD Biosciences, San Jose, CA), as previously described.8 Fluorescein isothiocyanate (FITC)–, R-phycoerythrin (PE)–, PE-cyanine 5 (PE-Cy5)–, or allophycocyanin (APC)–conjugated antibodies specific for CD4 (H129.19), CD8α (53-6.7), CD3ϵ (145-2C11), TCRβ (H57-597), Vβ6 TCR (RR4-7), TCRγδ (GL3), CD5 (53-7.3), CD24 (M1/69), CD25 (7D4), CD69 (H1.2F3), CD62L (MEL-14), CD117 (2B8), CD122 (TM-b1), CD127 (SB/199), B220 (RA3-6B2), and IgM (R6-60.2) (BD Biosciences) were used. The data were analyzed using CellQuest (BD Biosciences) and FlowJo (Tree Star, Ashland, OR) software.
Southern blotting and quantitative RT-PCR
Southern blot analysis on gDNA was performed as previously described.8 Total RNA was extracted from the indicated T cells from 6- to 8-week-old C57BL/6 mice or from TEL-JAK2 leukemic cells using TRIzol (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. Total RNA (5 μg) was reverse-transcribed using the First Strand cDNA Synthesis Kit (GE Healthcare, Chalfont St Giles, United Kingdom) and a NotI-(dT)18 primer, according to the manufacturer's instructions. Quantitative polymerase chain reaction (PCR) was performed on an iCycler iQ Multi-Color Real-Time PCR Detection System (Bio-Rad, Hercules, CA) using iQ SYBR Green Supermix (Bio-Rad) and 150 nM of Hprt, Ctsw, Prf1, and Runx3 primer pairs. The primer sequences are described in “Supplemental materials and methods” (available on the Blood website; see the Supplemental Materials link at the top of the online article). After an initial hold at 95°C for 90 seconds, triplicate cDNAs were amplified for 40 cycles at 95°C for 30 seconds and 60°C for 1 minute. Results were averaged after outlier exclusion and Ctsw, Prf1, or Runx3 mRNA levels relative to those of Hprt were determined for each cDNA sample by the 2−ΔΔCt method.15
Oligonucleotide array analysis
Microarray analyses were performed using total RNA from 25 samples (see “Results”) using the Murine Genome U74Av2 array (Affymetrix, Santa Clara, CA) as previously reported.16 Except where indicated, transcriptome analyses were carried out using either an assortment of R system software (v 1.9.0) packages including those of Bioconductor17 (V1.1.1) or original R code. R packages and versions are indicated when appropriate. Raw feature data were normalized and log2 intensity expression summary values for each probe set were calculated using robust multiarray average (RMA, package affy V1.4.32).18 A detailed description of the unsupervised hierarchical clustering analysis and supervised analysis is presented in “Supplemental materials and methods.”
Mouse array comparative genomic hybridization
The genome-wide CIT M3 Mus musculus 1K BAC comparative genomic hybridization (CGH) array was built in the laboratory. This array contains 1081 BACs that were isolated, validated by end-sequencing, and amplified by multiple displacement amplification. The collection was spotted on Corning Ultragaps (Corning, NY) slides. For a detailed description of the gDNA array hybridization and detection, see “Supplemental materials and methods.”
Results
TEL-JAK2 leukemic cells display an aberrant cell surface phenotype
As previously reported, EμSRα-TEL-JAK2 transgenic mice develop a CD8+, partially CD4+ fatal T-cell leukemia.8 To know whether TEL-JAK2 leukemic T cells are arrested at a particular developmental stage, they were characterized by flow cytometry using a panel of antibodies recognizing different cell surface markers. All leukemia cases analyzed showed cell surface expression of TCRβ and CD3ϵ, albeit with variable levels among individual tumors and lower than those found in normal mature thymocytes (Figure 1A top panels). Supporting the notion that a functional TCRαβ complex is present in TEL-JAK2 leukemic cells, we found that these were hypersensitive to TCR stimulation in vitro, because they increased in size, expressed the CD69 T-cell activation marker, and proliferated in response to a low dose of anti-CD3ϵ monoclonal antibody (Figure S1A-B). TEL-JAK2 leukemic cells also proliferated in response to phorbol-myristyl acetate (PMA) and ionomycin, which together mimic TCR signaling (Figure S1B). Strikingly, TEL-JAK2 leukemic cells proliferated robustly when treated with PMA alone, contrasting with control thymocytes that did not proliferate under the same conditions (Figure S1B and data not shown). Together, these data indicate that TEL-JAK2 leukemic cells originate from αβ lineage-committed T cells and express a functional TCRαβ complex.
Cell surface phenotypic characterization of TEL-JAK2 leukemic cells by flow cytometry. (A) Cell surface immunostaining of 3 representative TEL-JAK2 tumor cases (nos. 31, 35, and 58) using antibodies against CD3ϵ, TCRβ, CD24, and CD25. Leukemic cells were also stained with an isotypic control monoclonal antibody. CD3ϵ and TCRβ expression in normal CD8 SP thymocytes is also shown. (B) Leukemic cells from 2 representative TEL-JAK2 transgenic tumors (nos. 1115 and 1281) were stained with CD62L or an isotypic control antibody. (C) CD5 expression levels in CD4/CD8 DP, CD4 (CD4SP), and CD8 (CD8SP) wild-type thymocytes as compared to isotypic antibody-labeled thymocytes (top panel). Leukemic cells from 2 representative TEL-JAK2 transgenic tumors (nos. 1119 and 1129) were stained with CD5 or an isotypic control antibody (bottom panel). (D) Wild-type thymocytes (WT) and 3 representative TEL-JAK2 transgenic tumors (nos. 1531, 1551, and 1563) were double-stained with CD25 and CD122 antibodies. Cell percentages for each quadrant are indicated.
Cell surface phenotypic characterization of TEL-JAK2 leukemic cells by flow cytometry. (A) Cell surface immunostaining of 3 representative TEL-JAK2 tumor cases (nos. 31, 35, and 58) using antibodies against CD3ϵ, TCRβ, CD24, and CD25. Leukemic cells were also stained with an isotypic control monoclonal antibody. CD3ϵ and TCRβ expression in normal CD8 SP thymocytes is also shown. (B) Leukemic cells from 2 representative TEL-JAK2 transgenic tumors (nos. 1115 and 1281) were stained with CD62L or an isotypic control antibody. (C) CD5 expression levels in CD4/CD8 DP, CD4 (CD4SP), and CD8 (CD8SP) wild-type thymocytes as compared to isotypic antibody-labeled thymocytes (top panel). Leukemic cells from 2 representative TEL-JAK2 transgenic tumors (nos. 1119 and 1129) were stained with CD5 or an isotypic control antibody (bottom panel). (D) Wild-type thymocytes (WT) and 3 representative TEL-JAK2 transgenic tumors (nos. 1531, 1551, and 1563) were double-stained with CD25 and CD122 antibodies. Cell percentages for each quadrant are indicated.
