Severe congenital neutropenia (SCN) is an inborn disorder of granulopoiesis. Mutations of the ELA2 gene encoding neutrophil elastase (NE) are responsible for most cases of SCN and cyclic neutropenia (CN), a related but milder disorder of granulopoiesis. However, the mechanisms by which these mutations disrupt granulopoiesis are unclear. We hypothesize that the ELA2 mutations result in the production of misfolded NE protein, activation of the unfolded protein response (UPR), and ultimately apoptosis of granulocytic precursors. Expression of mutant NE but not wild-type NE strongly induced BiP/GRP78 mRNA expression and XBP1 mRNA splicing, 2 classic markers of the UPR. The magnitude of UPR activation by a specific ELA2 mutation correlated with its associated clinical phenotype. Consistent with the UPR model, expression of mutant NE in primary human granulocytic precursors increased expression of CHOP (DDITS) and induced apoptosis in a protease-independent fashion. Most strikingly, UPR activation and decreased NE protein expression were detected in primary granulocytic precursors from SCN patients. Collectively, these data provide strong support for a UPR model of SCN disease pathogenesis and place SCN in a growing list of human diseases caused by misfolded proteins.
Introduction
Severe congenital neutropenia (SCN) is a rare disorder characterized by severe neutropenia present at birth, an arrest of granulocytic differentiation at the promyelocyte or myelocyte stage, and a marked propensity to develop acute myeloid leukemia and myelodysplasia.1,2 Cyclic neutropenia (CN) is a related disorder of granulopoiesis, characterized by periodic oscillations in the numbers of circulating neutrophils and other peripheral blood cells.3 Mutations of the ELA2 gene encoding neutrophil elastase (NE) have been identified in nearly all patients with CN and in approximately 50% of cases of SCN.4,,,,–9 To date, more than 50 distinct mutations of the ELA2 gene have been reported in patients with CN or SCN.5,–7,10 Most of the mutations (∼80%) are missense mutations, although mutations that lead to splicing defects (∼10%) and premature stop codons (∼10%) also have been observed. With a few exceptions, specific ELA2 mutations are associated with SCN or CN, but not both, suggesting a genotype-phenotype correlation.
The molecular mechanisms by which ELA2 mutations disrupt granulopoiesis are unclear. Genetic studies provide 2 important clues. First, in all cases, the ELA2 mutations are heterozygous,5,–7 suggesting a dominant mechanism of action. Second, a case report of paternal mosaicism for an ELA2 mutation provides evidence that expression of mutant NE inhibits granulopoiesis in a cell intrinsic fashion, since no toxic paracrine effects of mutant NE protein on wild-type granulocytic cells in this mosaic individual were observed.11 NE is a serine protease expressed at extremely high levels at the promyelocyte stage of granulocytic differentiation.12 However, an extensive in vitro biochemical characterization of a large number of NE mutants failed to document a consistent effect of these mutations on NE proteolytic activity, substrate specificity, or serpin inhibition.13
The diversity of ELA2 mutations in SCN and the lack of any consistent effect of these mutations on NE function led us to hypothesize that structural rather than functional perturbations in the NE protein are responsible for the disruption in granulopoiesis. Specifically, we hypothesized that mutations in ELA2 result in the production of misfolded NE protein, induction of the unfolded protein response (UPR), and the subsequent UPR-dependent apoptosis of granulocytic precursors. Consistent with this hypothesis, Köllner et al recently showed that expression of mutant NE in a myeloid cell line induced expression of BiP/GRP78, a well-characterized marker of the UPR.14 Proteins destined for secretion (such as NE) are translocated cotranslationally to the lumen of the endoplasmic reticulum (ER). Protein folding in the specialized environment of the ER is a dynamic and imperfect process that is dependent upon a number of cellular factors, including nutrient availability, oxidation-reduction balance, and rate of secretory protein synthesis.15 Accumulation of misfolded proteins in the ER triggers the UPR, a coordinated adaptive program evolved to protect the cell from this type of ER stress.16 The canonical mammalian UPR pathway has 3 main branches that are regulated by the transmembrane ER resident proteins, AFT6, IRE1, and PERK. Under basal conditions, the ER chaperone protein BiP/GRP78 associates with and stabilizes the inactive state of each of these proteins. Under ER stress conditions, BiP preferentially associates with misfolded protein, permitting the activation of these ER sensors. Activation of AFT6, IRE1, and PERK lead to the attenuation of general translation initiation, induction of the expression of ER resident chaperones, and activation of ER-associated degradation (ERAD) pathways. If the ER stress is severe, these adaptive responses can be overwhelmed, in which case apoptosis is induced. There is accumulating evidence that the UPR may play key roles in the pathogenesis of a number of diseases, including diabetes mellitus, neurodegenerative disease, and cancer.17
The rationale for the hypothesis that ELA2 mutations disrupt granulopoiesis through induction of the UPR is based on the following observations. First, induction of apoptosis by the UPR requires high-level expression of misfolded protein. NE is expressed at extremely high levels in promyelocytes, the cell stage at which granulocytic differentiation is arrested in SCN.18 Moreover, several of the ELA2 mutations are predicted to strongly induce protein misfolding; 7 of 8 cysteine residues in NE are targeted by different ELA2 mutations, disrupting disulfide bonding required for the stability of the mature NE protein. Second, the UPR model of disease pathogenesis is consistent with genetic studies indicating that the ELA2 mutations act in a dominant and cell intrinsic fashion to disrupt granulopoiesis.11 Herein, we show that expression of mutant NE strongly activates the UPR, with the magnitude of UPR activation by a specific ELA2 mutation correlating with the severity of its associated clinical phenotype. Moreover, UPR activation was detected in primary SCN granulocytic precursors. Finally, expression of mutant NE induces apoptosis in myeloid cells in a protease-independent fashion. Together, these data provide strong support for a UPR model of SCN disease pathogenesis.
