Most mature follicular B cells circulate within the periphery in a quiescent state, without actively contributing to an acute immune response. Lasting B-cell persistence in the periphery is dependent on survival signals that are transduced by cell surface receptors. We recently demonstrated that cell surface CD74 controls mature B-cell survival. Stimulation of cell surface CD74 leads to NF-κB activation, which enables entry of the stimulated B cells into the S phase, induction of DNA synthesis, and cell division, and augments the expression of survival genes. In the present study, we investigated CD74 target genes to determine the identities of the molecules whose expression is modulated by CD74, thereby regulating B-cell survival. We report that CD74 activates the p65 member of the NF-κB family, which in turn up-regulates the expression of p53-related TAp63 proteins. TAp63 then binds and transactivates the Bcl-2gene and induces the production of Bcl-2 protein, thereby providing the cells with increased survival capacity. Thus, the CD74/NF-κB/TAp63 axis defines a novel antiapoptotic pathway in mature B cells, resulting in the shaping of both the B-cell repertoire and the immune response.
Introduction
In healthy individuals, the pool of peripheral lymphocytes is constant in size. The control of lymphoid homeostasis is the result of a very fine balance between lymphocyte production, survival, and proliferation. Survival factors have been shown to play a critical role in maintaining lymphocyte homeostasis
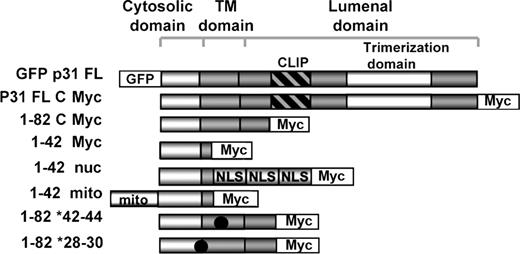
Recently, we demonstrated that CD74 (invariant chain, Ii) controls mature B-cell survival. CD74 is a nonpolymorphic type II integral membrane protein, containing a short N-terminal cytoplasmic tail of 28 amino acids (aa's), followed by a single 24-aa transmembrane region, and an approximately 150-aa lumenal domain. The CD74 chain was originally thought to function mainly as an MHC class II chaperone, promoting the exit of MHC class II molecules from the endoplasmic reticulum (ER), directing them to endocytic compartments, preventing peptide binding within the ER, and contributing to peptide editing in the MHC class II compartment.1
However, CD74 was recently found to play an additional role as an accessory signaling molecule. A small proportion of CD74 is modified by the addition of chondroitin sulfate (CD74-CS), and this form of CD74 is expressed on the surface of antigen-presenting cells together with CD44.2,3 In macrophages, CD74 was recently reported to demonstrate high-affinity binding to the proinflammatory cytokine, macrophage migration-inhibitory factor (MIF). MIF binds to the extracellular domain of CD74, and this interaction is required for MIF-mediated cell proliferation.4
In a previous series of studies, we showed that CD74 is directly involved in shaping the B-cell repertoire5 through a pathway leading to the activation of transcription mediated by the NF-κB p65/RelA homodimer and its coactivator, TAFII105.6 NF-κB activation is mediated by the cytosolic region of CD74 (CD74-ICD), which is liberated from the membrane.7 We demonstrated that following the removal of the CD74 lumenal domain, an intramembranal cleavage event at amino acid 42 occurs, resulting in the release of the CD74 cytosolic fragment (CD74-ICD; aa's 1-42). CD74-ICD, which has no NLS motif, then translocates to the nucleus and activates NF-κB.8 Thus, following processing, CD74 serves as a signaling molecule that enables accumulation of mature B cells. This signal is terminated by degradation of the active protein and its removal from the cytoplasm.7,9
Moreover, we previously demonstrated that CD74 stimulation with anti-CD74 antibody leads to NF-κB activation, enabling entry of the stimulated B cells into the S phase, elevation of DNA synthesis, cell division, and augmented expression of antiapoptotic members of the Bcl-2 protein family. These findings therefore confirm that surface CD74 functions as a survival receptor.10
The present study was designed to identify the specific molecules whose expression is modulated by CD74 to regulate B-cell survival. We report that CD74 stimulation and the resultant activation of the p65 member of the NF-κB family lead to enhanced expression of the α and γ isoforms of the p53-related TAp63 protein, which in turn transactivate the Bcl2gene and augment B-cell survival. The p63 gene exhibits a high sequence and structural homology to p53.11 Like p53, the p63gene encodes an N-terminal transactivation domain, a core DNA-binding domain, and a carboxy-terminal oligomerization domain. The p63 gene contains 2 transcriptional start sites that enable the generation of transcripts encoding proteins with or without an N-terminal transactivation domain. Proteins containing the transactivation domain are termed TAp63, and proteins lacking the transactivation domain are known as ΔNp63. p63 was shown to play a role in developmental regulation of limbs, skin, most epithelial tissues, and epidermal differentiation.12,13 Our findings suggest a novel role for p63 in modulating B-cell survival and shaping the immune response in a CD74-dependent manner.
Materials and methods
Cells and separation of B cells
Spleen cells were obtained from 6- to 8-week-old C57BL/6 or CD74−/− mice,14 as described in Shachar et al.15 All animal procedures were carried out in accordance with the Guidelines for the Care and Use of Research Animals at the Weizmann Institute of Science.
CD74−/− B cells were purified, using magnetic-activated cell sorting (MACS) CD19 beads (Miltenyi Biotec, Bergisch Gladbach, Germany). Control mature B cells were purified based on their expression of the IgD marker: Spleen cells were incubated with FITC-labeled anti-IgD antibody (BD Biosciences, San Diego, CA) for 30 minutes at 4°C. Cells were then incubated with anti-FITC magnetic beads and separated, using the MACS system (Miltenyi Biotec). The purity of each cell population was determined by flow cytometry.
Constructs and molecular cloning
Full-length and truncated CD74 constructs used in this study were described previously.6 For the GFP construct, FL CD74 was cloned into the EcoR1/BamH1 sites of the vector pEGFP N1 (Clontech, Mountain View, CA; GenBank accession no. 455762). The luciferase constructs for p63 were cloned into pGL2 vector (E1641; Promega, Madison, WI) into Mlu1/Bgl2 sites. The TAp63 promoter oligo for Mlu1 was 5′-CCGACGCGTGGCTTTAGATTGTCTCAC-3′, and for Bgl2, 5′-CTTCTAGACTGTAGGGTGGCACAC- 3′. The ΔNp63 promoter oligo for Mlu1 was 5′-CCGACGCGTGCCACTTACAACCTTTC-3′, and for Bgl2, 5′-CTTCTAGATCAATGCTGCTGTCC-3′.
Cell transfection
HEK-293 cells.
Cells were seeded in a 10-cm2 dish. Transfections were performed using the standard CaPO4 method, as previously described.7 A total of 5 μg plasmid DNA was used per dish.
B cells.
Freshly prepared splenocytes from CD74−/− mice were incubated for 48 hours in RPMI plus 10% FCS plus 25 μg/mL LPS (Salmonella; Sigma, St Louis, MO).7 Fluorescence-activated cell sorting (FACS) analysis subsequently revealed that 96% of the resulting cells were B cells. Cells were washed twice with medium lacking both serum and antibiotics. Washed cells (2 × 106 per transfection) were cultured in 6-well plates. Transfection was performed with Transfast reagent (E-2431; Promega), 3 μL/1 μg DNA. In these transfections, 100 ng of each DNA was supplemented with empty vector DNA to a total of 1 μg DNA/transfection. After 48 hours, cell maturation was determined by FACS following staining with anti-IgD.
For RNA preparation and gene evaluation, transfection into CD74−/− B cells was performed by electroporation according to the manu-facturer's instructions (program G-16) using AMAXA R solution (Gaithersburg, MD).
DNA chip arrays
The Affymetrix GeneChip expression analysis system was used for differential expression analysis (Santa Clara, CA). The Weizmann Institute has an Affymetrix-based service facility, which routinely performs RNA labeling, hybridization, and data analysis. RNA from GFP or CD74-GFP HEK-293–transfected cells was reverse-transcribed to cDNA in the presence of biotinylated nucleotides. The target cDNA was then hybridized to the GeneChip under stringent conditions. The hybridized probe array was stained with a streptavidin phycoerythrin conjugate, and scanned using the GeneArray scanner (Affymetrix).