The analysis of T-cell activation markers showed that no TEL-JAK2 leukemia case analyzed expressed CD44 (data not shown), whereas low CD69 expression was restricted to a small leukemic cell subset (5%-20%; Figure S2). CD62L expression, which decreases on T-cell activation, was absent or minimal in the majority of leukemic cells (Figure 1B bottom panel). CD5 expression, which is up-regulated on TCR signaling during positive selection,19 was detected in all tumors analyzed, but at lower levels as compared to control DP thymocytes (Figure 1C). These observations suggest that TEL-JAK2 leukemic cells do not express a full set of markers associated with TCR-dependent activation.
The DN thymocyte markers CD117 (c-Kit) and CD127 (IL-7Rα) were not detected in any TEL-JAK2 leukemic sample analyzed (data not shown). In contrast, intermediate to high expression levels of CD24 (HSA) and CD25 (IL-2Rα) were systematically detected (Figure 1A and data not shown). CD122 (IL-2Rβ) expression was also detected in TEL-JAK2 leukemic cells together with CD25, indicating that a functional IL-2 receptor may be expressed in these cells (Figure 1D). The cell surface phenotyping thus showed that TEL-JAK2 leukemic T cells present an aberrant differentiation phenotype, characterized by the simultaneous expression of CD8, CD4, CD3ϵ, TCRβ, CD24, CD25, CD122, and CD5.
TEL-JAK2 induces T-cell leukemia in the absence of TCR expression
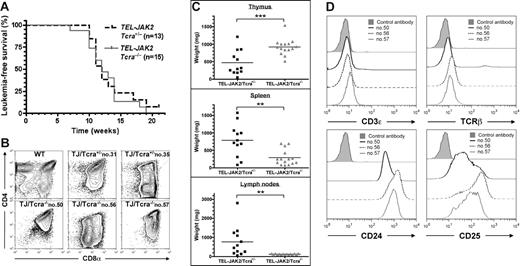
We next analyzed whether the TCR complex could play a role in TEL-JAK2–induced leukemogenesis. To this end, EμSRα-TEL-JAK2 transgenic mice were bred with TCRα-deficient mice, which are totally devoid of mature TCRαβ cells.10 As shown in Figure 2A, absence of TCRα did not alter the latency of leukemia development in TEL-JAK2/Tcra−/− mice, as compared to TEL-JAK2/Tcra+/− littermates (median survival of 12 weeks). All TEL-JAK2/Tcra−/− mice developed large thymic CD8+ lymphomas (Figure 2B), but, in contrast to TEL-JAK2/Tcra+/− littermates, showed relatively little involvement of spleen and lymph nodes (Figure 2C). Importantly, high white blood cell counts, bone marrow infiltration, and invasion of visceral organs, notably liver and lungs, was observed in both Tcra−/− and Tcra+/−TEL-JAK2 leukemic mice (data not shown), thus suggesting that Tcra inactivation did not prevent bloodborne leukemic cell dissemination to distant metastatic sites. The absence of a functional TCRαβ complex in TEL-JAK2/Tcra−/− leukemic cells was confirmed by the very low levels of CD3ϵ and TCRβ expression at their cell surface (Figure 2D top panels) and their unresponsiveness in vitro to anti-CD3ϵ stimulation (Figure S3). TEL-JAK2/Tcra−/− and TEL-JAK2/Tcra+/− leukemic cells expressed similarly CD4, CD8, CD24, and CD25 (Figures 1A and 2B,D) and were equally capable to transplant into recipient nu/nu or syngeneic mice (data not shown). Collectively, these findings support the notion that the T-cell lymphoma/leukemia in TEL-JAK2 transgenic mice originates in the thymus rather than in peripheral lymphoid organs, and that the DN/DP thymocyte subset is the main target for TEL-JAK2 transformation. In addition, these results suggest that TCRαβ signaling favors tumor load in peripheral lymphoid organs.
T-cell leukemia development in Tcra-deficient TEL-JAK2 transgenic mice. (A) Kaplan-Meier leukemia-free survival curves for EμSRα-TEL-JAK2 transgenic mice deficient or not for the Tcra gene show no statistically significant differences. Mice were monitored for disease as described in “Materials and methods.” The number of mice in each group is given in parentheses. (B) Three representative TEL-JAK2/Tcra−/− transgenic tumors (nos. 50, 56, and 57), 2 representative TEL-JAK2/Tcra+/− tumors (nos. 31 and 35), and control thymocytes were double-stained with CD4 and CD8α antibodies and analyzed by flow cytometry. (C) Thymus, spleen, and lymph node weights were determined for Tcra+/− (n = 12) and Tcra−/− (n = 15) TEL-JAK2 transgenic mice. **P < .01; ***P < .001 (unpaired t test). Average organ weight is indicated by horizontal bars. (D) Cell surface immunostaining of 3 representative TEL-JAK2 tumor cases (nos. 50, 56, and 57) using antibodies against CD3ϵ, TCRβ, CD24, and CD25 or an isotypic control monoclonal antibody.
T-cell leukemia development in Tcra-deficient TEL-JAK2 transgenic mice. (A) Kaplan-Meier leukemia-free survival curves for EμSRα-TEL-JAK2 transgenic mice deficient or not for the Tcra gene show no statistically significant differences. Mice were monitored for disease as described in “Materials and methods.” The number of mice in each group is given in parentheses. (B) Three representative TEL-JAK2/Tcra−/− transgenic tumors (nos. 50, 56, and 57), 2 representative TEL-JAK2/Tcra+/− tumors (nos. 31 and 35), and control thymocytes were double-stained with CD4 and CD8α antibodies and analyzed by flow cytometry. (C) Thymus, spleen, and lymph node weights were determined for Tcra+/− (n = 12) and Tcra−/− (n = 15) TEL-JAK2 transgenic mice. **P < .01; ***P < .001 (unpaired t test). Average organ weight is indicated by horizontal bars. (D) Cell surface immunostaining of 3 representative TEL-JAK2 tumor cases (nos. 50, 56, and 57) using antibodies against CD3ϵ, TCRβ, CD24, and CD25 or an isotypic control monoclonal antibody.