Patients, materials, and methods
NE constructs
The cDNAs for wild-type, V72M, P110L, and R191Q NE were kindly provided by Marshall Horowitz (University of Washington, Seattle, WA). The G185R, G192X, S173A, V72M-S173A, and P110L-S173A NE cDNAs were generated by site-directed mutagenesis and sequenced to verify the integrity of the open reading frame. All of the NE cDNAs were cloned into the LENpptIRES-GFP vector (kindly provided by Jeff Millbrandt, Washington University, St Louis, MO).19 Production of full-length NE protein from all of the ELA2 mutations in RBL-1 was comparable to wild-type ELA213 (and data not shown).
Human bone marrow samples
After informed consent was obtained in accordance with the Declaration of Helsinki, bone marrow samples were obtained from 9 healthy volunteers and 8 patients with SCN. Two of the healthy donors received G-CSF (5 μg/kg/day) for 5 days in preparation for stem cell harvesting. All 8 patients with SCN carried ELA2 mutations: 4 with S97L and one each with A28T, A32V/I31T, C194X, and R74L. The patients with the S97L ELA2 mutation have been previously described.20 All patients with SCN were treated with G-CSF at the time of bone marrow harvest. These studies were approved by the human studies committee at Washington University.
Primary human myeloid culture
Mononuclear cells were isolated from human cord blood by centrifugation over a Histopaque-1077 gradient per manufacturer's recommendations (Sigma, St Louis, MO). Cells were incubated with anti-human CD34 magnetic beads (Miltenyi Biotec, Auburn, CA), and CD34+ cells were isolated using the AutoMACS device (Miltenyi Biotec), as per manufacturer's recommendations. The purified CD34+ cells were placed in bone marrow culture media (Iscove essential media supplemented with 20% fetal calf serum, 2 mM l-glutamine, 50 nM β-mercaptoethanol, 200 μg/mL human transferrin, 100 μg/mL human insulin, 100 ng/mL granulocyte colony-stimulating factor [rhuG-CSF] and 10 ng/mL stem cell factor [rhuSCF]) at a density of 1 to 2 × 105 cells/mL. Cultures were harvested on day 5, at which time 74.8% plus or minus 6.6% of cells exhibited a promyelocyte or myelocyte morphology.
Transfections
The U937 and RBL-1 cell lines were obtained from the ATCC (Manassas, VA). 1 × 107 U937 cells were electroporated with 45 μg of DNA using an ECM 630 electroporator (BTX, Holliston, MA). Instrument settings were 225V, 350W, 600μF using a 2-mm gap cuvette.
Cultured human myeloid cells and RBL-1 cells were transfected using the Amaxa Nucleofector system (Amaxa Biosystems, Cologne, Germany). In brief, 1 to 2 × 106 cultured human myeloid cells in 100 μL human dendritic cell nucleofector solution were transfected with 5 μg DNA using the Amaxa Nucleofector electroporation device set to program X-001. For RBL-1 cells, the nucleofector cell line kit V solution and program T-30 were used.
Flow cytometry/cell sorting
Transfected cells were first incubated with 7-amino-actinomycin D (7AAD) to exclude nonviable cells. 7AAD-negative, GFP-positive cells were sorted using the MoFlo high-speed flow cytometer (Dako, Ft Collins, CO). To isolate human early granulocytic precursors, bone marrow samples were incubated with the following directly conjugated antibodies (all antibodies from BD Pharmingen, San Diego, CA): phycoerythrin (PE)–conjugated CD16, fluorescein isothiocyanate (FITC)–conjugated CD15, and allophycocyanin (APC)–conjugated CD14 or CD9. Isotype-matched antibodies were used as negative controls. CD15+ CD16lo CD14− CD9− cells were sorted using a MoFlo high-speed flow cytometer.
XBP1 splicing assays
XBP1 mRNA processing was measured using an assay described by Harding et al.21 In brief, cDNA was generated using the Eppendorf cMaster RTplusPCR System according to manufacturer's instructions. The following primers were used to amplify the XBP1 cDNA: forward 5′-AAACAGAGTAGCAGCTCAGACTGC-3′; reverse 5′-TCCTTCTGGGTAGACCTCTGGGAG-3′. The PCR products were digested with PstI and resolved on a 5% polyacrylamide gel.
Real-time RT-PCR
Total RNA was isolated from cells using the High Pure RNA Isolation Kit (Roche Diagnostics, Indianapolis, IN). Real-time quantitative RT-PCR was performed using the TaqMan One-Step RT PCR Master Mix Reagents Kit (Applied Biosystems, Foster City, CA) on a GeneAmp 7300 Sequence Detection System (Applied Biosystems). Reactions were repeated in the absence of reverse transcriptase to confirm the absence of DNA contamination. RNA content was normalized to human ribosomal RNA. The following primer and probe sets were used: hELA2 forward 5′-TTCGAAAACGGCTACGACCC-3′; hELA2 reverse 5′-CAGCTGGGCCACCTGCAC-3′; hELA2 probe 5′-[Fam]ATCGTGATTCTCCAGCTCAACGGGTCG[Tamra]-3′; hHSPA5 forward 5′-AGAAGACGGGCAAA-GATGTCA-3′; hHSPA5 reverse 5′-TGCTTGATGCTGAGAAGACAGG-3′; hHSPA5 probe 5′-[Fam]CGGCGCGAGGTAGAAAAGGCCA[Tamra]-3′; hrRNA forward 5′-CGCCGCTAGAGGTGAAATTCT-3′; hrRNA reverse 5′-CGAACCTCCGACTTTCGTTCT-3′; hrRNA probe 5′-[Fam]-AGACGGACCAGAGCGAAAGCATTTGC[Tamra]-3′; hCHOP forward 5′-GGAGCTGGAAGCCTGGTA TG-3′; hCHOP reverse 5′-GCTCTGGGAGGTGCTTGT-3′; hCHOP probe 5′-[Fam]TCTTCACCACTCTTGA-CCCTGCTTCTCT[Tamra]-3′.
Apoptosis assay
Cultured primary human myeloid cells were transfected as described and cultured for 24 hours in bone marrow culture media without G-CSF or SCF. Apoptotic cells were identified by flow cytometry using the Annexin V-PE Apoptosis Detection Kit 1 (BD Pharmingen) according to manufacturer's instructions.
