Abstract
To delineate the relationship between epigenetic modifications and hemoglobin switching, we compared the pattern of histone acetylation and pol II binding across the β-globin locus at fetal and adult stages of human development. To make this comparison possible, we introduced an external control into experimental samples in chromatin immunoprecipitation (ChIP) assays. Using this common standard, we found that the locus control region (LCR) was acetylated to the same level at all stages, whereas acetylation levels at the individual gene regions correlated with the state of transcription. In the active genes, the promoters were less acetylated compared with the coding regions. Furthermore, all globin promoters were acetylated to a similar level irrespective of the state of transcription. However, after correction for the loss of nucleosomes, the level of acetylation per histone at the active γ and β promoters was 5- to 7-fold greater than that at the inactive ε promoter. Although the histone acetylation level within the LCR was developmentally stable, pol II binding in fetal erythroblasts was 2- to 3-fold greater than that in adult erythroblasts. These results demonstrate that dynamic changes in histone acetylation and pol II take place as the human β-globin gene region undergoes its developmental switches.
Introduction
The human β-globin locus consists of around 100 kb found on chromosome 11 and is composed of 5 functional genes: ε, Gγ, Aγ, δ, and β, which are arranged in the order of their expression during development. The locus also contains a locus control region (LCR) that consists of 5 DNase I hypersensitive sites (HS). The LCR is essential for physiologic level expression in mice, although it is not required for chromatin opening activities.1,2 The tissue-specific expression of the embryonic, fetal, and adult globin genes is developmentally regulated and the globin genes undergo 2 switches in expression during development. The ε-globin gene is expressed in embryonic development at the blood island of the yolk sac. At approximately 6 to 8 weeks of gestation, ε-globin is silenced, whereas the Gγ- and Aγ-globin genes are activated in the fetal liver. The second switch occurs late in gestation when the fetal γ-globin genes are progressively silenced, although β-globin is eventually expressed at high levels after birth.3 The exact mechanism by which these complex switches occur is not yet fully understood.
Chromatin epigenetic changes have long been thought to play a role in the expression of genes.4,5 With regard to the core histones, the effects of covalent modifications are 2-fold. First, modifications such as acetylation, methylation, and phosphorylation help change the access of trans-acting factors to the genetic elements found within the chromatin and affect the binding specificity of certain trans-factors.6 Second, these modifications can affect the physical property of chromatin, such as compactness, stability, and flexibility.7-9 Generally, histone acetylation makes the chromatin more flexible and helps it adopt an open conformation.10,11 This process may be mediated through weakening of key interactions in the nucleosomes.12 All these alterations can influence the readiness of a gene or a gene cluster for transcription.
The involvement of histone modifications on globin gene expression has been analyzed in several species. In chicken embryo erythrocytes, the whole β-globin locus is in a defined domain of increased histone acetylation, and neighboring regions are found to be hypoacetylated.13,14 In the murine locus, a similar situation exists with the LCR and active genes being heavily acetylated while the inactive genes are only mildly acetylated.15,16 In mouse erythroleukemia (MEL) cells containing a human chromosome 11, the LCR was not required for general H3 and H4 acetylation at the β-globin locus.17 In transgenic mice containing a human β-globin YAC, acetylation is enriched at the LCR and the active gene.18 It is noteworthy that the α-globin locus also seems to have histone acetylation and methylation changes during differentiation.19 Because of the difficulty in obtaining primary human erythroid cells, an erythroleukemia cell line K562, which expresses the ε- and γ-globin genes, is often chosen as a model for study of the role of histone epigenetic changes in human β-globin gene regulation.20,21
Lower eukaryotic model organisms, particularly yeast, have greatly facilitated histone modification studies. Recently, most patterns and profiles regarding the relationship between gene activation/repression and histone modifications are based on studies in yeast.22 Although many established rules based on studies in lower eukaryotes seem universal, some of them are unlikely to be extrapolated to higher eukaryotes. For instance, dimethyl histone H3 K4 is an epigenetic mark for permissive state of transcription and is evenly distributed from the 5′- to 3′-transcribed regions in yeast.23 However, it was found in eight active genes in chicken erythroblasts that histone H3 K4 was heavily dimethylated in the 5′ transcribed region, whereas the 3′ region contained no or much less dimethylated histone H3 K4.24 A recent comprehensive study on the β-globin locus in K562 cells carried out by Kim et al20 highlighted the consistencies and differences between yeast and higher eukaryotes regarding the mono-, di-, and tri-methyl patterns at histone H3 K4, K9, and K36. Studies on higher eukaryotes involve additional difficulties, because in many cases, certain types of primary cells are not available. When transformed cell lines are used as a substitute, the results may or may not be a faithful representation of the in vivo events. For instance, trimethyl histone K4 is a mark for active transcription.25 An elevated level of this modification was measured in the highly transcribed γ gene, but not in the ε gene, which was also expressed, albeit at a lower rate, in K562 cells.20 In contrast, the ε gene was heavily trimethylated at histone H3 K4 in chicken primary embryonic erythroid cells.13 Although much work has been carried out on the β-globin locus and the involvement of histone acetylation in its regulation, no study has looked at human globin loci that undergo normal hemoglobin gene switching. We wanted to address the role of histone acetylation in the expression of the human β-globin locus in the natural setting of human erythroblasts. We used chromatin immunoprecipitation (ChIP), with different developmental stages of fetal and adult erythroblasts, against histone H3 acetylation to determine whether any changes were found between the 2 expression stages. To circumvent potential differences of histone acetylation in a reference gene in 2 different types of cells, we introduced an external control into experimental samples before ChIP assays. Using this common standard, we found that the LCR was acetylated to similar levels at all developmental stages, whereas the acetylation level in the globin regions correlated with the state of transcription. Our study indicated that acetylation per histone, instead of apparent histone acetylation level, was a better indication for correlation of gene activity and histone modifications. It remains to be determined whether these changes are causative for globin gene switching.