TEL-JAK2 transforms DN thymocytes
We next assessed whether TEL-JAK2 could transform cells lacking pre-TCR expression. To this end, we bred EμSRα-TEL-JAK2 transgenic mice with Rag2-deficient mice, which present strongly reduced thymocyte cellularity (∼0.5% of wild-type) due to a complete arrest in T-cell development at the DN3 stage.11 The resultant Rag2-deficient TEL-JAK2 mice developed T-cell leukemia/lymphoma with full penetrance, albeit with delayed onset (Figure 3A). B-cell lymphoma was never observed in these mice. Rag2-deficient tumors expressed similar cell surface markers (eg, CD4, CD8, CD24, and CD25) as Rag2-proficient TEL-JAK2 T-cell leukemia (Figure 3B). CD3ϵ deficiency results in the same differentiation block as Rag2 deficiency.20 The previously reported CD3ϵ-deficient TEL-JAK2 mice develop either B-cell leukemia9 or CD8+CD24+CD25+ T-cell leukemia with a latency similar to that of Rag2−/− TEL-JAK2 mice (Figure 3C and data not shown). The TEL-JAK2/Cd3e−/− T-cell leukemias were clonal/oligoclonal, as demonstrated by the presence of discrete TCRβ rearrangements (Figure 3D). In addition, EμSRα-TEL-JAK2 mice deficient in Rag2 or CD3ϵ presented a reduced infiltration of secondary lymphoid organs by leukemic cells (Figure 3E and 6D), likely due to the absence of TCRαβ expression (data not shown).
T-cell leukemia development in pre-TCR–deficient TEL-JAK2 transgenic mice. (A) Kaplan-Meier leukemia-free survival curves for EμSRα-TEL-JAK2 transgenic mice deficient or not for the Rag2 or Ptcra genes. The number of mice in each group is given in parentheses. The survival curves of Rag2- or Ptcra-deficient mice and the respective heterozygous littermates were significantly different (log-rank test, P < .001). Mice deceased from unrelated causes were censored in the analysis (tick marks). (B) Representative Rag2+/− (no. 25) and Rag2−/− (nos. 12, 22, and 1078) TEL-JAK2 leukemic cells and control thymocytes were double-stained with CD4 and CD8α antibodies (top panels) or stained with CD24 and CD25 antibodies (bottom panels) and analyzed by flow cytometry. (C) Kaplan-Meier T-cell leukemia survival curves for Cd3e−/− and Rag2−/− EμSRα-TEL-JAK2 mice. Mice deceased from B-cell lymphoma or other causes were censored in the analysis (tick marks). Note that no statistically significant difference in survival was observed between the 2 groups of mice (P = .98). (D) Tcrb locus Southern blot analysis of leukemic cell gDNA from Cd3e−/− (nos. 71, 74, and 58) and Cd3e+/+ (nos. 77 and 80) TEL-JAK2 thymic tumors. DNA obtained from wild-type liver (Liv), spleen (Spl), and thymus (Thy) was used as control. GL indicates the germline configuration and asterisks point to the distinct Tcrb locus clonal rearrangements. (E) Thymus, spleen, and lymph node weights were plotted for Cd3e+/− (n = 12) and Cd3e−/− (n = 10) TEL-JAK2 transgenic mice that developed T-cell leukemia/lymphoma. *P < .05 (unpaired t test). Average organ weight is indicated by horizontal bars.
T-cell leukemia development in pre-TCR–deficient TEL-JAK2 transgenic mice. (A) Kaplan-Meier leukemia-free survival curves for EμSRα-TEL-JAK2 transgenic mice deficient or not for the Rag2 or Ptcra genes. The number of mice in each group is given in parentheses. The survival curves of Rag2- or Ptcra-deficient mice and the respective heterozygous littermates were significantly different (log-rank test, P < .001). Mice deceased from unrelated causes were censored in the analysis (tick marks). (B) Representative Rag2+/− (no. 25) and Rag2−/− (nos. 12, 22, and 1078) TEL-JAK2 leukemic cells and control thymocytes were double-stained with CD4 and CD8α antibodies (top panels) or stained with CD24 and CD25 antibodies (bottom panels) and analyzed by flow cytometry. (C) Kaplan-Meier T-cell leukemia survival curves for Cd3e−/− and Rag2−/− EμSRα-TEL-JAK2 mice. Mice deceased from B-cell lymphoma or other causes were censored in the analysis (tick marks). Note that no statistically significant difference in survival was observed between the 2 groups of mice (P = .98). (D) Tcrb locus Southern blot analysis of leukemic cell gDNA from Cd3e−/− (nos. 71, 74, and 58) and Cd3e+/+ (nos. 77 and 80) TEL-JAK2 thymic tumors. DNA obtained from wild-type liver (Liv), spleen (Spl), and thymus (Thy) was used as control. GL indicates the germline configuration and asterisks point to the distinct Tcrb locus clonal rearrangements. (E) Thymus, spleen, and lymph node weights were plotted for Cd3e+/− (n = 12) and Cd3e−/− (n = 10) TEL-JAK2 transgenic mice that developed T-cell leukemia/lymphoma. *P < .05 (unpaired t test). Average organ weight is indicated by horizontal bars.
Pre-TCR expression cooperates with TEL-JAK2 to induce T-cell leukemogenesis
All Rag2-deficient TEL-JAK2 transgenic mice developed T-cell leukemia, but after longer latency than littermates (median survival time of 39 weeks for Rag2−/− versus 14 weeks for Rag2+/− littermates; Figure 3A). We reasoned that this increased latency could be due either (1) to the fact that post-DN3 cells are also targets for cellular transformation in TEL-JAK2-induced leukemogenesis, such cells being absent in Rag2−/− mice, or (2) to the fact that transformation of DN3 cells is assisted by pre-TCR signaling, which does not occur in Rag2−/− mice. To investigate further the role of pre-TCR in TEL-JAK2 leukemogenesis and to assess whether the presence of post-DN3 cells could affect leukemia latency, we bred EμSRα-TEL-JAK2 mice with pTα-deficient and therefore pre-TCR–deficient mice. Ptcra−/− mice present only a partial maturation arrest at the DN3 stage as evidenced by the presence of αβ-lineage DP thymocytes and a 10-fold increase in thymic cellularity as compared to Rag2-deficient mice.12,21 Except for one case of B-cell leukemia, all TEL-JAK2 transgenic mice lacking pTα developed T-cell leukemia with a similar latency as mice lacking Rag2 (median survival time of 39 weeks; Figure 3A). This result indicates that lack of pre-TCR signaling rather than reduced cellularity or lack of post-DN3 cells underlies the delay in T-cell transformation in Rag2-deficient mice as compared to pre-TCR-proficient TEL-JAK2 mice. We conclude from these experiments that pre-TCR expression cooperates with TEL-JAK2 to induce T-cell leukemogenesis.