Neutrophil elastase activity
Elastase activity was assayed using the chromogenic, NE-specific substrate N-methoxysuccinyl Ala-Ala-Pro-Val p-nitroanilide (Sigma), as previously described.22
Immunofluorescence/confocal microscopy
Cytospin preparations of bone marrow cells were fixed for 30 minutes at room temperature with BD Cytofix/Cytoperm Fixation and Permeabilization Solution (BD Biosciences), permeabilized with 10% DMSO in fetal calf serum at −80°C overnight, and incubated with BD Cytofix/Cytoperm Solution for 5 minutes at room temperature. Cells were then incubated with primary antibody for 2 hours at room temperature. Primary antibodies used were rabbit anti–human neutrophil elastase (EMD Biosciences-Calbiochem, La Jolla, CA), rabbit anti-calnexin (Sigma), and rabbit anti-myeloperoxidase (Abcam, Cambridge, MA). Rabbit normal immunoglobulin fraction (Dako A/S, Glostrup, Denmark) was used as an isotype control. Primary antibodies were fluorescently labeled with Zenon Rabbit IgG Antibody Labeling Kits (Invitrogen-Molecular Probes, Eugene, OR) according to manufacturer's instructions. Cells were counterstained with TO-PRO-3 iodide (Invitrogen-Molecular Probes). Immunofluorescence confocal microscopy was performed using a LSM 510 Laser Scanning Microscope (Carl Zeiss Advanced Imaging Microscopy, Jena, Germany) and analyzed with Zeiss LSM Image Browser software (Carl Zeiss Advanced Imaging Microscopy).
To estimate the relative abundance of neutrophil elastase in primary promyelocytes from healthy donor and SCN patient bone marrow aspirates, 3-dimensional reconstructions of representative promyelocytes were generated. In brief, single color images were converted into grayscale, and the average signal intensity per pixel (measured in arbitrary units, or AUs) within individual cells was measured using Scion Image software (Scion, Frederick, MD). The total AU within each cell was calculated as the product of the average AU per pixel and the number of pixels in the cell. To account for differences in magnification, the total AU per cell (based on area measured in pixels) was then multiplied by the number of square micrometers per pixel, yielding a final value of total AU per cell based on the objective area of the cell. Five to 12 representative promyelocytes from each sample were analyzed in this way.
Statistical analysis
Data are presented as mean plus or minus SEM unless otherwise stated. Statistical significance was assessed using a 2-sided Student t test.
Results
Expression of mutant NE in U937 cells induces the UPR
Based on their presence in 2 or more independent families and their specific association with SCN (V72M, G192X, and G185R), CN (R191Q), or both SCN and CN (P110L), 5 ELA2 mutations were chosen for study. Wild-type (WT) or mutant NE cDNA linked to the enhanced green fluorescent protein (GFP) by an internal ribosomal entry sequence motif were transiently transfected into the human myeloid cell line U937. At 12 hours after transfection, viable cells displaying at least a 1-log shift in GFP intensity were sorted by flow cytometry. Two classic biochemical assays of the UPR were performed on these cells: XBP1 (X-box binding protein-1) mRNA splicing and induction of BiP/GRP78 (HSPA5) mRNA expression.23
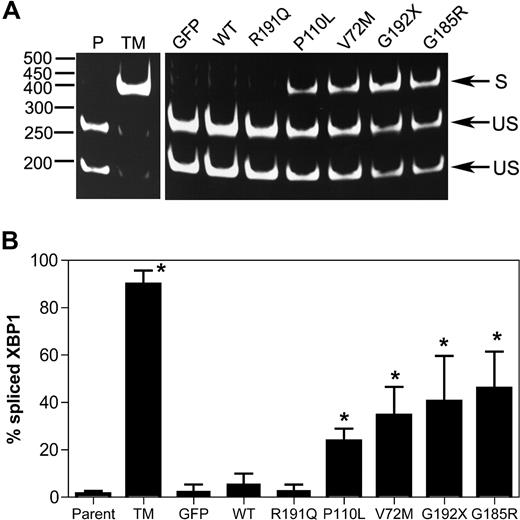
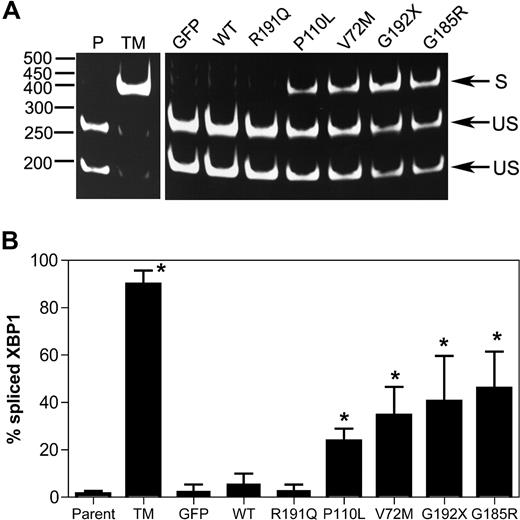
Upon activation of the UPR, unconventional splicing mediated by the IRE1α endonuclease removes a 26-nucleotide intron from the XBP1 mRNA, resulting in a translational frameshift that encodes for a potent transcriptional activator for many ER stress response genes.24 Thus, the determination of the relative levels of spliced and unspliced XBP1 mRNA transcripts can be used to assess the degree of UPR activation in a cell. As expected, treatment of U937 cells with tunicamycin (a protein glycosylation inhibitor and potent inducer of the UPR) strongly induced XBP-1 splicing (Figure 1). A significant increase in XBP1 splicing also was observed after expression of P110L, V72M, G192X, or G185R NE. In contrast, minimal changes were observed in cells expressing WT NE or the CN-associated R191Q NE.
Expression of mutant NE induces XBP1 splicing in U937 cells. U937 cells were transiently transfected with vector alone (GFP), wild-type (WT), or mutant NE cDNAs, and 12 hours later GFP + cells were sorted by flow cytometry. As controls, parent U937 cells (P) and cells treated with tunicamycin (TM) also were analyzed. (A) Representative gel showing spliced (S) and unspliced (US) XBP1 bands. (B) The percentage of spliced XBP1 is shown. Data represent the mean (± SD) of 5 independent experiments. *P < .05 compared with WT-transfected cells.