CD74 stimulation
First, 107 primary B cells were suspended in 1 mL RPMI medium containing 10% (vol/vol) FCS. Next, 1 μg antibody specific for mouse and human CD74 (C-16; Santa Cruz Technologies, Santa Cruz, CA) or control antibody (Santa Cruz Technologies) was added to cells that were then incubated for various time periods.
MIF stimulation
Primary B cells (107) were incubated in RPMI medium containing 0.1% (vol/vol) FCS, at 37°C for 3 hours. Cells were then resuspended in medium containing 100 ng/mL recombinant MIF and incubated at 37°C for various time points. Recombinant MIF was purified from an expression system as previously described, and contaminating endotoxin was removed by C8 chromatography.16
Cell lysis by hot SDS
Cell pellets (from 107 cells) were resuspended in a preboiled solution of 0.5% sodium dodecyl sulfate (SDS); 50 mM triethanolamine, pH 7.4; 0.1 mM NaCl; and 2 mM EDTA, heated to 99°C for 2 minutes, frozen in liquid nitrogen, boiled again for 2 minutes, and sonicated. The lysates obtained were supplemented with 2% Triton X-100, 5 mM iodoacetamide, and SDS loading buffer, and boiled prior to loading onto SDS–polyacrylamide gel electrophoresis (PAGE) gels.
Western blot analysis
To detect changes in protein expression, lysates were separated on 10% (wt/vol) or 5% to 15% (wt/vol) SDS-PAGE. The proteins then were transferred onto a nitrocellulose membrane and probed with anti-p63 (H137; Santa Cruz Technologies) followed by horseradish peroxidase–conjugated antirabbit (Dako, Glostrup, Denmark), or anti–Bcl-2 (C-2; Santa Cruz Technologies), followed by horseradish peroxidase–conjugated antimouse (Jackson Laboratories, West Grove, PA). The membrane was then stripped with 0.1% ponceau in 5% acetic acid for 10 minutes and reprobed with antitubulin antibody (Sigma) followed by peroxidase-conjugated antimouse (Jackson Laboratories).
RNA isolation and reverse transcription
Total RNA was isolated from cells using the Tri Reagent Kit (Molecular Research Center, Cincinnati, OH). Reverse transcription was carried out using Superscript II RT (Gibco-BRL, Grand Island, NY). Primers that were used for PCR reactions included the following: p63 human, (sense) 5′-GACAGAGTGTGCTGGTACCTTATG-3′, (antisense) 5′-TGAGTTCCAGGGACTCTTTGAT-3′; TA p63, (sense) 5′-GCCCATTGACTTGAACTTTGTG-3′, (antisense) 5′-GCTGGGCTGTGCGTAGG-3′; ΔN p63 (sense) 5′-TTGTACCTGGAAAACAATGC-3′, (antisense) 5′-GCTGGGCTGTGCGTAGG-3′; p63 alpha human, (antisense) 5′-GGAGCCCACACTGACTGTA-3′; p63 gamma human, (antisense) 5′-GGCTCCACA-AGCTCATTC-3′; p63 gamma mouse, (antisense) 5′-ATGGGTACACG-GAGTGGTTTGG-3′; p63 alpha mouse, (antisense) 5′-GAGCCCACAC-TGACGGTAGAGG-3′; p50 NFκB, (sense) 5′-AGATGGCCCATAC-CTTCAAAT-3′, (antisense) 5′-TCCGCCTTCTGCTTGTAGATAG-3′; p65 NFκB, (sense) 5′-GCCCAGACCGCAGTATCCATA-3′, (antisense) 5′-CAGCCTGGTCCCGTGAAATA-3′; Bcl-2, (sense) 5′-CAGGGCGATGTTGTCC-3′, (antisense) 5′-CTGGCATCTTCTCCTTCC-3′; HPRT, (sense) 5′-GAGGGTAGGCTGGCCTATGGCT-3′, (antisense) 5′-GTTGGATACAGGCCAGACTTTGTTG-3′; actin, (sense) 5′-TGAAGTGTGACGTGGACATCCG-3′, (antisense) 5′-GCTGTCACCTTCACCGTTCCAG-3′.
Real-time reverse-transcription–PCR analysis
Total RNA was extracted with the NucleoSpin kit (Macherey-Nagel, Düren Germany). From each RNA sample, 2μg was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Promega) using random hexamer primers. Real-time PCR was done on an ABI 7000 machine (Applied Biosystems, Foster City, CA) with Syber Green PCR Mastermix (Applied Biosystems). Primers used were as follows: human TAp63 (sense, AAGATGGTGCGACAAACAAG; antisense, AGAGAGCATCGAAGGTGGAG); human HPRT (sense, TGACACTGGCAAAACAATGCA antisense, GGTCCTTTTCACCAGCAAGCT).
Luciferase assay for monitoring p63 activation
Subconfluent HEK-293 cells were cotransfected in a 24-well plate, using a total of 1 μg plasmid, either empty or containing p65 (500 ng),6 or CD74 expression vectors (100 ng), in the presence or absence of I-κB,6 and together with either the TAp63 promoter sequence (75 ng) or ΔNp63 (100 ng) linked to the luciferase reporter, and 1 ng RSV Renilla luciferase RNA. The total amount of DNA was kept constant by adding an empty vector. Cells were harvested 24 hours later, and both luciferase and Renilla luciferase activities were measured.
Chromatin immunoprecipitation (ChIP)
The chromatin immunoprecipitation assay was performed on the basis of previously published methods but with some modifications.17 HEK-293 cells were transfected with empty TAαp63 or TAγp63 constructs. After 24 hours, the cells were cross-linked in vivo with 1% formaldehyde for 10 minutes at room temperature. The cells were washed once with cold phosphate-buffered saline (PBS), incubated with 1:5 trypsin for 10 minutes, then washed again with PBS and harvested. Cells were then resuspended in 1 mL lysis buffer (5 mM PIPES [pH 8.0], 85 mM KCl, and 0.5% NP-40) for 10 minutes. Nuclei were collected by centrifugation (3200g) for 8 minutes. Nuclei were lysed with 100 μL lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, and 1% SDS solution) for 10 minutes on ice and diluted with 1.5 mL 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl (pH 8.1), and 167 mM NaCl. The samples were then sonicated 10 times for 20 seconds each time. The extract was then clarified by centrifugation for 15 minutes in a microcentrifuge at 4°C and diluted 10-fold with dilution buffer solution to yield solubilized chromatin. Next, the extract was precleared by addition of 50 μL 50% suspension of protein A–Sepharose beads per milliliter and incubation for 30 minutes on a rotator wheel. After a 15-minute centrifugation at 4°C, the extract was transferred to a new tube. A 5-μL sample of the soluble extract was analyzed on a 1% agarose gel to confirm an average size of 2 kb for the DNA fragments and to normalize the relative amount of the input material for each sample. For immunoprecipitation, 1 μg anti-p63 (clone 4A4; BD Biosciences, Pharmingen) and 1 μg anti-p63 (clone H137; Santa Cruz Technologies), or 1 μg HA-probe (mouse monoclonal IgG 2a, F-7; Santa Cruz Technologies) and 1 μg HA-probe (rabbit polyclonal IgG, Y-11; Santa Cruz Technologies) as a control antibody were used. Antibodies were added together with 30 μL of a blocked 50% slurry of protein A–Sepharose beads and incubated for 16 hours at 4°C. The beads were then washed sequentially with washing buffer I (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], and 167 mM NaCl), once with washing buffer 2 (2 mM EDTA, 50 mM Tris-HCl [pH 8.0], 0.2% sarkosyl), twice with washing buffer 3 (100 mM Tris-HCl [pH 9.0], 500 mM LiCl, 1% NP-40, 1% deoxycholic acid), and once more with TE. The immune complexes were then eluted off the Sepharose beads by incubating the beads in elution buffer (1% SDS, 0.1 M NaHCO3, 20 μg glycogen per mL) at 65°C for 16 hours to reverse the formaldehyde cross-links. The eluates were purified in a PCR purification kit (Qiagen, Valencia, CA). A 2-μL sample was used for PCR. All solutions were complemented with protease inhibitor cocktail 9 (Sigma).