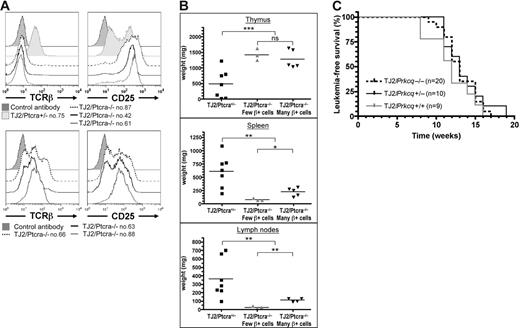
Similar to the original TEL-JAK2 leukemia,8 TEL-JAK2/Ptcra−/− tumors expressed CD8, in conjunction with CD4 or not, and high levels of CD25 (Figure 4A and data not shown). These cells expressed the TCRαβ complex at the cell surface at variable levels. Five cases presented 20% to 75% TCRβ+ cells (Figure 4A bottom panels and data not shown), whereas 3 presented only 4% to 5% (Figure 4A top panels). No correlation was found between TCRβ expression and leukemia onset, therefore indicating that endogenous TCRβ expression cannot compensate for the absence of pre-TCR to accelerate TEL-JAK2–induced leukemogenesis. The infiltration of secondary lymphoid organs by leukemic cells was reduced in EμSRα-TEL-JAK2/Ptcra−/− mice, as compared to Ptcra+/− littermates (Figure 4B). However, TEL-JAK2/Ptcra−/− leukemias containing a high percentage of TCRαβ-expressing cells were associated with statistically significant higher secondary lymphoid organ involvement than the other Ptcra−/− leukemias (Figure 4B). These results further support the notion that TCRαβ expression favors leukemic cell expansion in secondary lymphoid organs.
The pre-TCR cooperative role in TEL-JAK2 leukemogenesis cannot be replaced by TCRαβ expression and it does not require PKCθ. (A) Cell surface immunostaining of one TEL-JAK2/Ptcra+/− (no. 75) and 6 TEL-JAK2/Ptcra−/− (nos. 42, 61, 63, 66, 87, and 88) representative tumor cases using antibodies against CD25 and TCRβ or an isotypic control antibody. (B) Thymus, spleen, and lymph node weights were plotted for Ptcra+/− (n = 7) and Ptcra−/− TEL-JAK2 transgenic mice showing CD8+ T-cell leukemia containing a low (n = 3) or high (n = 5) percentage of TCRβ+ cells. NS indicates not significant; *P < .05; **P < .01; ***P < .001 (unpaired t test). Average organ weight is indicated by horizontal bars. (C) Kaplan-Meier leukemia-free survival curves for EμSRα-TEL-JAK2 transgenic mice deficient or not for Prkcq show no statistically significant differences. The number of mice in each group is given in parentheses.
The pre-TCR cooperative role in TEL-JAK2 leukemogenesis cannot be replaced by TCRαβ expression and it does not require PKCθ. (A) Cell surface immunostaining of one TEL-JAK2/Ptcra+/− (no. 75) and 6 TEL-JAK2/Ptcra−/− (nos. 42, 61, 63, 66, 87, and 88) representative tumor cases using antibodies against CD25 and TCRβ or an isotypic control antibody. (B) Thymus, spleen, and lymph node weights were plotted for Ptcra+/− (n = 7) and Ptcra−/− TEL-JAK2 transgenic mice showing CD8+ T-cell leukemia containing a low (n = 3) or high (n = 5) percentage of TCRβ+ cells. NS indicates not significant; *P < .05; **P < .01; ***P < .001 (unpaired t test). Average organ weight is indicated by horizontal bars. (C) Kaplan-Meier leukemia-free survival curves for EμSRα-TEL-JAK2 transgenic mice deficient or not for Prkcq show no statistically significant differences. The number of mice in each group is given in parentheses.
PKCθ does not contribute to TEL-JAK2–induced T-cell leukemia
A requirement for pre-TCR signaling in Notch3-induced murine T-cell leukemia has been recently reported.22 Genetic evidence indicated that PKCθ contributes to Notch3-induced T-cell leukemia and that PKCθ activation is dependent on pre-TCR signaling.23 To assess whether PKCθ activity could also be involved in TEL-JAK2–induced T-cell leukemia we bred EμSRα-TEL-JAK2 with Prkcq-deficient mice.13 Irrespective of the presence or absence of PKCθ, all transgenic mice developed fatal T-cell leukemia/lymphoma, affecting the thymus and peripheral lymphoid organs (median survival of 13 weeks; Figure 4C). These results indicate that PKCθ activity is not required for TEL-JAK2–induced leukemia and that other signaling components activated by the pre-TCR are required for the rapid induction of T-cell leukemia in TEL-JAK2 transgenic mice.