Materials and methods
Collection and culturing of cells
Human fetal liver samples were obtained from abortuses. The studies of fetal samples were carried out according to procedures approved by the Human Experimentation Committee at the University of Washington and after maternal consent. Cell suspensions were prepared by homogenization of the fetal liver in RPMI 1640 medium (Mediatech, Herndon, VA) using a Dounce homogenizer. At the time points used in this study, the majority of nucleated cells within the fetal liver are of the erythroid lineage. Adult CD34+ cells were obtained from peripheral blood and cultured in a 2-phase system for approximately 21 days.26 Cells were then processed for ChIP. K562 cells were maintained in RPMI 1640 medium containing 10% fetal calf serum at 37°C in 5% CO2. For induction, hemin was added to a final concentration of 50 μM to cells in logarithmic phase. Induced cells were harvested after 3 days for ChIP assay.
ChIP
Chromatin immunoprecipitation was carried out as described previously with minor modifications.27 Normal rabbit IgG(12-370), anti-acetylated histone H3K9, 14 (06-599) antibodies were obtained from Upstate (Charlottesville, VA). Pol II (sc-9001x) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-histone H3, H4, H2A, and H2B (ab1791, ab31827, ab18255, and ab1790) antibodies were obtained from Abcam (Cambridge, MA). Immunoprecipitations (IPs) were performed at least 3 times. In the later phase of this study, we added cells from the same batch of MEL cells into cell suspensions of fetal liver and adult erythroblasts simultaneously, and then processed the samples to fixation as described above. MEL cells were used as an external control against differences in acetylation of human control sequences at different developmental time points.
Quantitative PCR
Real-time quantitative polymerase chain reaction (PCR) was performed on the immunoprecipitated samples using the Opticon 2 (MJ Research, Watertown, MA) or LightCycler (Roche, Indianapolis, IN) PCR machines. PCR reactions were performed using SYBR green master mix according to the manufactors instructions (QIAGEN, Valencia, CA). Primer pairs were designed using Primer 3 software. Table S1 lists sequences of all the primers used in this study (available on the Blood website; see the Supplemental Materials link at the top of the online article). All data were expressed as a ratio of the PCR readings of a given primer set in IP and input DNAs over an IP/input ratio of a control gene, and the standard deviation was calculated.
RNase protection
RNA was purified from single cell suspensions of K562, 105-day-postcoitum (dpc) fetal livers or cultured adult erythroblasts using the Total RNA Isolation system (Promega, Madison, WI). Human globin mRNAs were quantified by RNase protection analysis using the RPA II kit (Ambion, Austin TX). RNA probes were synthesized using the MAXIscript T7 RNA polymerase transcription kit (Ambion). Template DNAs used to prepare probes to measure human α-, ε-, γ-, and β-globin mRNA were: pSp6Hα, pT7Hε, pT7Hγ, and pT7Hβ, respectively. Signals were quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Results
General profiles of gene expression and histone H3 acetylation within the β-globin gene cluster in human primary fetal and adult erythroblasts
RNase protection assays were performed to measure the levels of gene expression in primary fetal liver and adult erythroblasts. In approximately 100-dpc fetal liver samples, the predominant form of globin RNA was that of γ-globin, with a ratio of 90% of total β-like globin. In this stage of development, expression of the ε gene is halted; however, a trace amount of ε mRNA (<1%) was detectable (Figure 1A). In adult erythroblasts, β-globin mRNA was the predominant form (75%), whereas γ-globin contributed approximately 25% of total β-like globin mRNA. ε mRNA was undetectable at this stage (Figure 1B). To assess abundance of globin mRNA in primary erythroid cells, RNase protections were carried out at 3 different amounts of total RNA, and intensity of the protected bands were compared with those in induced K562 cells (Figure 1C). From this comparison, we estimated that globin mRNA in fetal or adult primary erythroid cells was 10- to 20-fold more abundant than that in K562 cells.