PCR was performed on 10 ng extracted DNA using oligomers specific for Bcl2 and GAPDH promoters.
The following oligomers were used: Bcl2 promoter, 5′-3750 TCTGGGCTCGAAGGATTCTC-3′ 4378 CAGACCTGTCTCAAGACTG; GAPDH promoter, 5′-GGCTCTCCAGAACATCATC-3′ AGCCAAATTCGTTGTCATAC.
siRNA transfection and analysis
First, 107 freshly prepared splenocytes were transfected with siRNA using Dharmafect no. 2 reagent (Dharmacon, Lafayette, CO). Cells were incubated in 0.6 mL Opti-mem medium (Gibco-BRL). Then, 100 pmol siRNA in 0.2 mL Opti-mem was added to 0.2 mL Opti-mem with 4 μL Dharmafect reagent. The solution was incubated for 15 minutes at room temperature. The oligo mix was added to cells and incubated for 6 hours at 37°C.
Medium was changed to 10% FCS in RPMI and chosen antibodies were added for an additional 24 hours: si LacZ (AAGTGACCAGCGAATACCTGT) and si p63 (CAGATCAAGGTGATGACCCCA).
Generation of chimeric mice
C57BL/6 WT mice were exposed to a single lethal dose (9.5 gray) of total body irradiation, followed by intravenous transfer of 3 × 106 (in 100 mL PBS) wild-type or p63−/−12 fetal liver cells. The mice were allowed to rest for 8 weeks before use.
Flow cytometry
Staining was performed as previously described.10 Cells were incubated with RA3–6B2 anti-CD45R (BD Biosciences) and annexin (BD Biosciences) with or without PI (Bender MedSystems, Vienna, Austria) for 15 minutes at room temperature.
Electrophoretic mobility shift assay
Nuclear extracts were prepared from HEK-293 cells after transfection of NF-κB subunits as indicated. A double-stranded oligonucleotide containing the NF-κB site was labeled with the Klenow fragment of DNA polymerase I as probe. The oligonucleotide sequence was 5′-GATCCAGAGGGGACTTTTCCGAGAG-3′; 5′-GATCCTCTCGGAAAGTCCCCTCTG-3′.
Accession numbers
Accession numbers were as follows: AF075430: TAp63 alpha human; AF075428: TAp63 gamma human; AF075431: ΔNp63 alpha human; AF075429: DNp63 gamma human; AF075436: TAp63 alpha mouse; AF075434: TAp63 gamma mouse; AF075439: ΔNp63 alpha mouse; AF075437: ΔNp63 gamma mouse; AH010181: p63 gene human; AY220759: BCL2 gene human (Homo sapiens); NC 000018.8: BCL2 gene human (Homo sapiens); NM 000633: BCL2 alpha isoform human; NM 000657: BCL2 beta isoform human; NM 177410: BCL2 var2 mouse; NM 009045: p65 NF-κB mouse; NM 008689: p50 (105) NF-κB mouse; NM 021975: p65 NF-κB human; NM 003998: p50 (105) NF-κB human.
Results
TAp63 is a target gene of CD74
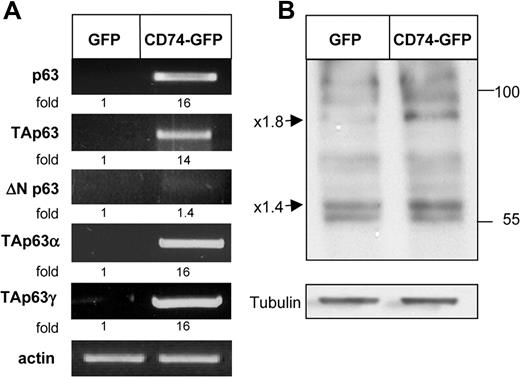
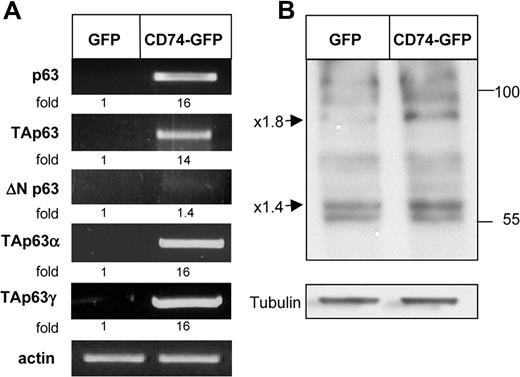
To identify target genes of CD74, we transfected HEK-293 cells with full-length (FL) CD74-GFP constructs or empty vector (Figure 1), and used the Affymetrix GeneChip expression analysis system to compare the expression patterns of RNA from these 2 populations. In this analysis, RNA from CD74+ HEK-293 cells was compared with RNA derived from CD74− cells. Many genes were found to be differentially expressed in these populations; one striking example was the p53-related gene p63, whose expression was markedly elevated in the CD74+ cells. To verify that p63 expression is indeed modulated by CD74, we first analyzed its transcript levels by RT-PCR in HEK-293 cells transfected with full-length CD74, in comparison with cells transfected with an empty vector. As seen in Figure 2A (top row), CD74 indeed dramatically elevated p63 mRNA levels.
CD74 up-regulates TAp63 α and γ expression. (A) HEK-293 cells were transfected with GFP or CD74-GFP constructs for 20 hours. Total RNA was isolated and reverse transcription was carried out using Superscript II RT. RT-PCR was performed to determine p63, TAp63, ΔNP63, TAp63α, and TAp63γ mRNA levels. The results presented are representative of 3 to 10 experiments. (B) HEK-293 cells were transfected with GFP or CD74-GFP constructs for 20 hours. Cells were collected and lysed as described in “Cell lysis by hot SDS,” and lysates were separated on 5% to 15% (wt/vol) gradient SDS-PAGE and blotted with anti-p63 antibody followed by HRP-conjugated antirabbit antibodies. The arrows indicate bands of 80 and 56 kDa, representing p63TAα and p63TAγ. The results presented are representative of at least 4 separate experiments. The intensity of the p63 band was divided by the intensity of the actin or tubulin band. The activation fold ratio in the GFP-transfected cells was normalized to 1, and the ratio for each transcription was calculated as the intensity of the sample, relative to 1.
CD74 up-regulates TAp63 α and γ expression. (A) HEK-293 cells were transfected with GFP or CD74-GFP constructs for 20 hours. Total RNA was isolated and reverse transcription was carried out using Superscript II RT. RT-PCR was performed to determine p63, TAp63, ΔNP63, TAp63α, and TAp63γ mRNA levels. The results presented are representative of 3 to 10 experiments. (B) HEK-293 cells were transfected with GFP or CD74-GFP constructs for 20 hours. Cells were collected and lysed as described in “Cell lysis by hot SDS,” and lysates were separated on 5% to 15% (wt/vol) gradient SDS-PAGE and blotted with anti-p63 antibody followed by HRP-conjugated antirabbit antibodies. The arrows indicate bands of 80 and 56 kDa, representing p63TAα and p63TAγ. The results presented are representative of at least 4 separate experiments. The intensity of the p63 band was divided by the intensity of the actin or tubulin band. The activation fold ratio in the GFP-transfected cells was normalized to 1, and the ratio for each transcription was calculated as the intensity of the sample, relative to 1.
The p63 gene exhibits a high sequence and structural homology to p53. The p63 gene contains 2 transcriptional start sites that are used to generate transcripts encoding proteins with (TAp63) or without an N-terminal transactivation domain (ΔNp63). Both TAp63 and ΔNp63 transcripts can be alternatively spliced, to generate proteins with different C termini, termed α, β, and γ. At least 6 variants can be generated from the 2 promoters of the p63 gene in combination with the 3 different C termini.11
To identify the specific p63 isoforms whose expression is up-regulated following CD74 expression, we analyzed mRNA levels of TAp63 and ΔNp63 in transfected HEK-293 cells. As shown in Figure 2A, while ΔNp63 transcription was hardly changed in response to CD74 expression, mRNA levels of the TAp63 α and γ isoforms were dramatically elevated in CD74-transfected cells. We next analyzed p63 protein levels following CD74 transfection. As shown in Figure 2B, the expression of 2 p63 isoforms (approximately 80 and 56 kDa), corresponding to the expected sizes of TAp63 α and γ, was up-regulated in the presence of CD74, whereas the other bands detected by the anti-p63 antibody were practically unaltered. Thus, CD74 induces the expression of the p63 TA isoforms at both RNA and protein levels.
