Binding of multiple myeloma (MM) cells to bone marrow stromal cells (BMSCs) triggers expression of adhesive molecules and secretion of interleukin-6 (IL-6), promoting MM cell growth, survival, drug resistance, and migration, which highlights the possibility of developing and validating novel anti-MM therapeutic strategies targeting MM cells–host BMSC interactions and their sequelae. Recently, we have found that expression of the peroxisome proliferator-activated receptor γ (PPARγ) and its ligands can potently inhibit IL-6–regulated MM cell growth. Here we demonstrate that PPARγ agonists 15-d-PGJ2 and troglitazone significantly suppress cell-cell adhesive events, including expression of adhesion molecules and IL-6 secretion from BMSCs triggered by adhesion of MM cells, as well as overcome drug resistance by a PPARγ-dependent mechanism. The synthetic and natural PPARγ agonists have diverging and overlapping mechanisms blocking transactivation of transcription factors NF-κB and 5′-CCAAT/enhancer–binding protein β (C/EBPβ). Both 15-d-PGJ2 and troglitazone blocked C/EBPβ transcriptional activity by forming PPARγ complexes with C/EBPβ. 15-d-PGJ2 and troglitazone also blocked NF-κB activation by recruiting the coactivator PGC-1 from p65/p50 complexes. In addition, 15-d-PGJ2 had a non–PPARγ-dependent effect by inactivation of phosphorylation of IKK and IκB. These studies provide the framework for PPARγ-based pharmacological strategies targeting adhesive interactions of MM cells with the bone marrow microenvironment.
Introduction
Multiple myeloma (MM) is a malignancy of differentiated B lymphocytes characterized by accumulation of clonal plasma cells in the bone marrow, accounts for 10% of all hematologic cancers, and remains an incurable hematologic malignancy.1,,,,,,–8 This highlights the urgent need for novel biologically based treatment strategies.9 Binding of MM cells to bone marrow stromal cells (BMSCs) triggers both adhesion- and cytokine-mediated MM cell growth, survival, drug resistance, and migration. The interaction of myeloma cells with the BM stromal cells is believed to be mediated by the cell surface antigens called adhesion molecules. Interactions between very late antigen 4 (VLA-4 [CD29-CD49d]) and its ligand vascular cellular adhesion molecule 1 (VCAM-1 [CD106]) and between lymphocyte function-associated antigen 1 (LFA-1 [CD11a-CD18]) and its ligand intercellular adhesion molecule (ICAM-1 [CD54]) play a role in the binding of multiple myeloma cells to BMSCs.10
MM cell binding to BMSCs up-regulates IL-6 secretion from BMSCs. IL-6 subsequently activates signal pathways and their downstream targets, including cytokines and antiapoptotic proteins in MM cells. IL-6 seems primarily involved in myeloma osteolysis, as well as in the growth and survival of malignant plasma cells. Clinically, serum IL-6 and IL-6 receptors are prognostic factors in MM, reflective of the proliferative fraction of tumor cells.11,12 Although some MM cells secrete IL-6 and grow in an autocrine fashion, IL-6 is primarily produced in BMSCs induced by either MM cell adhesion or cytokines and mediates paracrine MM cell growth.5 Thus, it should be advantageous to find new anti-MM agents that potentially target molecular consequences of the adhesive interaction between MM cells and BMSCs and related IL-6 secretion.
The peroxisome proliferator-activated receptor γ (PPARγ) is a prototypical member of the nuclear receptor super family, functions as a ligand-dependent transcription factor, and is activated by diverse synthetic and naturally occurring substances. Although most studies concern the regulation of glucose and lipid metabolism by PPARγ because of its abundant expression in adipocytes,13 recent research studies have suggested that this nuclear receptor might also play a number of additional roles in inflammation, atherosclerosis, and cancer.14,15 We have previously found expression of PPARγ in IL-6–responsive MM cells. The PPARγ agonist 15-deoxy-Δ12,14 -prostaglandin J2 (15-d-PGJ2) and troglitazone completely abolished IL-6–inducible MM cell growth through transcriptional inactivation of the IL-6/Stat3 signaling pathway.16 The PPARγ ligands also induced multiple myeloma cell apoptosis.16,–18 These data suggest PPARγ may serve as a significant molecular target for treatment of multiple myeloma.
In this study, we investigate the effect of PPARγ activation on adhesion of MM tumor cells to stromal cells and IL-6 production. The results show that PPARγ and its ligands effectively inhibit adhesive interaction between MM and BMSCs, overcome drug resistance, and also block induced IL-6 transcription and secretion from BMSCs through PPARγ competition for its coactivator PGC-1 recruiting NF-κB and direct association with C/EBPβ. The endogenous ligand 15-d-PGJ2 also had a direct effect on inactivation of NF-κB through decreasing phosphorylation of IKK and IκB.