Profiles of globin gene expression and histone H3 acetylation in fetal and adult erythroblasts. The expression profiles of the human globin genes (A-C) were measured by RNase protections assays. Total RNA was prepared from several fetal liver samples and adult erythroblasts as well as K562 cells. Probes for RNase protections were human α-globin, ε-globin, γ-globin, and human β-globin samples were 105d FL (A), adult erythroblasts (B), and K562 (C). The amount of RNA used in each reaction is shown at the top of the panels. Quantification was carried out using a PhosphorImager (GE Healthcare, Chalfont St. Giles, United Kingdom). Histone acetylation profiles within the β-globin locus are shown in D (fetal stage) and E (adult stage). Each data point represents enrichment of crosslinked chromatin immunoprecipitated using antibodies against histone H3 acetylation. The y-axis values measure relative enrichments relative to the endogenous amylase gene. The results of 3 to 9 experiments are shown (± SEM). The x-axis is the coordinate of the human β-globin locus with the cap site of the ε-globin gene being set at 1. The human β-globin locus is represented above the graphs. Open box, ε-globin gene; gray boxes, Gγ- and Aγ-globin genes; closed boxes, δ- and β-globin genes; striped boxes, olfactory receptor genes. Gray arrowheads on the x-axis mark the positions of the globin gene promoters. Vertical arrows indicate DNase I hypersensitive sites.
Profiles of globin gene expression and histone H3 acetylation in fetal and adult erythroblasts. The expression profiles of the human globin genes (A-C) were measured by RNase protections assays. Total RNA was prepared from several fetal liver samples and adult erythroblasts as well as K562 cells. Probes for RNase protections were human α-globin, ε-globin, γ-globin, and human β-globin samples were 105d FL (A), adult erythroblasts (B), and K562 (C). The amount of RNA used in each reaction is shown at the top of the panels. Quantification was carried out using a PhosphorImager (GE Healthcare, Chalfont St. Giles, United Kingdom). Histone acetylation profiles within the β-globin locus are shown in D (fetal stage) and E (adult stage). Each data point represents enrichment of crosslinked chromatin immunoprecipitated using antibodies against histone H3 acetylation. The y-axis values measure relative enrichments relative to the endogenous amylase gene. The results of 3 to 9 experiments are shown (± SEM). The x-axis is the coordinate of the human β-globin locus with the cap site of the ε-globin gene being set at 1. The human β-globin locus is represented above the graphs. Open box, ε-globin gene; gray boxes, Gγ- and Aγ-globin genes; closed boxes, δ- and β-globin genes; striped boxes, olfactory receptor genes. Gray arrowheads on the x-axis mark the positions of the globin gene promoters. Vertical arrows indicate DNase I hypersensitive sites.
To measure the level of histone acetylation across the human β-globin region ChIP was carried out on fetal and adult erythroblasts. Enrichments of acetylated histone H3 were expressed as an increase at a given globin primer set compared with the endogenous control amylase sequence. Histone H3 acetylation across approximately 100 kb of the globin locus is profiled in fetal liver (∼100 dpc; Figure 1D). In all ChIP assays, nonspecific binding, which was measured in each experiment using the normal IgG antibody, was less 5% of the control gene (Figure S2). At the 5′ olfactory receptor gene (which is not expressed in this tissue15 ), the acetylation level was near the background level. In the LCR region (−20 to −6 kb), the histone H3 acetylation was enriched by approximately 10-fold. The lack or low level of acetylation at the ε-globin region was consistent with the fact that this gene was silenced in the fetal livers we studied. When the acetylation was analyzed at the γ-globin genes, histone H3 acetylation was found at high levels (∼10- to 25-fold). The acetylation enrichment spread beyond the γ-globin gene in both directions. It is noteworthy that the acetylation at the γ promoter was the lowest within the whole gene region. The acetylation outside the γ region was slightly higher than the background. At the δ- and β-globin promoters, the acetylation was approximately 3-fold enriched. However, the acetylation within exon 1 of the β (and δ) gene was approximately 10-fold. The 3′HS1 site showed approximately 3-fold enrichment.
In adult erythrocytes, different gene expression occurs compared with the fetal stage: hemoglobin switching has occurred. To determine whether this switch in expression had any effect on acetylation, we analyzed the β-globin locus in adult erythroblasts derived from CD34+ cells in a 2-phase culture system. In Figure 1E, the H3 histone acetylation pattern is shown. As at the fetal stage, the LCR showed enrichment for acetylation of around 10-fold. The ε-globin was still found at background levels. At the γ-globin genes, there was a significant change in the adult stage: the γ-globin genes were less than 5-fold enriched and the promoter was only approximately 3-fold enriched (compared with 10- to 25-fold in the fetal samples). The δ- and β-globin genes were heavily enriched in the adult stage, with approximately 20 to 25-fold enrichment of the exons, whereas the promoters were enriched only to 2- to 4-fold.
Comparison of the levels of histone acetylation in the 2 developmental stages
The acetylation levels between fetal and adult erythroblasts depicted above cannot be directly compared because it is unknown whether the acetylation level of the endogenous amylase gene, which served as the control, remains unchanged in the 2 different stages of development. To address this question, MEL cells were added to human erythroblasts before the ChIP process. Because the same batch of MEL cells were used in mixtures of MEL/fetal and MEL/adult erythroblasts, the levels of histone acetylation of a mouse gene in the 2 mixtures should be identical. Thus, using an external mouse gene as a control, acetylation levels of a human gene in the 2 different types of human erythroblasts can be directly compared.