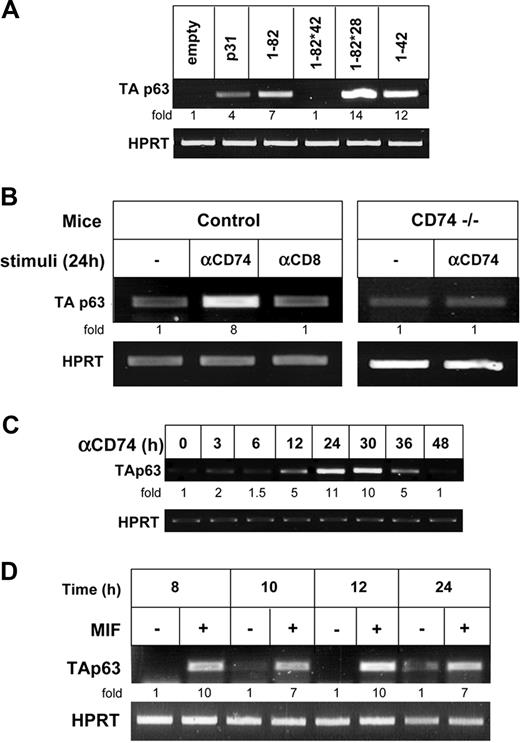
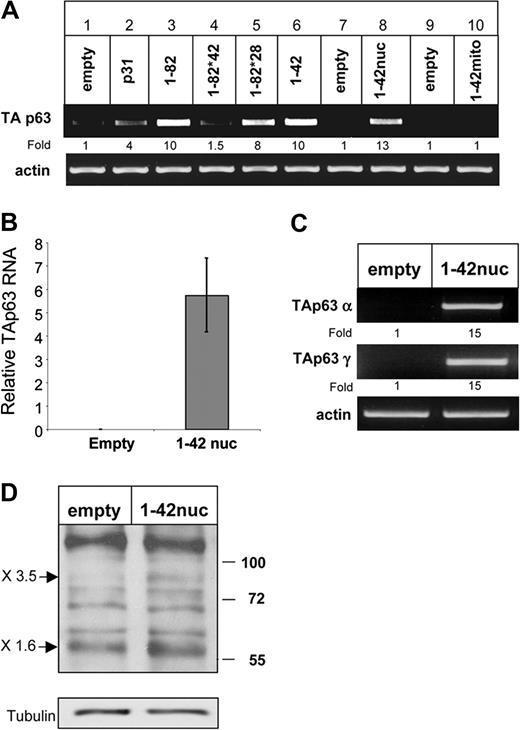
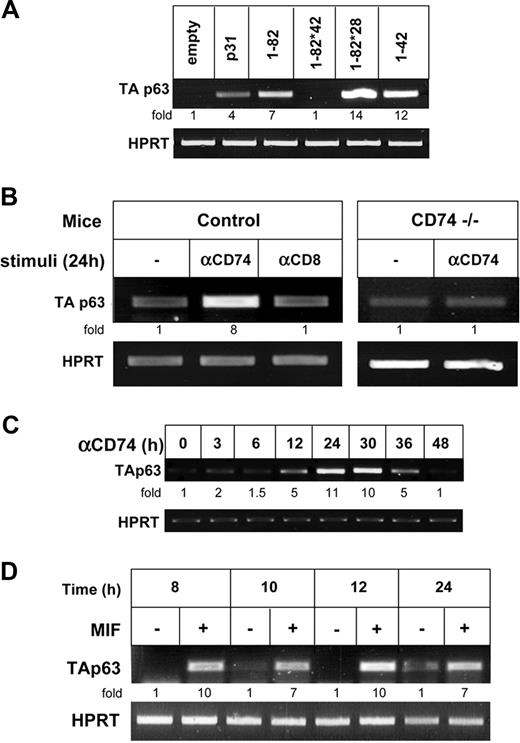
CD74 is cleaved in the plane of the membrane and releases its cytosolic domain, which activates NF-κB in the nucleus. To study whether the release of the CD74 cytosolic fragment (CD74-ICD) regulates p63 expression, we transfected HEK-293 cells with various constructs of CD74 and monitored TAp63 transcription. As shown in Figure 3A, while full-length p31 CD74 elevated TAp63 expression (lane 2), truncated CD74 (aa's 1-82; Figure 1), which lacks most of the lumenal domain, did so significantly better, probably due to its ability to release CD74-ICD more efficiently.8 To determine whether CD74-ICD itself controls TAp63 expression, we analyzed TAp63 mRNA levels in cells transfected either with CD74-ICD (aa's 1-42; Figure 1) or with the 1-82 construct whose cleavage site is mutated at amino acids 42-44, and therefore cannot be cleaved to release CD74-ICD (1-82 *42; Figure 1).7 As shown in Figure 3A lane 6, CD74-ICD induced expression of TAp63, whereas uncleavable CD74 did not (Figure 3A lane 4). In addition, mutated long-lived CD74-ICD (1-82 *28; Matza et al7 ), whose degradation is inhibited due to the absence of a destruction box motif, elevated TAp63 mRNA levels. The induction of TAp63 mRNA by CD74-ICD was also confirmed by quantitative real-time RT-PCR analysis (Figure 3B). We therefore propose that the liberated CD74 cytosolic domain regulates TAp63 transcription.
Translocation of CD74-ICD to the nucleus is required for TAp63 transcription. (A) HEK-293 cells were transfected for 20 hours with various constructs of CD74. Transfection efficiency was determined by analyzing the mRNA levels of a sequence that appears in all CD74 constructs. RNA was isolated and TAp63 transcription was determined by RT-PCR. The results presented are representative of 3 different experiments. (B) HEK-293 cells were transfected with empty vector or a construct containing CD74-ICD conjugated to a nuclear localization signal (1–42 nuc). Total RNA was extracted and real-time PCR was performed as described in “Real-time reverse-transciption–PCR analysis.” mRNA levels are shown after normalization for the HPRT control. Error bars represent SD. (C,D) HEK-293 cells were transfected with empty vector or a construct containing CD74-ICD conjugated to a nuclear localization signal (1-42 nuc). TAp63α and γ transcription was followed by RT-PCR. The results presented are representative of 8 independent experiments (C). Western blot showing protein expression of p63 isotypes. Cells were collected and lysed as described in “Cell lysis by hot SDS,” and lysates were separated on a 5% to 15% (wt/vol) gradient SDS-PAGE and blotted with anti-p63 antibody, followed by HRP-conjugated antirabbit antibodies. Arrows indicate bands of 80 and 56 kDa, representing p63TAα and p63TAγ. The results presented are representative of 5 different experiments (D). The intensity of the p63 band was calculated as described in Figure 2.
Translocation of CD74-ICD to the nucleus is required for TAp63 transcription. (A) HEK-293 cells were transfected for 20 hours with various constructs of CD74. Transfection efficiency was determined by analyzing the mRNA levels of a sequence that appears in all CD74 constructs. RNA was isolated and TAp63 transcription was determined by RT-PCR. The results presented are representative of 3 different experiments. (B) HEK-293 cells were transfected with empty vector or a construct containing CD74-ICD conjugated to a nuclear localization signal (1–42 nuc). Total RNA was extracted and real-time PCR was performed as described in “Real-time reverse-transciption–PCR analysis.” mRNA levels are shown after normalization for the HPRT control. Error bars represent SD. (C,D) HEK-293 cells were transfected with empty vector or a construct containing CD74-ICD conjugated to a nuclear localization signal (1-42 nuc). TAp63α and γ transcription was followed by RT-PCR. The results presented are representative of 8 independent experiments (C). Western blot showing protein expression of p63 isotypes. Cells were collected and lysed as described in “Cell lysis by hot SDS,” and lysates were separated on a 5% to 15% (wt/vol) gradient SDS-PAGE and blotted with anti-p63 antibody, followed by HRP-conjugated antirabbit antibodies. Arrows indicate bands of 80 and 56 kDa, representing p63TAα and p63TAγ. The results presented are representative of 5 different experiments (D). The intensity of the p63 band was calculated as described in Figure 2.