Expression of mutant NE induces XBP1 splicing in U937 cells. U937 cells were transiently transfected with vector alone (GFP), wild-type (WT), or mutant NE cDNAs, and 12 hours later GFP + cells were sorted by flow cytometry. As controls, parent U937 cells (P) and cells treated with tunicamycin (TM) also were analyzed. (A) Representative gel showing spliced (S) and unspliced (US) XBP1 bands. (B) The percentage of spliced XBP1 is shown. Data represent the mean (± SD) of 5 independent experiments. *P < .05 compared with WT-transfected cells.
BiP is an ER-resident chaperone involved in protein folding. Under conditions of ER stress, the transcription of BiP is up-regulated, and thus changes in the expression of this gene are widely used as a marker of ER stress. Transient expression of the NE mutants associated with SCN (P110L, V72M, G192X, or G185R) strongly induced BiP mRNA expression, whereas it was unchanged in cells expressing WT or R191Q NE (Figure 2A). Indeed, after normalizing for NE mRNA expression, BiP mRNA expression was 6- to 8-fold higher in cells expressing the SCN-related NE mutants compared with WT or R191Q NE (Figure 2B). Collectively, these data show that transient expression of mutant NE associated with SCN activates the UPR in U937 cells.
Expression of mutant NE induces BiP expression in U937 cells. U937 cells were transiently transfected with vector alone (GFP), wild-type (WT), or mutant NE cDNAs, and 12 hours later GFP + cells were sorted by flow cytometry. BiP, NE, and ribosomal RNA (rRNA) were quantified by real-time RT-PCR. (A) Expression of BiP mRNA relative to rRNA is shown. (B) Expression of BiP mRNA to NE mRNA is shown. Data are normalized to WT-transfected cells, where the BiP to NE ratio in each experiment was defined as 1. Data represent the mean (± SD) of 5 independent experiments. *P < .05 compared with WT-transfected cells.
Expression of mutant NE induces BiP expression in U937 cells. U937 cells were transiently transfected with vector alone (GFP), wild-type (WT), or mutant NE cDNAs, and 12 hours later GFP + cells were sorted by flow cytometry. BiP, NE, and ribosomal RNA (rRNA) were quantified by real-time RT-PCR. (A) Expression of BiP mRNA relative to rRNA is shown. (B) Expression of BiP mRNA to NE mRNA is shown. Data are normalized to WT-transfected cells, where the BiP to NE ratio in each experiment was defined as 1. Data represent the mean (± SD) of 5 independent experiments. *P < .05 compared with WT-transfected cells.
Expression of mutant NE in U937 cells reduces their clonogenic capacity
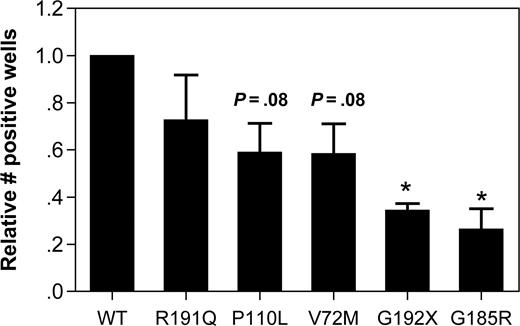
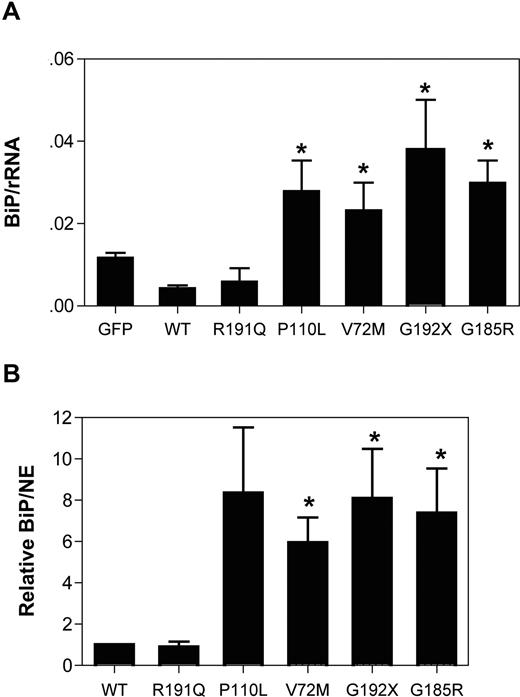
The effect of mutant NE expression on cell viability was assessed by measuring the clonogenic capacity of transfected U937 cells. In U937 transfected with WT NE, 10.6% plus or minus 6.1% of sorted GFP + cells were able to form colonies after plating at limiting dilution. Since considerable interexperiment variability in the clonogenic potential was observed, the data for the other NE mutants was normalized to WT NE–transduced cells for each experiment (Figure 3). Expression of the G192X and G185R NE mutants significantly reduced the clonogenic capacity of U937 cells, with a borderline significant decrease in V72M- and P110L-transfected cells. No significant difference in clonogenic capacity was observed in R191Q-transfected cells.
Expression of mutant NE reduces the clonogenic capacity of U937 cells. U937 cells transfected with the indicated NE cDNA were sorted based on GFP expression and plated at limiting dilution in 96-well plates. The number of positive wells on day 14 of culture is shown. Data are normalized to WT-transfected cells, where the number of positive wells was defined as 1. Data represent the mean (± SD) of 3 independent experiments. *P < .05 compared with WT-transfected cells.
Expression of mutant NE reduces the clonogenic capacity of U937 cells. U937 cells transfected with the indicated NE cDNA were sorted based on GFP expression and plated at limiting dilution in 96-well plates. The number of positive wells on day 14 of culture is shown. Data are normalized to WT-transfected cells, where the number of positive wells was defined as 1. Data represent the mean (± SD) of 3 independent experiments. *P < .05 compared with WT-transfected cells.