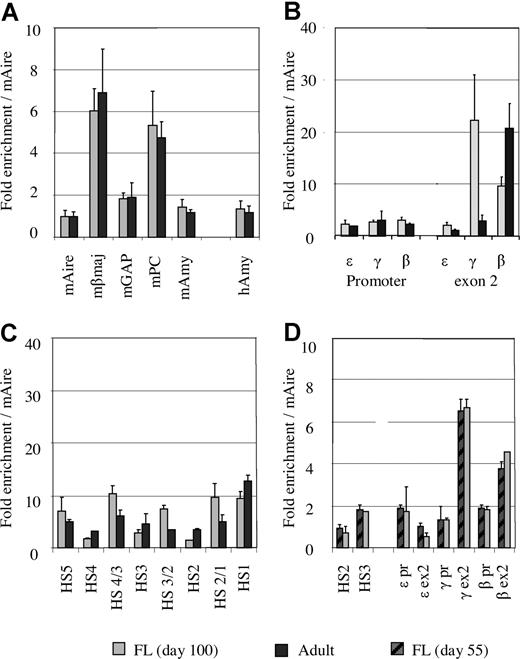
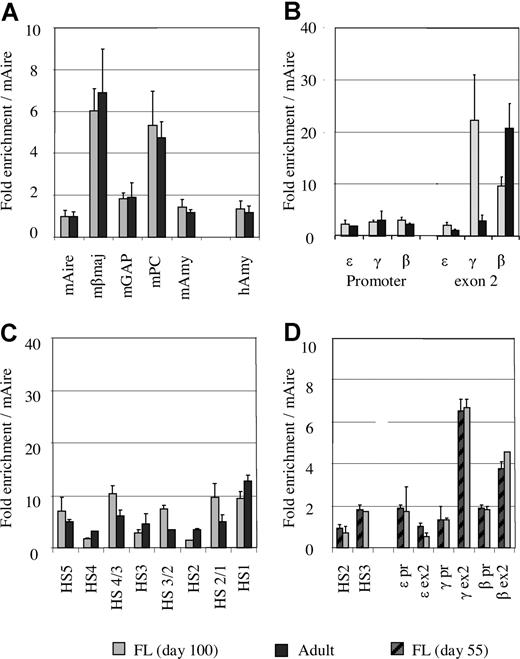
To validate this approach, we measured acetylation levels at 5 mouse genes: Aire (autoimmune regulator gene), βmaj, glyceraldehyde-3-phosphate dehydrogenase, pyruvate carboxylase, and amylase. As expected, the acetylation level of each the mouse gene in the 2 mixtures (MEL/human fetal erythroblasts and MEL/adult erythroblasts) was identical (Figure 2A). When the mAire gene is used as a reference, the acetylation level of human amylase was 1.4 (± 0.37) in fetal erythroblasts and 1.2 (± 0.35) in adult erythroblasts, suggesting that the acetylation level at the human amylase gene is slightly different at the 2 developmental stages.
The levels of histone acetylation corrected by an external mouse reference gene. (A) Comparison of acetylation levels of the mouse genes between the mixtures of MEL/fetal liver and MEL/adult erythroblasts. (B) Comparison of histone acetylation levels between fetal and adult erythroblasts at the promoters and exon 2 of the ε-, γ-, and β-globin genes using the mouse Aire gene as an external control. (C) Comparison of histone acetylation levels between fetal liver (∼day 100) and adult erythroblasts in the LCR region. (D) Comparison of histone acetylation levels between day 55 and day 100 fetal livers using the mouse Aire gene as control.
The levels of histone acetylation corrected by an external mouse reference gene. (A) Comparison of acetylation levels of the mouse genes between the mixtures of MEL/fetal liver and MEL/adult erythroblasts. (B) Comparison of histone acetylation levels between fetal and adult erythroblasts at the promoters and exon 2 of the ε-, γ-, and β-globin genes using the mouse Aire gene as an external control. (C) Comparison of histone acetylation levels between fetal liver (∼day 100) and adult erythroblasts in the LCR region. (D) Comparison of histone acetylation levels between day 55 and day 100 fetal livers using the mouse Aire gene as control.
Having introduced the external mouse control, direct comparison of the acetylation levels in fetal and adult erythroblasts in the human globin locus becomes possible. Figure 2B shows the comparison of acetylation levels of the ε-, γ-, and β-globin genes based on the mouse Aire gene in fetal and adult erythroblasts. It is noteworthy that the levels of histone acetylation at the promoters of the 3 genes were comparable regardless of the state of gene transcription. As shown in Figure 1A,B, expression profiles of the globin genes in the 2 types of erythroblasts were distinct. In fetal erythroid cells, the majority of globin mRNA was transcribed from the γ gene (90%), and a small amount was from the β gene (10%). Adult erythroblasts predominantly expressed the β gene, whereas the γ gene was transcribed at a low level. ε-Globin mRNA was barely detectable in either fetal or adult erythroblasts. Despite such differences, histone acetylation at all the promoters was maintained at similar levels in fetal and adult erythroblasts (Figure 2B).