To determine whether the translocation of CD74-ICD into the nucleus is essential for TAp63 transcriptional regulation, we analyzed the ability of the CD74-ICD fragment conjugated to a nuclear localization signal (1-42 nuc; Figure 1)8 to induce TAp63 expression, relative to a CD74 1-42 fragment linked to a mitochondrial targeting signal (1-42 mito; Figure 1).8 As shown in Figure 3A, lanes 8 and 10, and in Figure 3B, translocation of CD74-ICD to the nucleus resulted in the augmentation of TAp63 expression. CD74-ICD specifically elevated the mRNA levels of the TAp63 α and γ isoforms (Figure 3C). In addition, translocation of the CD74 cytosolic domain to the nuclei of HEK-293 cells elevated the expression of the 56- and 80-kDa forms of p63 proteins, corresponding in size to TAp63 α and γ (Figure 3D). Thus, the activity of CD74-ICD in the nucleus is essential for TAp63 α and γ expression.
We next studied whether CD74 similarly regulates TAp63 expression in primary B cells; to this end, we analyzed its expression in CD74−/− primary B splenocytes transfected with various constructs of CD74. As shown in Figure 4A, expression of full-length and truncated (aa's 1-82; Figure 1) CD74 induced TAp63 transcription in primary B cells. The release of CD74 cytsolic domain was shown to be essential for maintaining TAp63 mRNA levels, since CD74-ICD (1-42) and a construct with mutated long-lived CD74-ICD (1-82*28) induced elevated mRNA levels, while in cells transfected with CD74 that cannot release its cytosolic domain (1-82*42), TAp63 was significantly reduced, indicating that TAp63 is a target gene of CD74 in primary B cells as well. We previously demonstrated that stimulation of CD74 expressed on the surface of B cells initiates a signaling cascade that results in NF-κB activation.10 To determine whether activation of cell surface CD74 induces p63 expression, primary B cells from control and CD74−/− mice were stimulated with anti-CD74 or control anti-CD8 antibodies. As demonstrated in Figure 4B, stimulation of CD74 specifically augmented TAp63 transcription in control cells, but not in CD74−/− B cells. The elevation of TAp63 mRNA levels was time dependent; TAp63 expression was first observed 12 hours following stimulation, and reached a peak at 30 hours (Figure 4C). Finally, since MIF was shown to be a ligand of CD74,4 we tested whether MIF could induce expression of TAp63. B splenocytes were incubated in the presence or absence of MIF (100 μg/mL), and TAp63 mRNA levels were monitored at different time points. As shown in Figure 4D, higher TAp63 mRNA levels were detected in the MIF-treated cells, implying that MIF induces transcription of TAp63.
Activation of CD74 expressed on primary B cells controls TAp63 transcription. (A) Primary B cells were transfected for 8 hours with various constructs of CD74 using AMAXA reagents, as described in “Cell transfection.” Transfection efficiency was determined by analyzing the mRNA levels of a sequence that is shared in all CD74 constructs. RNA was isolated and TAp63 transcription was determined by RT-PCR. (B) Control IgD+ or CD74−/− B cells were incubated in the presence or absence of anti-CD74 antibody or control anti-CD8 antibody for 24 hours. Total RNA was isolated and RT-PCR was performed, as described in “RNA isolation and reverse transciption.” The results presented are representative of 3 different experiments. (C) Control IgD+ cells were incubated in the presence or absence of anti-CD74 antibody, for various lengths of time. Total RNA was isolated and RT-PCR was performed, as described in “RNA isolation and reverse transciption.” The results presented are representative of 3 different experiments. (D) Primary B cells were incubated with MIF 100 μg/mL for various lengths of time. Total RNA was isolated and RT-PCR was performed, as described in “RNA isolation and reverse transcription.” The results presented are representative of 3 different experiments. The intensity of the p63 band was calculated as described in Figure 2.
Activation of CD74 expressed on primary B cells controls TAp63 transcription. (A) Primary B cells were transfected for 8 hours with various constructs of CD74 using AMAXA reagents, as described in “Cell transfection.” Transfection efficiency was determined by analyzing the mRNA levels of a sequence that is shared in all CD74 constructs. RNA was isolated and TAp63 transcription was determined by RT-PCR. (B) Control IgD+ or CD74−/− B cells were incubated in the presence or absence of anti-CD74 antibody or control anti-CD8 antibody for 24 hours. Total RNA was isolated and RT-PCR was performed, as described in “RNA isolation and reverse transciption.” The results presented are representative of 3 different experiments. (C) Control IgD+ cells were incubated in the presence or absence of anti-CD74 antibody, for various lengths of time. Total RNA was isolated and RT-PCR was performed, as described in “RNA isolation and reverse transciption.” The results presented are representative of 3 different experiments. (D) Primary B cells were incubated with MIF 100 μg/mL for various lengths of time. Total RNA was isolated and RT-PCR was performed, as described in “RNA isolation and reverse transcription.” The results presented are representative of 3 different experiments. The intensity of the p63 band was calculated as described in Figure 2.
Activation of the p65 member of the NF-κB family by CD74 induces p63 expression
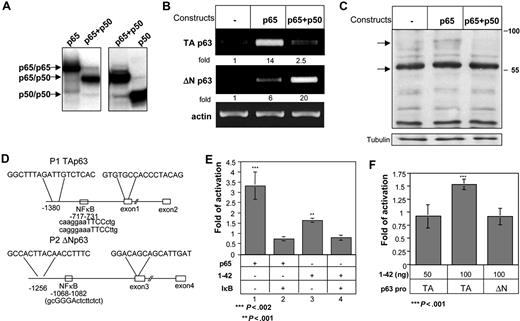
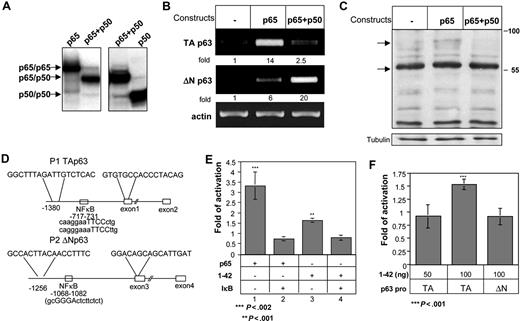
Analysis of the p63 TA and ΔN promoter regions revealed NF-κB–binding sites in both (Figure 5C). Since the p65 subunit of NF-κB is activated by CD74,6,10 we next wished to determine whether p65 specifically regulates TAp63 transcription. Accordingly, HEK-293 cells were transfected with empty vector, or expression plasmids for p65 alone or p65 together with the p50 subunit.6 Formation of p65-p65 homodimer or p65-p50 heterodimer in transfected cells was verified by electrophoresis mobility shift assay (EMSA) using labeled NF-κB–binding site as a probe (Figure 5A). As shown in Figure 5B, while expression of p65 alone resulted in elevation of TAp63 mRNA levels, coexpression of both p65 and p50 preferentially augmented the transcription of ΔNp63. Parallel protein analysis revealed elevated expression of the TAp63 isoforms (Figure 5C) in p65-transfected cells. These observations suggest that whereas p65/p50 NF-κB heterodimers promote the expression of ΔNp63 isoforms, p65/p65 homodimers preferentially induce the transcription of TAp63.