Transcriptome analysis of Rag2-deficient and Rag2-proficient TEL-JAK2 leukemias
To know to what extent the T-cell leukemia arising in Rag2−/− TEL-JAK2 mice is similar to TEL-JAK2 T-cell leukemia arising in a wild-type background, we compared their respective transcriptome using 12k Affymetrix U74Av2 oligonucleotide arrays. Three independent EμSRα-TEL-JAK2/Rag2−/− thymic lymphomas were compared to 8 EμSRα-TEL-JAK2 thymic lymphomas, 6 thymic lymphomas that developed in Ikaros-defective (IkL/L) mice,16 and thymocytes from 6 wild-type and 2 preleukemic IkL/L mice. IkL/L tumors are phenotypically similar to TEL-JAK2 tumors in that both develop with similar latency, affect the thymus and other lymphoid organs, their cells express CD8 and TCRβ with intermediate to high levels, and present clonal/oligoclonal TCRβ rearrangements.16 To classify our sample population based solely on their expression profiles, we performed an extensive unsupervised hierarchical clustering analysis. For this we compared a series of sample dendrograms obtained using different gene lists and clustering parameters (see “Supplemental materials and methods”). The gene lists were comprised of the top 36 to 2641 most varying probe sets across all 25 samples. The resulting sample topology was highly consistent across the different dendrograms. A representative dendrogram is shown in Figure 5 and illustrates 3 major groups of samples: a tight cluster of TEL-JAK2/Rag2−/− samples, which also grouped with the TEL-JAK2 leukemic samples (Figure 5, cluster C1), and apart from wild-type thymocytes and IkL/L tumors (Figure 5, clusters C2 and C3). This sample topology proved to be extremely robust using different clustering parameters and when confronted with data perturbation (average stability score = 0.99; see “Supplemental materials and methods” and Table S1). We conclude from these experiments that TEL-JAK2 and TEL-JAK2/Rag2−/− leukemias form a biologic entity clearly distinct from IkL/L leukemia and normal T cells.
Global gene expression and genomic analyses of pre-TCR–deficient and pre-TCR–proficient TEL-JAK2 T-cell leukemias. (A) Representative unsupervised hierarchical clustering of TEL-JAK2 (TJ, light-blue), TEL-JAK2/Rag2−/− (TJR, dark-blue), and IkarosL/L (T, yellow) tumors and wild-type (WT, pink) and preleukemic IkarosL/L (Ik, red) thymocytes using the top 5% most varying probe sets (n = 354) across all 25 samples, resulting in 3 major groups (C1, C2, and C3). Hues of red and blue indicate higher and lower levels of relative fluorescence signal, respectively. Samples and genes were clustered using the Ward linkage and 1-Pearson coefficient of correlation as the distance metric. Sample annotations indicating the tumor samples (T in black box) and nontumor (W in white box) samples are shown below the sample dendrogram. P values correspond to a global Fisher exact test. Gene clusters are labeled a to e. (B) Array-based CGH ideograms of 5 TEL-JAK2 (TJ2), 3 TEL-JAK2/Rag2−/− (TJ2/Rag2−/−), and 6 TEL-JAK2/Ptcra−/− (TJ2/Ptcra−/−) tumors. Chromosomes are listed in numerical order from left to right. Red squares above zero on the y-axis represent genomic gains (ratio > 1.2), whereas green squares below zero indicate losses (ratio < 0.8) relative to the diploid control. The yellow squares indicate the absence of numerical alterations relative to the diploid control. X chromosome profiles should be discounted because some lymphomas were derived from female mice and the reference DNA was obtained from a wild-type C57BL/6 male mouse.
Global gene expression and genomic analyses of pre-TCR–deficient and pre-TCR–proficient TEL-JAK2 T-cell leukemias. (A) Representative unsupervised hierarchical clustering of TEL-JAK2 (TJ, light-blue), TEL-JAK2/Rag2−/− (TJR, dark-blue), and IkarosL/L (T, yellow) tumors and wild-type (WT, pink) and preleukemic IkarosL/L (Ik, red) thymocytes using the top 5% most varying probe sets (n = 354) across all 25 samples, resulting in 3 major groups (C1, C2, and C3). Hues of red and blue indicate higher and lower levels of relative fluorescence signal, respectively. Samples and genes were clustered using the Ward linkage and 1-Pearson coefficient of correlation as the distance metric. Sample annotations indicating the tumor samples (T in black box) and nontumor (W in white box) samples are shown below the sample dendrogram. P values correspond to a global Fisher exact test. Gene clusters are labeled a to e. (B) Array-based CGH ideograms of 5 TEL-JAK2 (TJ2), 3 TEL-JAK2/Rag2−/− (TJ2/Rag2−/−), and 6 TEL-JAK2/Ptcra−/− (TJ2/Ptcra−/−) tumors. Chromosomes are listed in numerical order from left to right. Red squares above zero on the y-axis represent genomic gains (ratio > 1.2), whereas green squares below zero indicate losses (ratio < 0.8) relative to the diploid control. The yellow squares indicate the absence of numerical alterations relative to the diploid control. X chromosome profiles should be discounted because some lymphomas were derived from female mice and the reference DNA was obtained from a wild-type C57BL/6 male mouse.
Pre-TCR–deficient TEL-JAK2 leukemia displays frequent chromosomal alterations
To search for molecular differences that could correlate with the difference in leukemia latency between TEL-JAK2 and TEL-JAK2/Rag2−/− mice, we next performed a supervised analysis of the transcriptome of these 2 sets of samples (see “Supplemental materials and methods”). We identified 527 probe sets that significantly distinguish Rag2+/+ and Rag2−/− leukemias (P < .01; Table S2). These include genes known to be expressed after thymocyte β-selection (eg, Tcra, Gimap4, Cd6, Cd53, and Gimap1) and found to be down-regulated in TEL-JAK2/Rag2−/− leukemic cells. Among the 274 genes up-regulated in TEL-JAK2/Rag2−/− as compared to TEL-JAK2 leukemic cells, a significant enrichment of genes (51 of 274, P = 2.64 e−20 Fisher exact test) located on chromosome 15 was observed, including genes implicated in oncogenesis (eg, Myc, Nov, Angpt1, Hsf1, Mtdh, Bop1, and Xrcc3/Ku70).
To investigate whether TEL-JAK2/Rag2−/− tumors acquired specific genetic gains or losses, we performed array-based CGH. As depicted in Figure 5B, tumors from the parental TEL-JAK2 transgenic line rarely exhibited numerical chromosomal alterations, as these were detected in only one tumor out of 5, which presented a gain of chromosome 10. In contrast, all 3 TEL-JAK2/Rag2−/− tumors analyzed showed chromosomal gains, including a recurrent gain of chromosome 15. As shown in Figure 5B, TEL-JAK2/Ptcra−/− leukemias also acquired recurrent chromosomal alterations, including gain of chromosome 15 in 3 cases (nos. 63, 66, and 88) and a chromosome 4 deletion encompassing the Cdkn2a and Cdkn2b loci in 2 cases (nos. 66 and 87; Figure 5B). These results show that pre-TCR–deficient TEL-JAK2 leukemias are frequently associated with specific chromosomal alterations, suggesting that the selection for these aberrations may compensate for the absence of pre-TCR to facilitate the leukemogenic process.