By contrast, the levels of histone acetylation in the exon regions were associated with the state of transcription of the globin genes (Figure 2B). Exon 2 of the ε-globin gene, which is not expressed in fetal or adult erythroblasts, was poorly acetylated. The γ gene exon 2 was heavily acetylated in fetal erythroid cells, which correlated to the high level of expression of the gene at this stage. When transcription of the γ gene declined in adult erythroid cells, the acetylation level in γ exon 2 was decreased. For the β-globin gene, a reciprocal trend was observed (ie, the β exon 2 region was heavily acetylated in adult erythroid cells and acetylated to a lesser extent in fetal cells).
Figure 2C shows the comparison of histone acetylation in the LCR region of fetal and adult erythroblasts. In both types of erythroblasts, the LCR region was acetylated to a similar level, suggesting that histone acetylation in the LCR is developmentally stable.
We analyzed fetal erythroid samples for histone acetylation at 2 time points: days 55 and approximately 100. Nine regions in the locus were selected for this assessment. As shown in Figure 2D, the levels of histone acetylation of fetal liver were undistinguishable in all the measured regions in the 2 developmental ages, suggesting that the pattern and level of histone H3 acetylation remains unchanged when the fetus grows from day 55 to day 100.
Taken together, these results indicate that the acetylation pattern and level in the LCR are stable during development; the promoters of the globin genes are always lightly acetylated in erythroid cells irrespective of their state of transcription, whereas histone acetylation in the gene coding regions is correlated with the state of transcription.
Acetylation per histone at the promoters correlated with gene activity
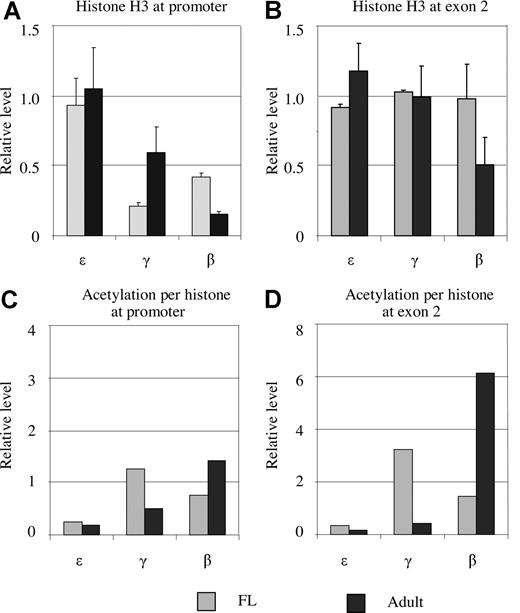
The low levels of acetylated histone H3 observed at the activated globin promoters could reflect the real situation or could be due to the loss of histone H3 at the regions. To address this question, we assessed the amount of histone H3 in the globin promoters using an antibody against both acetylated and nonacetylated forms. The amount of histone H3 bound at the ε gene promoter was equal in fetal and adult erythroblasts when corrected to mAire (Figure 3A). The γ gene promoter bound approximately 20% and 60% of histone H3 compared with the amount of H3 at the ε gene promoter in fetal and adult erythroblasts, respectively. The levels of histone H3 at the β gene promoter corresponded to 40% and 16% of those in the ε gene promoter in the fetal and adult stages, respectively. We repeated the experiments using antibodies against histone H2A, H2B, or H4, and found the histone H2A/H2B/H3/H4 ratio to be 1:1:1:1 (Figure S1). These results indicate that the γ and β gene promoter sequences lose contact with histones when they are activated.
Histone H3 occupancy at the promoters and exon 2 of the ε-, γ-, and β-globin genes in fetal and adult erythroblasts. (A) Histone H3 occupancy at the ε-, γ-, and β-globin gene promoters. The y-axis is the H3 amount relative to the external mouse Aire gene. The ε gene promoter contains the same amount of H3 as the Aire gene. In comparison with the ε promoter, the occupancy of H3 at the γ and β gene promoters is reduced in fetal and adult erythroblasts. (B) H3 occupancy in exon 2 of the ε-, γ-, and β-globin genes in fetal and adult erythroblasts. Exon 2 of the γ and β genes has the same amount of histone H3 as that at ε gene at both stages except β exon 2 in adult erythroblasts. (C,D) Acetylation levels per histone H3 between fetal and adult erythroblasts at the promoters and exons.
Histone H3 occupancy at the promoters and exon 2 of the ε-, γ-, and β-globin genes in fetal and adult erythroblasts. (A) Histone H3 occupancy at the ε-, γ-, and β-globin gene promoters. The y-axis is the H3 amount relative to the external mouse Aire gene. The ε gene promoter contains the same amount of H3 as the Aire gene. In comparison with the ε promoter, the occupancy of H3 at the γ and β gene promoters is reduced in fetal and adult erythroblasts. (B) H3 occupancy in exon 2 of the ε-, γ-, and β-globin genes in fetal and adult erythroblasts. Exon 2 of the γ and β genes has the same amount of histone H3 as that at ε gene at both stages except β exon 2 in adult erythroblasts. (C,D) Acetylation levels per histone H3 between fetal and adult erythroblasts at the promoters and exons.