CD74 induces activation of the p65 member of the NF-κB family, which in turn activates TAp63 transcription. HEK-293 cells were transfected with empty, p65, or p65 + p50 constructs for 20 hours. (A) Analysis of HEK-293 cells transfected with NF-κB proteins by electrophoresis mobility shift assay using the NF-κB–binding site as DNA probe. (B) Nuclear extracts from p65-, p65 + p50–, and p50-transfected cells were used for DNA-binding reactions as indicated at the top the lanes. Total RNA was isolated, and RT-PCR was performed as described in “RNA isolation and reverse transciption.” (C) Cells were then collected and lysed, as described in “Cell lysis by hot SDS.” Lysates were separated on 5% to 15% (wt/vol) gradient SDS-PAGE, and blotted with anti-p63 antibody, followed by HRP-conjugated antirabbit antibodies. Arrows indicate bands of 80 and 56 kDa, representing p63TAα and p63TAγ. (D) Schematic representation of the p63 promoters. The sequences indicated are the PCR primers used for generating the luciferase constructs. (E,F) TAp63 activation was analyzed by luciferase assay, as described in “Luciferase assay for monitoring p63 activation.” (E) The HEK-293 cells were transfected with a luciferase construct containing the TAp63 promoter, together with a p65 construct or CD74-ICD (aa's 1-42), in the absence or presence of IκB. Cells were lysed, and luciferase activity was measured. Luciferase activities were normalized to the activity of the cotransfected RSV promoter–driven Renilla reporter luciferase, which was used to correct for differences in transfection efficiencies. Fold activation was calculated as the activity of p65 or CD74 constructs relative to the activity of the empty plasmid. The results presented are representative of 3 different experiments. (F) HEK-293 cells were transfected for 20 hours with luciferase constructs containing the TAp63 or ΔNp63 promoters, together with CD74-ICD (aa 1-42's). Cells were then lysed and luciferase activities were normalized to the activity of cotransfected RSV promoter–driven Renilla reporter luciferase, which was used to correct for differences in transfection efficiencies. Fold activation was calculated as the activity of the CD74-ICD construct, relative to the activity of cells transfected with an empty plasmid. The graph represents the average of 5 independent experiments. The results presented are representative of 3 different experiments. The intensity of the p63 band was calculated as described in Figure 2. Error bars represent SD.
CD74 induces activation of the p65 member of the NF-κB family, which in turn activates TAp63 transcription. HEK-293 cells were transfected with empty, p65, or p65 + p50 constructs for 20 hours. (A) Analysis of HEK-293 cells transfected with NF-κB proteins by electrophoresis mobility shift assay using the NF-κB–binding site as DNA probe. (B) Nuclear extracts from p65-, p65 + p50–, and p50-transfected cells were used for DNA-binding reactions as indicated at the top the lanes. Total RNA was isolated, and RT-PCR was performed as described in “RNA isolation and reverse transciption.” (C) Cells were then collected and lysed, as described in “Cell lysis by hot SDS.” Lysates were separated on 5% to 15% (wt/vol) gradient SDS-PAGE, and blotted with anti-p63 antibody, followed by HRP-conjugated antirabbit antibodies. Arrows indicate bands of 80 and 56 kDa, representing p63TAα and p63TAγ. (D) Schematic representation of the p63 promoters. The sequences indicated are the PCR primers used for generating the luciferase constructs. (E,F) TAp63 activation was analyzed by luciferase assay, as described in “Luciferase assay for monitoring p63 activation.” (E) The HEK-293 cells were transfected with a luciferase construct containing the TAp63 promoter, together with a p65 construct or CD74-ICD (aa's 1-42), in the absence or presence of IκB. Cells were lysed, and luciferase activity was measured. Luciferase activities were normalized to the activity of the cotransfected RSV promoter–driven Renilla reporter luciferase, which was used to correct for differences in transfection efficiencies. Fold activation was calculated as the activity of p65 or CD74 constructs relative to the activity of the empty plasmid. The results presented are representative of 3 different experiments. (F) HEK-293 cells were transfected for 20 hours with luciferase constructs containing the TAp63 or ΔNp63 promoters, together with CD74-ICD (aa 1-42's). Cells were then lysed and luciferase activities were normalized to the activity of cotransfected RSV promoter–driven Renilla reporter luciferase, which was used to correct for differences in transfection efficiencies. Fold activation was calculated as the activity of the CD74-ICD construct, relative to the activity of cells transfected with an empty plasmid. The graph represents the average of 5 independent experiments. The results presented are representative of 3 different experiments. The intensity of the p63 band was calculated as described in Figure 2. Error bars represent SD.
To directly demonstrate that the cleaved CD74 peptide (1-42 nuc) is able to induce p63 expression by elevating NF-κB transcriptional activity, we generated expression plasmids carrying a luciferase reporter downstream to the TAp63 or ΔNp63 promoters, which contain NF-κB–binding sites at positions −717 to −731 and −1068 to −1082, respectively (Figure 5D). The reporter plasmids were transfected into HEK-293 cells along with p65, I-κB, or CD74-ICD plasmids, and the RSV promoter, used as a reference. As shown in Figure 5E, p65 activated the TAp63 promoter (lane 1), and this was down-regulated by overexpression of I-κB (lane 2). To determine whether CD74 can induce p63 transcription, CD74-ICD (1-42 nuc) was transfected into HEK-293 cells in the presence or absence of I-κB. As demonstrated in Figure 5D (lanes 3-4), CD74 could induce TAp63 promoter activity, albeit to a lesser extent than p65 activation, while I-κB inhibited this activation. This suggests that CD74 stimulates TAp63 transcription through NF-κB activation. To further illustrate the specificity of CD74 activation, we compared its ability to activate the TAp63 and ΔNp63 promoters. While CD74-ICD induced TAp63 promoter activity, it had no effect on the ΔNp63 promoter, indicating that CD74-ICD specifically activates TAp63 transcription (Figure 5F).
TAp63 induces a survival cascade in B cells
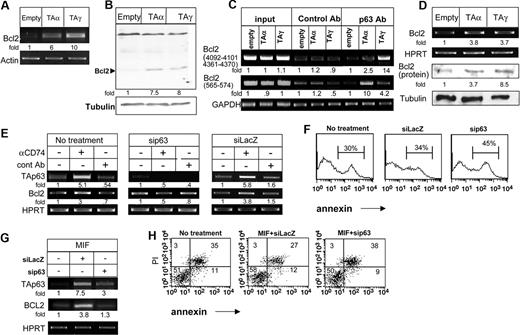
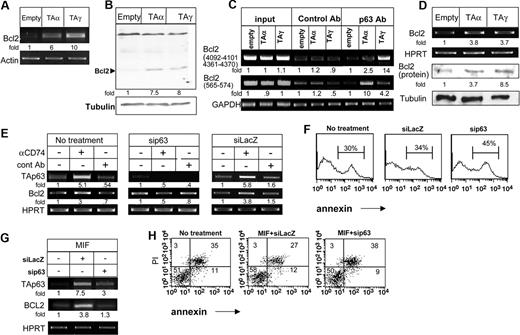
We recently demonstrated that stimulation of cell surface CD74 leads to initiation of a survival cascade.10 To determine whether this survival cascade is mediated through TAp63 activity, expression plasmids encoding the TAp63 α and γ isoforms were transfected into HEK-293 cells, and the transcription and translation of the antiapoptotic protein Bcl-2 was then analyzed. As shown in Figure 6A,B, both TAp63 isoforms up-regulated Bcl-2 mRNA (Figure 6A) and protein (Figure 6B), confirming that, indeed, TAp63 can induce a survival cascade. To gain further insight into the regulation of Bcl-2 expression, we analyzed whether p63 binds directly to the Bcl-2 promoter. Sequence analysis of the promoter revealed 3 potential p63-binding sites.18 We therefore designed oligonucleotides spanning these potential sites, and analyzed p63 binding by chromatin immunoprecipitation (ChIP). As seen in Figure 6C, TAp63γ and TAp63α were indeed found to be associated with the Bcl-2 promoter (Figure 6C).