Expression of mutant NE induces the UPR and apoptosis in primary human promyelocytes and is associated with increased CHOP mRNA expression
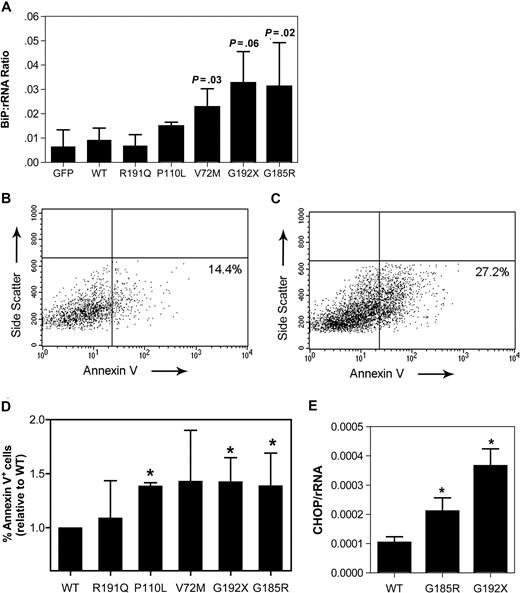
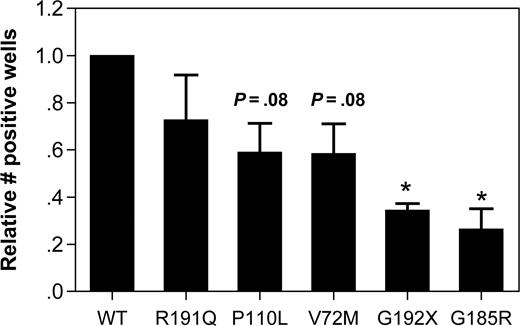
To extend the U937 cell line data to a more physiologic model, we developed an assay to characterize the effect of mutant NE expression in primary human early granulocytic precursors. CD34-positive cells isolated from umbilical cord blood were cultured in media containing G-CSF and stem-cell factor. After 5 days in culture, the majority of cells in culture displayed a promyelocyte or myelocyte morphology (74.8% ± 6.6%; data not shown). These cells were transiently transfected using the Amaxa Nucleofector system, with transduction efficiencies of 28% to 46% obtained (data not shown). Expression of the SCN-associated NE mutants in these cells strongly induced BiP mRNA expression, achieving statistical significance with V72M and G185R (Figure 4A).
Expression of mutant NE activates UPR-induced apoptosis in promyelocytes. Cultured human promyelocytes were transiently transfected with vector alone (GFP) or the indicated NE cDNA. (A) BiP mRNA expression relative to rRNA was determined on sorted GFP+ cells 12 hours after transfection. (B,C) Representative scatter plots of annexin V expression on cells transfected with WT (B) or G185R NE (C); data are gated on GFP+ 7AAD− cells. Gates were set based on untransfected cells (not shown). (D) The percentage of GFP+ 7AAD− cells that were annexin V+ at 24 hours after transfection is shown. Data are normalized to WT-transfected cells, where the percentage of annexin V+ cells was defined as 1. (E) CHOP mRNA expression relative to rRNA was determined on sorted GFP+ cells 12 hours after transfection. Data represent the mean (± SD) of 4 to 6 independent experiments * P < .05 compared with WT-transfected cells.
Expression of mutant NE activates UPR-induced apoptosis in promyelocytes. Cultured human promyelocytes were transiently transfected with vector alone (GFP) or the indicated NE cDNA. (A) BiP mRNA expression relative to rRNA was determined on sorted GFP+ cells 12 hours after transfection. (B,C) Representative scatter plots of annexin V expression on cells transfected with WT (B) or G185R NE (C); data are gated on GFP+ 7AAD− cells. Gates were set based on untransfected cells (not shown). (D) The percentage of GFP+ 7AAD− cells that were annexin V+ at 24 hours after transfection is shown. Data are normalized to WT-transfected cells, where the percentage of annexin V+ cells was defined as 1. (E) CHOP mRNA expression relative to rRNA was determined on sorted GFP+ cells 12 hours after transfection. Data represent the mean (± SD) of 4 to 6 independent experiments * P < .05 compared with WT-transfected cells.
Persistent and severe ER stress can lead to UPR-induced apoptosis. Consistent with this observation, an increased susceptibility to apoptosis has been detected in granulocytic precursors from patients with SCN.25,26 To more directly test this possibility, apoptosis was assessed in primary cultured promyelocytes/myelocytes after transfection with the various NE mutants. Specifically, surface expression of annexin V, an early and sensitive marker of apoptosis, was measured in transduced (GFP+) cells 24 hours after transfection (Figure 4B,C). In cells transfected with WT NE, 19.5% plus or minus 8.5% were annexin V+. Again, since considerable interexperiment variability in the percentage of apoptotic cells was observed, the data for the other NE mutants was normalized to WT NE for each experiment. Expression of the P110L, G192X, and G185R NE mutants was associated with a significant increase in annexin V+ cells (Figure 4D). Though not achieving statistical significance, a trend toward increased apoptosis also was observed in V72M NE–transfected cells. In contrast, no significant change in R191Q NE–transfected cells was observed. Collectively, these data show that expression of SCN-associated NE mutants activates the UPR and induces apoptosis in primary granulocytic precursors.
UPR-induced apoptosis is associated with transcriptional activation of CCAAP/enhancer binding protein (C/EBP) homologous protein (CHOP, also known as DDITS or GADD153).27 Chop−/− mice are resistant to UPR-induced apoptosis, suggesting that induction of CHOP mRNA expression is a key step in UPR-induced apoptosis.28 Consequently, we next determined whether expression of mutant NE induced CHOP mRNA expression in primary cultured promyelocytes. Indeed, compared with wild-type NE, a 2.7- and 4.4-fold increase in CHOP mRNA was observed in cells transfected with G185R or G192X NE, respectively (Figure 4E).