Having established the levels of both acetylated and total histone H3, we calculated the acetylation level per histone H3 (the degree of acetylation). The degree of acetylation at the ε gene promoter was approximately 2-fold higher than that at the mAire gene in fetal and adult erythroblasts (Figure 3C). At the γ gene promoter, acetylation levels per histone H3 in fetal and adult erythroblasts were 12- and 5-fold higher, respectively, than those at mAire. All the differences were statistically significant (P < .05). For the β gene promoter, the corresponding acetylation degrees were 7- and 15-fold higher than the reference gene in fetal and adult erythroblasts, respectively. Thus, gene activation is associated with a higher level of acetylation per histone, instead of the total acetylation level, which is affected by the loss of histones from the promoter areas.
As described above, when a globin gene was actively transcribed, the exon regions of the gene were heavily acetylated. Different from the promoter regions, total histone H3 in the exon 2 regions of the ε, γ, and β genes remained at similar levels in both fetal and adult erythroid cells with the exception of β exon 2 at the adult stage (Figure 3B). The estimated acetylation density in exon 2 of the ε gene was low in both fetal and adult erythroblasts, which was at a similar level as that at the ε promoter. Acetylation density in exon 2 region of the actively transcribed genes was found at a high level: it was 30-fold higher than that in mAire at γ exon 2 in fetal liver and 60-fold higher at the β exon 2 in adult erythroid cells (Figure 3D).
In summary, these results suggest that gene activation led to the loss of histone H3 at the promoters. As a result, apparent acetylation levels at the active promoters seem low; however, each of the remaining histone H3 at the promoters is heavily acetylated.
Pol II binding changes during development
To determine whether stage-specific changes in pol II binding occur, ChIP was carried out on fetal liver and adult erythroblasts (Figure 4). In approximately 100-dpc fetal samples, pol II was enriched 8-fold at the HS 3 and 4 core regions, and 2- to 4-fold at HS 1 and 2; the HS 5 site was not substantially enriched. Pol II was barely recruited to sequences between the HS cores. There was no continuous spread of pol II from the LCR region toward the ε-globin gene (Figure 4A). These results suggest that only the binding motifs in the HS 2, 3, and 4 cores are responsible for pol II recruitment. A similar pattern of pol II recruitment remained in adult erythroblasts (ie, the highest level of recruitment occurred in the HS 2, 3, and 4 cores with lower levels of recruitment in the intervening regions). However, a marked difference between the 2 stages was that the levels of pol II recruitment at the HS cores in adult erythroblasts were reduced to between one half to one third of that in fetal liver.
Comparison of pol II recruitment at the human β-globin locus between fetal and adult erythroblasts. (A) Comparison of pol II recruitment in the LCR between fetal and adult erythroblasts. Each data point represents enrichment relative to the external mouse Aire gene of cross-linked chromatin from approximately 100 dpc fetal liver (▒) or adult erythroblasts (■), immunoprecipitated using antibodies against Pol II. The y-axis values measure relative enrichments. Notice that the HS cores of the LCR in fetal erythroblasts bind pol II 2 to 3 times more than in adult erythroblasts, whereas the intervening regions between the HS sites bind similar amounts of pol II at the fetal and adult stages. (B) Comparison of pol II recruitment in fetal and adult erythroblasts at the promoters and exons of the ε-, γ-, and β-globin genes.
Comparison of pol II recruitment at the human β-globin locus between fetal and adult erythroblasts. (A) Comparison of pol II recruitment in the LCR between fetal and adult erythroblasts. Each data point represents enrichment relative to the external mouse Aire gene of cross-linked chromatin from approximately 100 dpc fetal liver (▒) or adult erythroblasts (■), immunoprecipitated using antibodies against Pol II. The y-axis values measure relative enrichments. Notice that the HS cores of the LCR in fetal erythroblasts bind pol II 2 to 3 times more than in adult erythroblasts, whereas the intervening regions between the HS sites bind similar amounts of pol II at the fetal and adult stages. (B) Comparison of pol II recruitment in fetal and adult erythroblasts at the promoters and exons of the ε-, γ-, and β-globin genes.
Pol II was enriched approximately 20-fold at the γ-globin promoter, and it was at a lower level in the γ-globin exon 2 region in fetal liver; at this stage there was moderate pol II recruitment to the β promoter and exon 2 (Figure 4B). In adult erythroblasts, the β gene promoter was heavily enriched for pol II. It is noteworthy that pol II was enriched to an extremely high level in the β exon 2 region (40-fold). The ε gene promoter did not bind any significant amount of pol II at the fetal and adult stages of development.