TAp63 regulates Bcl-2 expression. (A-B) HEK-293 cells were transfected with empty, p63TAα, and p63TAγ constructs for 24 hours. (A) Total RNA was isolated, and RT-PCR for Bcl-2 was performed as described in “RNA isolation and reverse transciption.” The results presented are representative of 4 different experiments. (B) Cells were collected and lysed as described in “Cell lysis by hot SDS,” and lysates were separated on 10% (wt/vol) SDS-PAGE and blotted with anti–Bcl-2 antibody, followed by HRP-conjugated antimouse antibodies. The membrane was stripped and reblotted with antitubulin. The arrow indicates the Bcl-2 band. The results presented are representative of 3 separate experiments. (C) ChIP analysis of p63 binding to the Bcl-2 promoter. HEK-293 cells were transfected with empty, p63TAα, and p63TAγ constructs for 24 hours. Chromatin prepared from these cells was immunoprecipitated with control and anti-p63 antibodies. Presence of the promoter sequence was then quantified by RT-PCR. (D) Primary B cells were transfected for 8 hours with various constructs of p63 using AMAXA reagents, as described in “Cell transfection.” RNA was isolated and Bcl-2 transcription was determined by RT-PCR. The results presented are representative of 3 different experiments. (E-H) siRNA for p63 or lacZ (control) was transfected into primary B cells in the presence or absence of anti-CD74 (E) or MIF (G,H) stimulation. Total RNA was isolated, and RT-PCR was performed as described in “RNA isolation and reverse transciption.” The results presented are representative of 4 different experiments (E,G). Annexin staining of B220+ cells was performed (F). Annexin and PI staining of B220+ cells was performed (H). The intensity of the p63 band was calculated as described in Figure 2. Numbers on the plots in panel H are percentages of total cells.
TAp63 regulates Bcl-2 expression. (A-B) HEK-293 cells were transfected with empty, p63TAα, and p63TAγ constructs for 24 hours. (A) Total RNA was isolated, and RT-PCR for Bcl-2 was performed as described in “RNA isolation and reverse transciption.” The results presented are representative of 4 different experiments. (B) Cells were collected and lysed as described in “Cell lysis by hot SDS,” and lysates were separated on 10% (wt/vol) SDS-PAGE and blotted with anti–Bcl-2 antibody, followed by HRP-conjugated antimouse antibodies. The membrane was stripped and reblotted with antitubulin. The arrow indicates the Bcl-2 band. The results presented are representative of 3 separate experiments. (C) ChIP analysis of p63 binding to the Bcl-2 promoter. HEK-293 cells were transfected with empty, p63TAα, and p63TAγ constructs for 24 hours. Chromatin prepared from these cells was immunoprecipitated with control and anti-p63 antibodies. Presence of the promoter sequence was then quantified by RT-PCR. (D) Primary B cells were transfected for 8 hours with various constructs of p63 using AMAXA reagents, as described in “Cell transfection.” RNA was isolated and Bcl-2 transcription was determined by RT-PCR. The results presented are representative of 3 different experiments. (E-H) siRNA for p63 or lacZ (control) was transfected into primary B cells in the presence or absence of anti-CD74 (E) or MIF (G,H) stimulation. Total RNA was isolated, and RT-PCR was performed as described in “RNA isolation and reverse transciption.” The results presented are representative of 4 different experiments (E,G). Annexin staining of B220+ cells was performed (F). Annexin and PI staining of B220+ cells was performed (H). The intensity of the p63 band was calculated as described in Figure 2. Numbers on the plots in panel H are percentages of total cells.
We previously demonstrated that CD74 stimulation induces the survival of a mature B-cell population in an in vitro culture.10 To show that TAp63 induces a survival cascade in B cells, immature B cells deficient in CD74 were transfected with TAp63α or TAp63γ expression plasmids, and Bcl-2 expression was analyzed. As shown in Figure 6D, both TAp63α and TAp63γ up-regulated Bcl-2 mRNA and protein levels, indicative of a survival signal. To validate that CD74 induces Bcl-2 expression in a p63-dependent manner, splenocytes were treated with small interfering RNA (siRNA) specific for p63, and Bcl-2 mRNA levels were then analyzed. Down-regulation of p63 expression eliminated the elevation of Bcl-2 mRNA levels following anti-CD74 (Figure 6G) or MIF (Figure 6E) stimulation. In parallel, the knock-down of p63 increased the apoptotic subpopulation, in unstimulated (Figure 6F) or MIF stimulated (Figure 6H) cells, consistent with an antiapoptotic role of p63 in this cell system, presumably mediated at least in part through its ability to up-regulate Bcl2 expression.
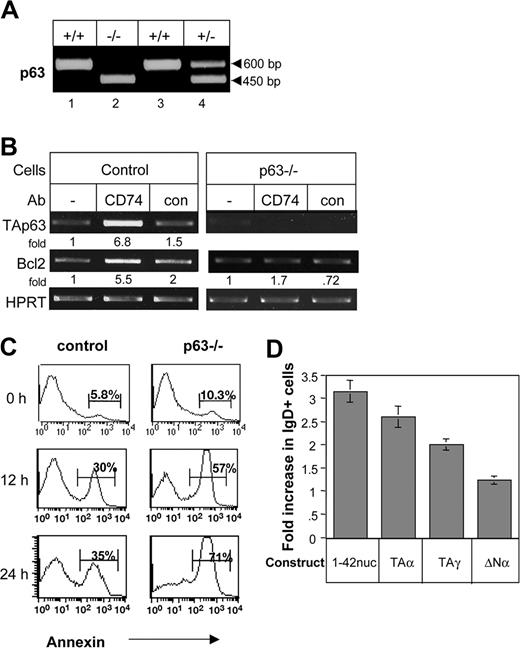
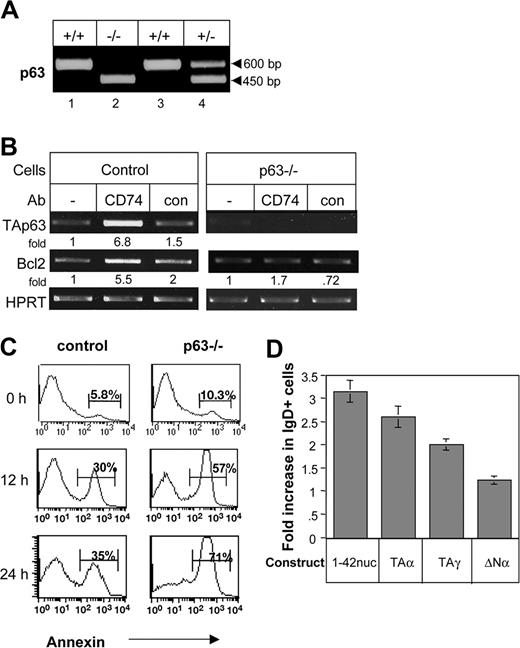
To determine whether TAp63 regulates the survival of B cells by maintenance of the mature B-cell population, we used chimeric mice. To that end, adult C57BL/6 mice were irradiated and injected with fetal liver cells from wild-type (wt) or p63-deficient mice12 (Figure 7A). B cells from chimeric mice were purified and stimulated with anti-CD74 or control antibody, and their Bcl-2 mRNA levels were analyzed. Lower levels of Bcl-2 mRNA were observed in B cells lacking p63 (Figure 7B), accompanied by an elevation in the apoptotic B-cell population (Figure 7C). In addition, while CD74 stimulation up-regulated Bcl-2 mRNA in control B cells, it did not affect Bcl-2 mRNA levels in p63−/− B cells (Figure 7B). Thus, p63 regulates Bcl-2 levels and is required for Bcl2 up-regulation following CD74 stimulation.
p63 regulates B-cell survival. (A-C) DNA screening of the wild-type (+/+), heterozygotic (+/−), and p63−/− (−/−) fetal liver cells (as previously described12 ) that were injected into irradiated mice (A). Splenocytes derived from the chimeric mice were stimulated with or without anti-CD74, and TAp63 or Bcl-2 mRNA levels were determined by RT-PCR (B). Annexin staining was performed on freshly isolated splenocytes (time 0) and on cells cultured for 12 or 24 hours. Histograms show the annexin-positive population of B220+ cells. The results presented are representative of 4 different experiments (C). (D) Primary CD74−/− B cells were transfected with empty plasmid, CD74-ICD, TAp63α, TAp63γ, or ΔNp63 constructs as described in “Cell transfection.” After 48 hours, the IgD+ population was analyzed by FACS. The results presented are the mean of 3 independent experiments. Error bars represent SD. The percentage increase of IgD+ cells was calculated by subtracting staining of empty expression plasmid from the percentage of IgD+ cells in each treatment, and dividing it into the same increased value, multiplied by 100%. The results presented are representative of 5 different experiments. The intensity of the p63 band was calculated as described in Figure 2.
p63 regulates B-cell survival. (A-C) DNA screening of the wild-type (+/+), heterozygotic (+/−), and p63−/− (−/−) fetal liver cells (as previously described12 ) that were injected into irradiated mice (A). Splenocytes derived from the chimeric mice were stimulated with or without anti-CD74, and TAp63 or Bcl-2 mRNA levels were determined by RT-PCR (B). Annexin staining was performed on freshly isolated splenocytes (time 0) and on cells cultured for 12 or 24 hours. Histograms show the annexin-positive population of B220+ cells. The results presented are representative of 4 different experiments (C). (D) Primary CD74−/− B cells were transfected with empty plasmid, CD74-ICD, TAp63α, TAp63γ, or ΔNp63 constructs as described in “Cell transfection.” After 48 hours, the IgD+ population was analyzed by FACS. The results presented are the mean of 3 independent experiments. Error bars represent SD. The percentage increase of IgD+ cells was calculated by subtracting staining of empty expression plasmid from the percentage of IgD+ cells in each treatment, and dividing it into the same increased value, multiplied by 100%. The results presented are representative of 5 different experiments. The intensity of the p63 band was calculated as described in Figure 2.