Transgenic TCRαβ constitutive expression rescues the leukemia delay induced by pre-TCR deficiency
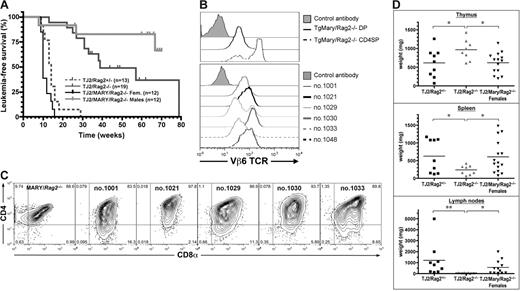
Previous studies have shown that transgenic TCRαβ overexpression can bypass the DN3 block induced by Rag1/2 deficiency, thus allowing the generation of DP and mature SP thymocytes.24 We thus analyzed the leukemogenic properties of TEL-JAK2 in mice in which the Rag2 deficiency was rescued by the expression of a transgenic TCRαβ complex. Thymocytes from the transgenic Marylin strain express an MHC class II-restricted TCRαβ (Vα1.1Vβ6) recognizing specifically the H-Y male antigen.14 When bred on a Rag2−/− genetic background, Marylin female mice present a high proportion of DN4 and normal numbers of DP thymocytes, an indication that the Rag2−/− DN3 block is bypassed. Furthermore, due to MHC class II restriction, these mice present only CD4 but not CD8 mature T cells.14 In contrast, male Marylin mice present few thymocytes due to H-Y–induced negative selection. EμSRα-TEL-JAK2/Marylin double-transgenic Rag2−/− female mice (hereafter named TMR) developed T-cell leukemia with short latency, similar to that observed for the original TEL-JAK2 transgenic line, thus indicating that the delayed disease onset linked to Rag2 deficiency was rescued under these conditions (Figure 6A). Male TMR mice rarely developed T-cell leukemia (Figure 6A). This is likely due to self-antigen–induced negative selection as female TMR leukemic cells undergo apoptosis when grafted into male mice (data not shown). TMR leukemic cells from female mice expressed the TCR-Vβ6 chain at the cell surface, at expression levels intermediate between those found in non-transformed Marylin Rag2−/− DP thymocytes and CD4SP mature thymocytes (Figure 6B). Remarkably, leukemic T cells from female TMR mice expressed CD8, in conjunction with CD4 or not, indicating that the MHC class II-restricted TCR-induced differentiation into the CD4 lineage was perturbed during leukemogenesis (Figure 6C). Additionally, expression of the Marylin TCRαβ transgene rescued the defective expansion of leukemic cells in the peripheral lymphoid organs caused by the Rag2 deficiency (Figure 6D), thus further supporting the notion that TCRαβ expression is critical for this property. Together, these data indicate that, in contrast to endogenous TCRαβ, a transgenic TCRαβ can replace the pre-TCR to cooperate with TEL-JAK2 in T-cell leukemogenesis.
Transgenic TCRαβ overexpression rescues the delay in T-cell leukemia development induced by Rag2 deficiency in TEL-JAK2 transgenic mice. (A) Kaplan-Meier leukemia-free survival curves for Rag2-deficient EμSRα-TEL-JAK2 Marylin TCR double-transgenic female (Fem) and male mice. The survival curves for EμSRα-TEL-JAK2 transgenic mice bred with Rag2-deficient mice (shown in Figure 3A) are depicted for comparison. The number of mice in each group is given in parentheses. Mice deceased from unrelated causes were censored in the analysis (tick marks). (B) Vβ6 TCR expression in representative Rag2-deficient EμSRα-TEL-JAK2 Marylin TCR double-transgenic females (bottom panel). As a control, Vβ6 TCR expression levels in CD4/CD8 double-positive (DP) and CD4 single-positive (CD4SP) thymocytes from Rag2-deficient Marylin transgenic female are shown (top panel). (C) CD4/CD8 costaining in representative Rag2-deficient EμSRα-TEL-JAK2 Marylin TCR double-transgenic females (nos. 1001, 1021, 1029, 1030, and 1033) and a Marylin/Rag2−/− female. (D) Thymus, spleen, and lymph node weights were determined for Rag2+/− (n = 9), Rag2−/− (n = 8), and Marylin/Rag2−/− (n = 14) TEL-JAK2 transgenic mice. *P < .05; **P < .01 (unpaired t test). Average organ weight is indicated by horizontal bars.
Transgenic TCRαβ overexpression rescues the delay in T-cell leukemia development induced by Rag2 deficiency in TEL-JAK2 transgenic mice. (A) Kaplan-Meier leukemia-free survival curves for Rag2-deficient EμSRα-TEL-JAK2 Marylin TCR double-transgenic female (Fem) and male mice. The survival curves for EμSRα-TEL-JAK2 transgenic mice bred with Rag2-deficient mice (shown in Figure 3A) are depicted for comparison. The number of mice in each group is given in parentheses. Mice deceased from unrelated causes were censored in the analysis (tick marks). (B) Vβ6 TCR expression in representative Rag2-deficient EμSRα-TEL-JAK2 Marylin TCR double-transgenic females (bottom panel). As a control, Vβ6 TCR expression levels in CD4/CD8 double-positive (DP) and CD4 single-positive (CD4SP) thymocytes from Rag2-deficient Marylin transgenic female are shown (top panel). (C) CD4/CD8 costaining in representative Rag2-deficient EμSRα-TEL-JAK2 Marylin TCR double-transgenic females (nos. 1001, 1021, 1029, 1030, and 1033) and a Marylin/Rag2−/− female. (D) Thymus, spleen, and lymph node weights were determined for Rag2+/− (n = 9), Rag2−/− (n = 8), and Marylin/Rag2−/− (n = 14) TEL-JAK2 transgenic mice. *P < .05; **P < .01 (unpaired t test). Average organ weight is indicated by horizontal bars.