Comparison of histone acetylation in K562 cells and fetal erythroblasts
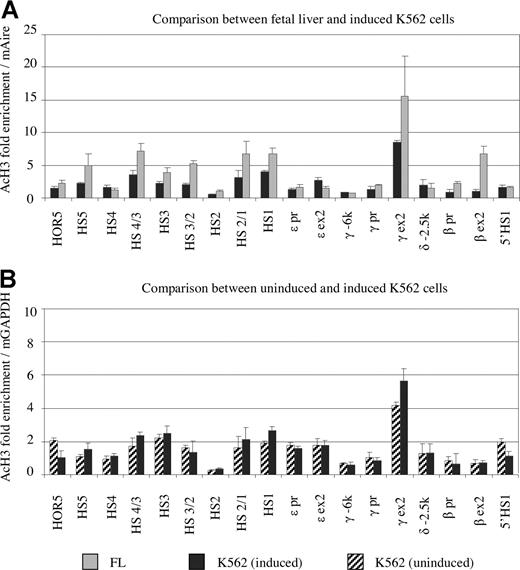
It is impossible to collect human primary embryonic erythroid cells. Instead, we used K562 erythroleukemia cell line as ε gene-expressing cells. In K562 cells, ε mRNA and γ mRNA account for 25% and 75% of the total β-like globin mRNA, respectively, and the β-globin gene is not expressed (Figure 1C). Hemin induction increased total amount of globin mRNA by approximately 2.5-fold, whereas the relative ratio between ε and γ genes remained unchanged. The abundance of globin mRNA between primary erythroid cells and erythroleukemia cells was significantly different. Production of globin mRNA in K562 cells (and MEL cells, data not shown) was 10- to 20-fold lower than that in primary erythroid cells (Figure 1C). It is believed that erythroleukemia cell lines are useful for evaluation of the correlation between histone acetylation and globin gene expression, but they are not suitable for study of the mechanism of hemoglobin switching.
Figure 5A shows the acetylation profiles of the β-globin cluster in K562 cells along with fetal liver. To make the comparison possible, ChIP enrichment was calculated on the basis of the mouse amylase gene in MEL cells, which were added before the ChIP assay. Acetylated histone H3 was enriched 2- to 3-fold in HS 1, 3, and 4 of the LCR with comparison to the reference gene. Histone acetylation was slightly enriched in the promoters of the ε- and γ-globin gene regions, which were active in K562 cells. The exon regions of these 2 genes were acetylated to a higher level than that in the promoter regions. The general profile of acetylation in K562 cells was similar to that in fetal primary erythroid cells except the β gene region. A notable difference between K562 and fetal liver was that the magnitude of the acetylation level in K562 cells on average is 2-fold lower than that in primary fetal liver cells.
Comparison of histone acetylation profiles of K562 and fetal liver cells. (A) Comparison of histone acetylation in fetal liver (∼day 100; ▒) and hemin-induced K562 cells (■). Notice that acetylation level of histone H3 in K562 cells generally is lower than that in primary fetal erythroid cells when using a common reference gene (mAmy). (B) Comparison of histone acetylation between uninduced (hatched) and induced (black) K562 cells. The levels of histone acetylation are unchanged upon hemin induction, except that exon 2 of the γ gene is acetylated to a slightly higher level after induction.
Comparison of histone acetylation profiles of K562 and fetal liver cells. (A) Comparison of histone acetylation in fetal liver (∼day 100; ▒) and hemin-induced K562 cells (■). Notice that acetylation level of histone H3 in K562 cells generally is lower than that in primary fetal erythroid cells when using a common reference gene (mAmy). (B) Comparison of histone acetylation between uninduced (hatched) and induced (black) K562 cells. The levels of histone acetylation are unchanged upon hemin induction, except that exon 2 of the γ gene is acetylated to a slightly higher level after induction.
Hemin increases globin gene expression in K562 cells. The mechanism of the induction remains to be determined. We sought to determine whether this increase of globin gene expression due to hemin was associated with a change in histone acetylation. Figure 5B shows that the pattern and magnitude of histone H3 acetylation within the entire β-globin cluster maintained unchanged before and after hemin induction with the exemption of a slight increase in the γ exon 2 region. These results suggest that the hemin-induced globin gene activation in K562 cells involves a mechanism other than histone acetylation.
Discussion
In this study, we compared the histone acetylation profiles of the β-globin cluster between the fetal and adult stages of development in human primary erythroblasts. To make this comparison possible, we introduced an external control into experimental samples in ChIP assays. Thus, a common standard could be used for comparisons of different types of cells. Using this approach, we found that the LCR was acetylated to the same level in both fetal and adult erythroblasts, whereas the acetylation in the globin gene regions correlated to the state of transcription. Our studies also demonstrated that the promoter in an active globin gene was acetylated to a lower level compared with the coding region. Moreover, all promoters, irrespective of the state of transcription, were acetylated to the same steady-state level in both fetal and adult primary erythroblasts. Thus, in fetal or adult erythroblasts, the acetylation level at the ε-globin gene promoter was similar to that at the γ- and β-globin gene promoters. This phenomenon is contrary to the general perception that the promoter of an active gene is invariably associated with an elevated level of histone acetylation.28 However, further studies indicated that this discrepancy can be reconciled by the fact that nucleosomes at the active promoter are depleted to a certain extent. The occupancy of histone H3 (and H2A, H2B, and H4) in the γ and β gene promoters was approximately 80% less than that at the ε gene in fetal and adult erythroblasts, respectively. Therefore, with regard to acetylation levels per histone, the active γ and β promoters are indeed acetylated 5- to 7-fold higher than the inactive ε promoter. It has been shown that histones at the yeast PHO5 promoter are first hyperacetylated and then lose contact with nucleosomes in the activated promoter.29 The loss of nucleosomes at active promoters is likely to be a genome-wide phenomenon in yeast.30 Our results suggest that this perspective could be extended to the human globin gene cluster. It has been proposed that during gene activation, a dynamic change occurs at the promoter chromatin.31 Enhanced histone acetylation might be a trigger leading to the eviction of nucleosomes at active promoters.32 Again, all these results indicate that it is better to use modification per histone, instead of level of modifications, in studying the biologic role of histone modifications in vivo.