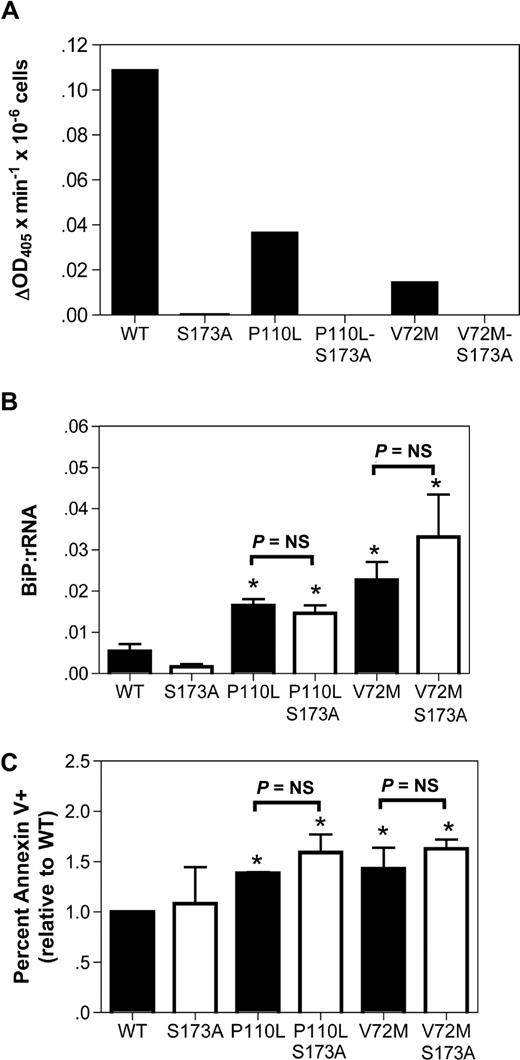
Induction of apoptosis by mutant NE is not dependent upon its protease activity
A prevailing competing hypothesis in the field is that ELA2 mutations disrupt granulopoiesis in a protease-dependent fashion.29,–31 In contrast, the UPR hypothesis predicts that the protease activity is dispensable. To address this issue, we generated NE double mutants carrying the SCN-related mutations P110L or V72M and an alanine substitution of the catalytic serine 173 residue; the S173A mutation has been shown to markedly attenuate the protease activity of NE.13 As reported previously, the P110L and V72M NE mutants displayed reduced protease activity compared with WT NE (Figure 5A).13 The S173A substitution abolished all detectable protease activity. We next transiently expressed the NE S173A or double mutants in cultured human granulocytic precursors. Induction of BiP mRNA expression by the protease-deficient double mutants was comparable to the observed in the corresponding SCN-related NE mutants (Figure 5B). Moreover, a similar percentage of annexin V+ cells was observed in cells expressing the single and double NE mutants (Figure 5C). These data show that the protease activity of the NE mutants is not required to activate the UPR or induce apoptosis.
The protease activity of mutant NE is not required to activate UPR-induced apoptosis. (A) The indicated NE cDNA was transiently transfected into RBL cells, and 24 hours later cell extracts were prepared from bulk cultures. NE protease activity was quantified using a chromogenic, NE-sensitive substrate. Shown is NE protease activity after correcting for the number of GFP+ cells analyzed. (B,C) Cultured human granulocytic precursors were transiently transfected with the indicated NE cDNA. (B) BiP mRNA expression relative to rRNA was determined on sorted GFP+ cells 12 hours after transfection. (C) The percentage of GFP+ 7AAD− cells that were annexin V+ at 24 hours after transfection is shown. Data represent the mean (± SD) of 4-5 independent experiments. *P < .05 compared with WT-transfected cells. NS indicates nonsignificant.
The protease activity of mutant NE is not required to activate UPR-induced apoptosis. (A) The indicated NE cDNA was transiently transfected into RBL cells, and 24 hours later cell extracts were prepared from bulk cultures. NE protease activity was quantified using a chromogenic, NE-sensitive substrate. Shown is NE protease activity after correcting for the number of GFP+ cells analyzed. (B,C) Cultured human granulocytic precursors were transiently transfected with the indicated NE cDNA. (B) BiP mRNA expression relative to rRNA was determined on sorted GFP+ cells 12 hours after transfection. (C) The percentage of GFP+ 7AAD− cells that were annexin V+ at 24 hours after transfection is shown. Data represent the mean (± SD) of 4-5 independent experiments. *P < .05 compared with WT-transfected cells. NS indicates nonsignificant.
The UPR is activated in promyelocytes/myelocytes from patients with SCN
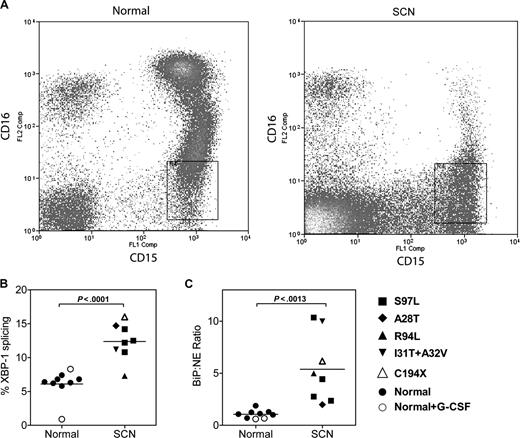
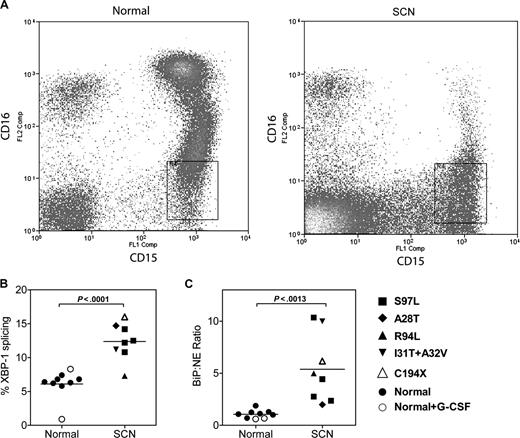
We next determined whether the UPR was activated in granulocytic precursors from patients with SCN. Fresh bone marrow samples were obtained from 8 patients with SCN carrying an ELA2 mutation, 7 healthy volunteers, and finally 2 healthy volunteers who had been treated with G-CSF (5 μg/kg per day) for 5 days. To ensure that the appropriate target cell population was analyzed, we developed a strategy to isolate early granulocytic precursors directly from the bone marrow. Specifically, after excluding monocytes and eosinophils, CD15+ CD16lo cells were sorted by flow cytometry (Figure 6A). This cell population is highly enriched for promyelocytes and myelocytes (87% ± 2.0% and 96% ± 0.8% from SCN and normal bone marrow, respectively). Representative scatter plots from a normal and SCN BM readily illustrate the block in granulocytic differentiation, with a nearly complete absence of the CD15+ CD16hi mature neutrophil population in the SCN sample (Figure 6A). Of note, expression of NE mRNA (relative to ribosomal RNA) in granulocytic precursors isolated from patients with SCN was comparable to healthy controls (relative NE mRNA expression in arbitrary units: 0.016 ± .007 arbitrary units versus 0.015 ± 0.005; P = NS). Consistent with UPR activation, a significant increase in XBP1 splicing and BiP mRNA expression (relative to NE mRNA expression) was observed in the SCN samples (Figure 6B,C); a trend to increased BiP mRNA expression relative to ribosomal RNA also was observed (relative BiP expression: 0.021 ± 0.004 [normal] versus 0.041 ± 0.010 [SCN], P = .08). Of note, G-CSF treatment of healthy volunteers had no effect on XBP-1 splicing or BiP mRNA expression. Together, these data suggest that the UPR is activated in early granulocytic precursors from patients with SCN carrying ELA2 mutations.