TEL-JAK2 leukemic T cells express CD8-lineage marker genes
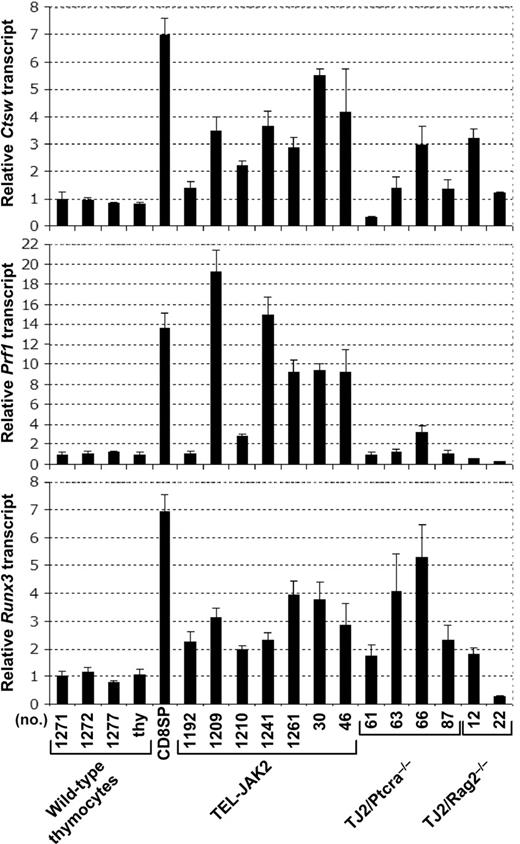
The CD8 expression observed in leukemic cells lacking TCR or pre-TCR (Figures 2B and 3D) or expressing the MHC class II-restricted Marylin TCR transgene (Figure 6C) suggests that TEL-JAK2 leukemic cells may partially differentiate into the CD8 lineage independently of TCR signaling. Expression of the Prf1, Ctsw, and Runx3 genes has been linked to DP thymocyte commitment into the CD8 lineage during MHC class I-mediated positive selection.25 To know whether CD8 expression in TEL-JAK2 leukemic cells reflects commitment to the CD8 lineage, we evaluated the expression status of Prf1, Ctsw, and Runx3 genes by quantitative reverse transcription-PCR (RT-PCR) analysis. As shown in Figure 7, these genes were expressed in TEL-JAK2 leukemic cells at levels higher than those found in thymocytes, but in most instances lower than those found in mature CD8 T cells. Furthermore, Ptcra−/− and Rag2−/− TEL-JAK2 tumors generally expressed Ctsw and Runx3, but were mostly negative for Prf1 expression (Figure 7). Together, these findings indicate that TEL-JAK2 leukemic cells express markers of cells committed to differentiation along the CD8 lineage in a partially TCR-independent manner.
Leukemic cells from TEL-JAK2 transgenic mice express CD8 lineage-associated genes. Quantitative RT-PCR analysis of Ctsw, Prf1, and Runx3 mRNA levels normalized for Hprt expression performed for 4 independent wild-type thymocyte samples, wild-type CD8 SP splenocytes, and thymic leukemic cells from the indicated transgenic TEL-JAK2, TEL-JAK2/Ptcra−/− and TEL-JAK2/Rag2−/− mice. Means and standard deviations were calculated as described in “Materials and methods.”
Leukemic cells from TEL-JAK2 transgenic mice express CD8 lineage-associated genes. Quantitative RT-PCR analysis of Ctsw, Prf1, and Runx3 mRNA levels normalized for Hprt expression performed for 4 independent wild-type thymocyte samples, wild-type CD8 SP splenocytes, and thymic leukemic cells from the indicated transgenic TEL-JAK2, TEL-JAK2/Ptcra−/− and TEL-JAK2/Rag2−/− mice. Means and standard deviations were calculated as described in “Materials and methods.”
Discussion
Using gene-targeted mouse strains exhibiting T-cell developmental defects, we demonstrate in the present report that (1) TEL-JAK2–induced T-cell leukemia develops from immature thymocytes, (2) the pre-TCR complex cooperates with TEL-JAK2 to induce rapid T-cell leukemia, (3) specific genetic alterations are acquired to compensate for the absence of pre-TCR, (4) endogenous TCRαβ signaling is not required for TEL-JAK2–induced leukemogenesis, and (5) TCRαβ expression induces leukemic cell expansion in secondary lymphoid organs.
Evidence that DN thymocytes can be efficient targets for TEL-JAK2 transformation is provided by 2 observations: (1) all Rag2−/− and Ptcra−/− TEL-JAK2 transgenic mice developed T-cell leukemia, and (2) the leukemias arising in these compound mice were similar to the original TEL-JAK2 leukemia, as demonstrated by cell surface and unsupervised gene expression profiling studies.
Thymocyte development is driven by the action of shifting combinations of transcription factors that drive new gene expression and developmental states.26 Differentiation from the DN1 to DN3 stages depends on c-Kit and common cytokine receptor γ chain (γc) stimulation. After rearranging their TCRβ locus at these stages, thymocytes expressing a functional pre-TCR complex lose their dependency on γc signaling to survive, proliferate, and differentiate.27 It has been shown that TEL-JAK2 signaling in cell lines can bypass cytokine signaling to induce survival and proliferation.4,6–8 More recently, we have found that TEL-JAK2 can transform to IL-7 independence an immature thymocyte CD4−CD8− DN cell line (J.G., unpublished observations, February 2004), which normally depends on IL-7 signaling to survive and proliferate in vitro.28 Taken together, these data indicate that the constitutive tyrosine kinase activity of TEL-JAK2 during the preleukemic phase of the disease likely perturbs the fine-tuned switch from γc to pre-TCR signaling to favor cell proliferation and the accumulation of additional pro-oncogenic mutations.
Pre-TCR signaling confers an advantage to TEL-JAK2–induced leukemogenesis, as demonstrated by the delayed leukemia onset in mice lacking an endogenous pre-TCR complex. The pre-TCR cooperative effect likely occurs early during the course of the disease since pTα surface expression was only found in a small percentage of the leukemic cells from terminally ill mice (data not shown). Array-based CGH analyses showed that pre-TCR–proficient TEL-JAK2 leukemia rarely displays chromosomal numerical abnormalities. In contrast, pre-TCR–deficient TEL-JAK2 leukemias frequently present chromosomal abnormalities, in particular a recurrent gain of chromosome 15. Supervised expression profiling analysis showed that over 50 genes located on chromosome 15 are up-regulated in TEL-JAK2/Rag2−/− leukemias as compared to pre-TCR–proficient TEL-JAK2 leukemias. These results suggest that there is a selective pressure for compensatory genetic alterations in the absence of pre-TCR. The requirement for such alterations to occur likely explains the delayed T-cell leukemogenesis in pre-TCR–deficient TEL-JAK2 mice. A role for pre-TCR in assisting T-cell leukemogenesis has also been observed for other T-ALL mouse models, namely, those driven by Notch activation,22,29 c-Myc overexpression,30 or Ikaros deficiency.31 In contrast, other T-cell leukemia mouse models (eg Trp53- or ATM-deficient mice) do not show such pre-TCR dependency.32,33 The delay in T-cell leukemia development in TEL-JAK2 transgenic mice lacking Rag2 is indistinguishable from that seen in the Ptcra−/− genetic background. Unlike Rag2−/− mice, which are completely devoid of post-DN3 cells, Ptcra−/− mice present αβ-lineage DP thymocytes.12,21 This indicates that lack of pre-TCR signaling rather than lack of post-DN3 cells hinders T-cell leukemia development. Pre-TCR signaling is associated with a proliferative burst of DN3 thymocytes accompanying differentiation into DN4 and DP stages,1 so it is plausible that the pre-TCR–assisted proliferation in TEL-JAK2 preleukemic cells increases the probability to acquire secondary oncogenic events ultimately leading to a clonal disease.