We noticed that our results were contradictory to those of 2 previous studies in baboon and human primary erythroid cells.33,34 The 2 groups reported that the levels of histone H3 acetylation at the globin promoters were correlated to gene activation (ie, the γ gene promoter was heavily acetylated at the fetal stage of development, a low level of acetylation was measured at the β gene promoter, and a reciprocal correlation was observed in adult erythroid cells). Our results demonstrate that excellent correlation between gene activation and hyperacetylation occurs only in the exon regions. Although what caused the differences between these studies remains unknown, the discrepancy might be derived from the different methods used in the calculation of amplicons enriched by specific antibodies.
Histone modifications that are associated with gene activity are not homogeneously distributed along promoter and coding regions. Several genome wide studies in yeast showed that acetylated histone H3 is enriched at promoter sequences compared with coding regions.22,35 The lower acetylation in the coding regions is proposed to be a mechanism to prevent illegitimate initiation of transcription beyond promoters.36 In contrast, our results showed that after correcting for the occupancy of histone H3, the coding region of the γ gene was acetylated 2-fold higher than of the γ promoter in fetal erythroblasts, and the acetylation level of exon 2 of the β gene was 5-fold higher than that of the β promoter in adult erythroblasts. It is possible that the elevated acetylation in transcribing regions facilitates dissociation of the nucleosomes in front of pol II. After passage of pol II, a normal nucleosome would reassociate.8 We speculate that the nucleosome density would be normal in the coding region of a gene with a low transcription rate, whereas the coding region of a gene with a high transcription rate would have a lower nucleosome density. This hypothesis is supported by the fact that the β-globin gene is transcribed at a rate 3- to 4-fold higher than that of the γ gene in transgenic mice.37 On the other hand, the level of acetylation is not quantitatively correlated to transcription rate.36 Reducing the average overall acetylation level in the coding region to about half of that of wild type in yeast showed no relationship with reduced transcription; however, a 4- to 5-fold reduction introduced at some genes correlated with a dramatic transcription effect.38 We observed a similar situation in the globin locus. For instance, the β gene is transcribed at a level approximately 10% of the γ gene in fetal erythroblasts, whereas the acetylation level at the β gene is approximately 50% of that at the γ gene.
The LCR is known to be able to recruit pol II with this ability positioned over the cores of the HSs 1-4.18,20,39,40 These results were confirmed in this study in human primary erythroblasts. However, we observed distinct pol II distributions in fetal and adult erythroblasts. In fetal cells, pol II bound to the LCR region was 2- to 3-fold greater than that in adult cells, and the majority of pol II was recruited to the LCR region compared with the gene region. On the other hand, in adult erythroblasts, the majority of pol II was recruited to the gene regions, particularly in the β-globin gene. It is noteworthy that the total amount of pol II measured in the entire locus was identical in fetal and adult erythroblasts (73 vs. 76 arbitrary units by totaling the levels of enrichment of pol II at all primer sets for each cell type). Although the molecular function of the pol II recruited to the LCR was unclear, this feature prompts speculation. The long-range polymerase transfer model proposes that pol II bound to the LCR is “loaded” onto the gene promoter for activation.39 We have proposed that in erythroid cells, the LCR is able to form an active transcription factory and the globin gene that is looped into the factory will be transcribed.18,41 The new results regarding the total amount of pol II at the locus implies that the transcription factory contains the same amount of pol II in both fetal and adult erythroid cells; however, the distribution of pol II between the LCR γ- and β-globin genes can be changed at different stages of development. This change might be carried out by varying the probability of loop formation between the LCR and the promoters.41 Nevertheless, the exact role for the LCR in the different stages of development still needs to be elucidated.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grants DK61805 and HL73439 (Q.L.) and DK45365 (G.S.).
National Institutes of Health
Authorship
Contribution: W.Y. and G.B. designed and performed research, collected and analyzed data, and wrote and revised the manuscript. X.F. and P.X. performed research and collected and analyzed data. H.C. performed research and provided cultured adult erythroblasts. G.S. designed research, analyzed data, and revised the draft manuscript. Q.L. designed research, analyzed data, and wrote and revised the manuscript. W.Y. and G.B. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Qiliang Li, Medical Genetics, Box 357720, University of Washington, Seattle, WA 98195; e-mail: li111640@u.washington.edu.