The UPR is activated in SCN promyelocytes/granulocytes. (A) Representative scatter plots of bone marrow cells from a healthy donor (normal) and a patient with ELA2-mutation positive SCN (A28T) stained for CD15 and CD16; data are gated to exclude CD14+ monocytes and CD9+ eosinophils. The boxed region shows the sorted CD15+ CD16lo CD14− CD9− cell population. (B) The percentage of spliced XBP-1 mRNA in the sorted cell population is shown. (C) Expression of BiP mRNA relative to NE mRNA in the sorted cell population is shown.
The UPR is activated in SCN promyelocytes/granulocytes. (A) Representative scatter plots of bone marrow cells from a healthy donor (normal) and a patient with ELA2-mutation positive SCN (A28T) stained for CD15 and CD16; data are gated to exclude CD14+ monocytes and CD9+ eosinophils. The boxed region shows the sorted CD15+ CD16lo CD14− CD9− cell population. (B) The percentage of spliced XBP-1 mRNA in the sorted cell population is shown. (C) Expression of BiP mRNA relative to NE mRNA in the sorted cell population is shown.
NE protein expression is markedly reduced in SCN granulocytic precursors
The UPR model of disease pathogenesis predicts that NE protein expression in SCN granulocytic precursors may be decreased. First, misfolded NE protein would be degraded by the ERAD system. Second, activation of the UPR would result in a general inhibition of protein translation. To test this prediction, confocal microscopy was performed on normal and ELA2 mutation-positive SCN bone marrow cells stained for NE and either calnexin, an ER resident protein, or myeloperoxidase, a primary granule constituent (Figure 7; Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). NE protein expression appeared to be consistently reduced in SCN cells compared with normal cells. Indeed, quantitative image analysis showed a significant reduction in NE signal (41.2 ± 5.9 arbitrary units [normal] versus 19.4 ± 2.0 [SCN]; P < .001). However, no ER accumulation of NE was noted; instead, the residual NE signal colocalized with MPO, suggesting a primary granule localization. Interestingly, MPO expression was consistently noted to be modestly decreased in SCN cells, perhaps reflecting a general decrease in protein translation. Collectively, these data show that NE protein expression is markedly reduced in ELA2 mutation-positive SCN granulocytic precursors and suggest that the residual mutant NE protein is trafficked normally to primary granules.
NE and MPO protein expression is decreased in SCN granulocytic precursors. Confocal microscopy of 2 different healthy donors (normal) and 3 ELA2-mutant SCN samples stained with NE and myeloperoxidase (MPO; a primary granule constituent). Isotype controls for NE and MPO are shown. Data are representative of 6 normal and 6 ELA2 mutation-positive SCN bone marrow samples.
NE and MPO protein expression is decreased in SCN granulocytic precursors. Confocal microscopy of 2 different healthy donors (normal) and 3 ELA2-mutant SCN samples stained with NE and myeloperoxidase (MPO; a primary granule constituent). Isotype controls for NE and MPO are shown. Data are representative of 6 normal and 6 ELA2 mutation-positive SCN bone marrow samples.
Discussion
Although the genetic evidence implicating ELA2 mutations in the pathogenesis of CN and most cases of SCN is compelling, the molecular mechanism by which these mutations disrupt granulopoiesis is unclear. In particular, it has been difficult to account for the diversity of ELA2 mutations (more than 50 mutations have been reported) and the lack of a consistent effect of these mutations on NE protease activity, serpin inhibition, or subcellular localization. In this study, we provide evidence in support of a model in which ELA2 mutations lead to the production of misfolded NE protein, an activation of the UPR, and ultimately the induction of apoptosis of granulocytic precursors. The following observations support this model. First, expression of mutant but not wild-type NE activated the UPR in a myeloid cell line and in primary granulocytic precursors. Two classic markers of the UPR, XBP1 splicing, and BiP mRNA expression were significantly increased. Second, compared with normal cells, the UPR was activated in granulocytic precursors from patients with SCN. Third, consistent with the UPR, expression of mutant NE induced apoptosis in a protease-independent fashion. In agreement with this observation, a preliminary report showed that inhibition of NE protease activity in SCN progenitor cells failed to rescue the block in granulocytic differentiation.32 Of note, no increase in annexin V-positive (apoptotic) granulocytic precursors was observed in freshly isolated SCN bone marrow (data not shown), perhaps reflecting the rapid clearance of apoptotic cells in vivo. Finally, the UPR model is consistent with genetic evidence that ELA2 mutations act in a dominant and cell-intrinsic fashion.11 Kollner and colleagues independently generated data supporting the UPR model of SCN disease pathogenesis.14 They showed that most of the tested ELA2 mutations displayed impaired protein processing with an accumulation of mutant NE in the cytoplasm. Moreover, they showed that expression of mutant NE in a myeloid cell line induced apoptosis and expression of BiP.