TEL-JAK2 leukemic T cells express CD8 and CD8 lineage-associated genes, even in the Rag2−/− or Cd3e−/− background. This finding suggests that circumvention of the DN3 developmental block occurs during leukemogenesis. Because to our knowledge JAK-STAT signaling is not able to replace pre-TCR signaling to induce thymocyte differentiation, we hypothesize that secondary alterations involving other signaling pathways are involved in the DN-to-DP transition. Several alterations impinging on proliferation or survival in mouse T-cell leukemias are known to bypass the DN3 block induced by Rag deficiency, such as Egr1, Notch3, Id1, or RasGRP1 overexpression22,34–36 or deficiency in p53, Ikaros, or E2A proteins.31,37,38 Further investigation should address the question whether alterations in these pathways occur during TEL-JAK2–induced T-cell leukemogenesis.
It has been proposed that Rag recombinase activity contributes to leukemia development in Atm−/− mice and in mice deficient in both Trp53 and TCRβ gene enhancer (Trp53−/−EβR/R) because their T-cell tumors present recurrent Rag-dependent chromosomal translocations involving the TCR loci that likely contribute to leukemogenesis.33,39 Our present data indicate that Rag activity does not play a major role in TEL-JAK2–induced T-cell leukemia because TEL-JAK2 mice lacking Rag2 are not less tumor-prone than TEL-JAK2 mice lacking Cd3e. Moreover, expression of a positively selecting H-Y TCR transgene on either a Rag2-deficient or a Rag2-proficient (data not shown) genetic background led to TEL-JAK2–induced leukemia with similar latency, thus demonstrating that Rag2 activity is not essential for this disease.
A role for persistent TCRαβ signaling in T-cell leukemia development is supported by the observation that expression of different TCRαβ transgenes accelerated T-cell lymphoma development in Trp53−/−,32 transgenic Stat5b,40 and transgenic Rasgrp134 mice. We did not observe an acceleration of TEL-JAK2-induced leukemia development by the sporadic expression of endogenously formed TCRαβ complexes in Ptcra−/− TEL-JAK2 transgenic mice or by H-Y TCRαβ expression on a Rag2-proficient background (data not shown). Leukemia onset was also not delayed by TCRα deficiency. Thus, although TEL-JAK2 leukemic cells express functional TCRαβ complexes at their cell surface, TCR signaling does not play a determinant role in leukemia development. However, in keeping with recent results obtained with other mouse models30,39,41 and with data showing that TEL-JAK2 leukemic cells are hypersensitive to TCR signaling, these results indicate that TCRαβ expression plays a role in leukemic cell expansion in peripheral lymphoid organs.
TEL-JAK2 leukemic cells express CD8 at the cell surface, independently of the presence of a pre-TCR or TCR. The relatively low levels of TCRβ and high levels of CD24 expression suggest that these cells may be arrested at the CD8+ ISP stage. Our results show, however, that TEL-JAK2 leukemic T cells resemble mature CD8 T cells with respect to the expression of genes normally associated with early CD8 lineage commitment,25 including Prf1, Ctsw, and Runx3. Interestingly, Ctsw and Runx3 are still expressed in Rag2-deficient TEL-JAK2 leukemic cells albeit at lower levels, indicating that expression of these genes is at least partially TCR-independent during the leukemogenic process. STAT5 overexpression or activation in transgenic T cells leads to an increase in CD8+ T-cell numbers.42–44 Conversely, mice deficient in both Stat5a and Stat5b present a severe reduction in CD8+ T cells.45,46 Recently, it has been reported that IL-2 receptor/STAT5 signaling contributes to establish a CD8 effector differentiation program following low-avidity TCR stimulation.47 These observations suggest that the STAT5 constitutive activation observed in TEL-JAK2 leukemic cells45,46 may have an impact on their differentiation phenotype.
Authorship
N.R.d.S. performed research, analyzed data, and wrote the paper; D.S.R. analyzed microarray data; A.d.R. analyzed microarray data; F.C. performed research and analyzed data; M.W. performed mouse genotyping; C.B. performed mouse array-CGH analysis; M.H.S. constructed the array and supervised the array-CGH study; and J.G. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jacques Ghysdael, Institut Curie, CNRS UMR146, Centre Universitaire, Bat 110, F-91405 Orsay, France; e-mail: jacques.ghysdael@curie.u-psud.fr.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported by funds from the Ligue Nationale contre le Cancer (Équipe labelisée Ligue and Programme Cartes d'Identité des Tumeurs [CIT]), Association for International Cancer Research (AICR; grant no. 03-029), Institut National du Cancer (INCA), Centre National de la Recherche Scientifique (CNRS), and Institut Curie. The construction of the mouse BAC-array was supported by grants from the CIT program of the Ligue Nationale Contre le Cancer.
N.R.d.S. is a recipient of a Marie Curie Fellowship from the European Community program “Improving the Human Research Potential and the Socio-Economic Knowledge Base” (no. HPMF-CT-2000-00481) and a fellowship from AICR.
We thank Yveline Bourgeois, Christophe Alberti, Josiane Ropers, Josiane Gaillard, and Noura Mebirouk for help with mouse husbandry; Nathalie Rocques for technical advice on Q-PCR; and Fabien Petel for help in data submission to EBI. We thank Philippe Kastner, Susan Chan, Emile van dan Akker, and Celio Pouponnot for critically reading the manuscript and Hind Medyouf and other Ghysdael laboratory members for helpful discussions.