Finally, CD74-deficient immature B cells were transfected with empty plasmid, CD74-ICD, TAp63α, TAp63γ, or ΔNp63 constructs and grown in vitro as previously described.6 After 48 hours, the cells were analyzed for their mature population. A specific increase in the mature (IgD high, IgM low) B-cell population was observed in the CD74-ICD–, as well as the TAp63α- and TAp63γ-, transfected cells (Figure 7D) but not ΔNp63-transfected cells. This, elevation resulted from the survival of the mature population, since no change was detected in the total cell number (data not shown). These findings suggest that CD74, by inducing a cascade that involves TAp63, controls the survival of the mature B-cell population, allowing their accumulation.
Discussion
The pool of peripheral B cells in mice consists of approximately 100 million cells, the majority of which are mature, follicular B cells with a lifespan ranging from several weeks to months. Most of these cells circulate in the periphery in a quiescent state, without actively contributing to an acute immune response. This vast number of resting cells must be maintained to preserve a diverse B-cell repertoire. Lasting B-cell persistence in the periphery is dependent on survival signals that are transduced by cell surface receptors.19
In B cells, an indispensable survival function has been established for at least 2 individual receptors, the B-cell receptor (BCR) complex and the receptor for the cytokine B-cell activating factor of the TNF family (BAFF). A shared outcome of both BCR and BAFF-R signaling is NF-κB activation. NF-κB is one of the principal transcription factors that regulates B-cell development, maintenance, and stimulation.20 We show here that CD74 is an additional survival receptor expressed on B cells, which induces survival by augmenting NF-κB activation and TAp63 expression. Currently, the interplay between these 2 survival cascades is not clear. BAFF-deficient animals show almost complete loss of the mature and marginal zone B cells. The remaining B lymphocytes are mostly the T1 transitional B cells, which appear in almost normal numbers, while there are almost no cells of a T2 phenotype. Bone marrow cells are present in nearly normal numbers; only the normal population of recirculating mature B lymphocytes is absent.21 This phenotype is similar but not identical to CD74-deficient mice. In these mice, there is an accumulation of the T1 population with a dramatic reduction of mature cells. However, CD74-deficient mice exhibit an accumulation of the marginal zone population.5,22 We suggest that for generation of a complete repertoire of peripheral B cells, both pathways must operate simultaneously.
We previously demonstrated that CD74 expressed on the surface of mature B cells initiates a signaling pathway that activates NF-κB and subsequent cell survival. In the current study, we followed the genes whose expression is induced by the activation of CD74. We showed that stimulation of CD74 on the cell surface induces the activation of the p65/RelA member of the NF-κB family, which in turn up-regulates TAp63 transcription and expression. TAp63 binds to the Bcl-2 promoter and induces the transcription of Bcl-2 mRNA and production of the Bcl-2 antiapoptotic protein, which enhances cell survival. Thus, TAp63 regulates an antiapoptotic pathway in mature B cells, thereby controlling their survival.
Deletion of p63 results in embryos with severe limb truncations, craniofacial malformations, and, most importantly, the absence of skin and most epithelial tissues, as well as defective epidermal differentiation.12,13 Thus, p63 plays a role in developmental regulation. In view of the severe effects of p63 loss on epithelia, the role of p63 has so far been studied primarily in epithelial cells. Notably, it was shown that basal cells of normal human epithelium, including the epidermis, strongly express p63 proteins, predominantly the ΔNp63 isoform (at a ratio of approximately 100 ΔNp63 to 1 TAp63;11 ) but lose expression of these proteins as soon as the cells withdraw from the early progenitor compartment.23 In addition, ΔNp63 isoforms are abundantly expressed in the progenitor cell layers of skin, breast, and prostate, while TAp63 proteins are barely detectable, indicating a switch in expression of p63 isoforms during normal cellular differentiation (for a review, see McKeon24 TAp63 isoforms are detected in differentiated cells in normal stratified epithelia, in the differentiated luminal cells in breast and prostate, and in the epithelium of the colon.25
We now report that p63, and more specifically its TA isoforms, also plays an important role in the B-cell lineage, particularly in maintaining the population of mature B cells following their terminal differentiation. These results extend the observation that TAp63 isoforms are detected in differentiated cells.24 Overall, our findings suggest that TAp63 controls the survival of the mature population and might regulate the immune response by shaping the B-cell repertoire.
In most cell types studied thus far, TA isoforms of p63 were shown to possess proapoptotic functions, whereas delta-N isoforms were often found to exert an opposing effect and thus display features consistent with a pro-oncogenic role.26,–28 This is also reflected in gene expression profiling analysis, where TAp63 was found to activate a variety of proapoptotic genes.29 Surprisingly, we show here that TAp63 can also exhibit a very different behavior, favoring cell survival rather than cell death. It thus appears that the outcome of p63 induction is strongly dependent on the cellular context, and therefore can vary greatly among different cell types. Specifically, our data suggest that TAp63 may subserve a prosurvival program in B cells, as part of a broader antiapoptotic response induced by cell surface receptors such as CD74, and mediated by a distinct pattern of NF-κB activation.
There is increasing evidence that p63 may play a role in human cancers; in fibroblasts, ectopic p63 expression induces anchorage-independent growth and tumor formation in nude mice.30 Of particular note, overexpression of p63 TA isoforms is frequently detected in B-cell malignancies, such as diffuse large B-cell lymphoma31,32 and follicular lymphoma.33 Moreover, high levels of TAp63 RNA and protein were found to correlate with higher grade, more aggressive disease, and a higher proliferative index. This is consistent with the conclusion that TAp63 may be involved in the progression of B-cell–derived malignancies. It is tempting to speculate that a TAp63-mediated transcriptional program, which in normal B cells is triggered only in response to environmental cues, becomes constitutively activated in malignant B cells. This might enable illegitimate survival and perhaps also excessive proliferation of such B cells, thereby driving disease progression. It will be interesting to determine whether the CD74-p65/RelA-TAp63 axis is indeed continuously active in these hematopoietic malignancies, and whether it contributes to their pathogenesis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by The Israel Science Foundation (founded by the Academy of Sciences and Humanities), Abisch Frankel Foundation, the Israel Cancer Association, Minerva foundation, and a kind donation from Mrs Nancy and Mr Peter Brown, as well as a grant from the Robert Bosch Foundation (M.O.). I.S. is the incumbent of the Dr Morton and Anne Kleiman Professorial Chair.
The authors thank Dr Frank McKeon for his generous gift of the p63−/− mice. The authors gratefully acknowledge members of the Shachar laboratory for helpful discussions.
Authorship
Contribution: F.L. designed and performed research, analyzed data, and wrote the paper; D.S. designed research and analyzed data; Y.G., L.F., and A.Y.-H. performed research and analyzed data; R.D., M.O., and I.S. designed research, analyzed data, and wrote the paper; L.L. contributed reagents; R.B. contributed reagents and wrote the paper; Y.M. performed research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Idit Shachar, Department of Immunology, Weizmann Institute of Science, Rehovot 76100, Israel; e-mail:idit.shachar@weizmann.ac.il.