Previous studies suggested that altered intracellular trafficking of mutant NE may contribute to the pathogenesis of SCN. Benson et al showed that several SCN-related NE mutants were localized primarily in the plasma membrane in RBL cells.33 Likewise, G185R NE expressed in HL60 cells localized primarily to the plasma and nuclear membranes.26 In contrast, a recent report showed that in primary neutrophils from patients with SCN, NE was predominantly localized to the cytoplasm.14 In the present study, we show that, even though NE mRNA expression is normal, NE protein expression is markedly reduced in primary granulocyte precursors from patients with SCN. Moreover, the residual NE protein is targeted correctly to primary granules, as indicated by colocalization with myeloperoxidase. Thus, our data argue against a primary defect in the intracellular trafficking of mutant NE. Instead, our findings are consistent with a UPR mechanism, since misfolded NE is predicted to be degraded by the ERAD pathway, and activation of the UPR would attenuate NE mRNA translation.
While both SCN and CN are disorders of granulopoiesis, CN is a milder syndrome, with less severe and intermittent neutropenia and no increased propensity to develop AML/MDS. There is an imperfect correlation between the ELA2 genotype and clinical phenotype. With a few exceptions, most of the ELA2 mutations are associated with SCN or CN but not both. The molecular basis for this genotype-phenotype association is unclear. Benson et al reported that SCN and CN NE mutants may be differentially trafficked in the cell. Whereas most SCN-related NE mutants were localized primarily to the plasma membrane, CN-related mutants were trafficked normally to primary granules.33 However, not all of the ELA2 mutants exhibited this pattern, and a mechanism by which mistargeted NE disrupts granulopoiesis was not identified. In the present study, we show that the magnitude of UPR activation and apoptosis induced in vitro by the different ELA2 mutations correlates with the severity of their associated clinical phenotype. The highest degree of BiP mRNA expression and XBP1 splicing was observed in cells transfected with the G185R or G192X ELA2 mutants. Though data for most of the ELA2 mutations is limited, there is evidence that the clinical phenotype of SCN associated with the G185R ELA2 mutation is particularly severe.6 In contrast, the CN-related R191Q ELA2 mutation did not induce detectable UPR activation. Of note, a much larger sample size (including samples from patients with CN) will be required to confirm the correlation between UPR activation and disease severity in vivo. Nonetheless, these data suggest that the propensity of individual NE mutants to misfold may determine the magnitude of UPR-induced apoptosis and ultimately the clinical phenotype. Though it is possible that the R191Q (and other CN-related) ELA2 mutants disrupt granulopoiesis through a distinct mechanism, we suggest that the R191Q ELA2 mutant likely induces low level UPR activation that is difficult to detect experimentally. For those ELA2 mutations associated with both SCN and CN (eg, P110L), it is possible that the genetic modifiers of the UPR pathway may contribute to the phenotype. For example, nonsynonymous polymorphisms in ATF6, an important ER stress sensor, have been linked to the development of type 2 diabetes mellitus, a disease in which ER stress is thought to play an important role.34
These observations place SCN in a group of newly described human diseases caused by the production of misfolded proteins. While the role of misfolded proteins in cystic fibrosis35 and osteogenesis imperfecta36 has long been appreciated, these diseases result not from the intrinsic ER stress caused by the production of these misfolded proteins but rather from the loss of functional CFTR and collagen. A second class of human diseases, including α1-antitrypsin deficiency and autosomal recessive juvenile Parkinson disease (AR-PD), are caused at least in part by misfolded protein-associated cell death. In the case of α1-antitrypsin deficiency, misfolded protein accumulates in hepatocytes, leading eventually to hepatic failure, while patients with AR-PD lack a ubiquitin ligase required for the degradation of misfolded proteins that accumulate in dopaminergic neurons of the substantia nigra.37 Finally, mutations in the UPR pathway itself have been identified. For example, patients with Wolcott-Rallison syndrome lack a functional PERK allele. These patients are unable to attenuate translation in the face of increased ER stress, resulting in abnormally high accumulations of misfolded proteins. Accordingly, numerous tissues, including pancreatic β-cells, undergo premature apoptosis. Our data suggest that SCN may represent a novel mechanism for UPR-associated disease pathogenesis. Specifically, our data suggest that the accumulation of misfolded NE protein is so severe that it results in the congenital loss of a cell type, namely, mature neutrophils.
The UPR model of SCN disease pathogenesis has several clinical and biological implications. First, it suggests that mutations of other genes expressed selectively and highly in promyelocytes may be responsible for cases of SCN that do not carry an ELA2 (or other known) mutation. Second, this model provides a potential mechanism by which dominant-negative mutations of GFI1 induce an SCN-like phenotype in humans.38 GFI1 is a transcriptional repressor for ELA2, and loss of GFI1 function leads to increased NE expression. Even though the rate of misfolding may be low, sufficient overexpression of wild-type NE protein would be predicted to induce the UPR. Finally, the UPR model predicts that agents that modulate the UPR might be effective therapeutics in SCN. Indeed, a recent report showed that treatment with salubrinal, a selective eIF2α phosphatase inhibitor, suppressed UPR-induced apoptosis.39
The online version of this article contains a data supplement.
Presented in part at the 47th annual meeting of the American Society of Hematology, Atlanta, GA, December 10-13, 2005.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Audrey Anna Bolyard for help in the collection and analysis of clinical data for the patients with SCN. We also thank Elin Rodger, Andrew Stein, and Steve Stein for their sequencing of the ELA2 gene.
This work was supported by National Institutes of Health grants #RO1 HL079562 (D.C.L.), #P01 CA101937 (D.C.L.), and #T32 HL07088-23 (D.S.G.). M.M. participated in the Howard Hughes Medical Institute Research Training Fellowship.
National Institutes of Health
Authorship
Contribution: D.S.G., M.M., J.G., and J.X. designed, performed, and analyzed the experiments. L.A.B., D.D., and M.C.D. provided critical reagents. D.C.L. supervised all of the research and edited the manuscript.
D.S.G. and M.M. contributed equally to the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel C. Link, Division of Oncology, Department of Medicine, 660 S Euclid Ave, Campus Box 8007, St Louis, MO 63110; e-mail:dlink@im.wustl.edu.