Materials and methods
Materials
Troglitazone, 15-d-PGJ2, and WY16463 were purchased from Biomol Research Laboratories (Plymouth Meeting, PA). Dexamethasone was from Sigma (St Louis, MO). Tissue culture materials were from Life Technologies (Gaithersburg, MD). Human IL-6 was obtained from PeproTech (Rocky Hill, NJ). IκBα, phospho-IκBα, IKKα, and phospho-IKKα antibodies were purchased from Upstate Biotechnology (Lake Placid, NY). Fluorochrome- and phytoerythrin-conjugated antibodies were purchased from BD BioSciences (San Jose, CA). Calcein-acetoxymethyl (calcein-AM) was obtained from Invitrogen Molecular Probes (Carlsbad, CA).
Cell lines and cell culture
The human marrow stromal cell line HS-5, human multiple myeloma cell lines MM.1S and MM.1R, and mouse MM cell line AP were grown in RPMI-1640 medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, and penicillin-streptomycin (50 IU/mL and 50 μg/mL, respectively). The IL-6–dependent MM cell lines, KAS6/1 and ANBL-6, kindly provided by Dr Diane Jelinek, Mayo Clinic, were maintained in RPMI-1640 medium containing 10% fetal calf serum, 2 mM l-glutamine, penicillin-streptomycin (50 IU/mL and 50 μg/mL, respectively), and IL-6 (1 ng/mL).
BMSC cultures
Bone marrow specimens were obtained from a healthy donor after informed consent was obtained in accordance with the Declaration of Helsinki and under a protocol approved by the National Cancer Institute Institutional Review Board for the use of samples for the purpose of research, and mononuclear cells separated by Ficoll-Hypaque density sedimentation was used to establish long-term BM cultures, as previously described by Hideshima et al.19
Cytokine measurements
IL-6 concentrations in culture supernatants were determined by an enzyme-linked immunosorbent assay (R&D Systems, Minneapolis, MN).
Ribonuclease protection assays
Total RNA was isolated using TRIzol from treated and control cells. The mRNA was examined by RNase protection assay.20
Adhesion assay21
Bone marrow stromal cells were grown in 96-well plates and starved in RPMI-1640 medium supplemented with 0.5% fetal calf serum overnight. Myeloma cells were incubated with calcein-AM for 1 hour at 37°C and 5% CO2. The fluorescence-labeled myeloma cells were added into stromal cell–coated 96-well plates and incubated at indicated time points. Nonadherent calcein-labeled cells were removed by gently washing twice with RPMI-1640 by inversion of the plates. Adherent cells were quantitated in a fluorescence multiwell plate reader (Molecular Devices, Sunnyvale, CA).
EMSA
Transfection of SiRNA for PPARγ
HS-5 cells were grown overnight in normal growth medium containing serum without antibiotics. The constructs of SiRNA-PPARγ or control vector (Panomics, Redwood City, CA) were diluted with Opti-MEM I Reduced Serum Medium (Invitrogen), then combined with the diluted FuGene-6, and incubated for 20 minutes at room temperature to allow DNA/FuGene-6 complexes to form. The above DNA/FuGene-6 complexes were added into the plate or flask containing the cells and cultured at 37°C in a CO2 incubator for 96 hours. Efficacy of the constructs was tested through transfection into HS-5 cells and Western blot analysis of the respective target PPARγ in transfected cells.
Transfection of luciferase reporter plasmids
FuGene-6 (Roche, Indianapolis, IN) was used to transfect the C/EBP-luciferase (C/EBP-luc) or NF-κB-luciferase (NF-κB-Luc) gene reporters, wild-type, or various mutant IL-6 promoter luciferase reporter plasmids,22,–24 kindly provided by Dr Bernd Stein at Signal Pharmaceuticals (San Diego, CA), into HS-5 cells.
Coimmunoprecipitation assays
Cells were lysed in 10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, and 0.5% Nonidet P-40. Immunoprecipitation was carried out using anti-PPARγ or anti-PGC1 antibody. Western blots were performed by anti–NF-κB (p50 or p65), anti-PPARγ, anti–PGC-1, or anti-C/EBP antibody (Santa Cruz Biotechnology, Santa Cruz, CA) as previously described.21
Flow cytometric analysis
Cells were incubated with mouse Fc blocking buffer (BD Pharmingen, San Diego, CA) before immunolabeling for 30 minutes at 4°C with fluorescence-conjugated antibodies. Cells were analyzed using a fluorescence-activated cell sorting (FACS) Calibur flow cytometer (BD Biosciences). Data were acquired from 10 000 gated events and staining was compared with isotype control antibodies.
Statistical analysis
Data are presented as the mean plus or minus SE. Treatment groups were compared by Student t test, with P value reported in each case.
Results
PPARγ agonists inhibit MM cell adherence to bone marrow stromal cells
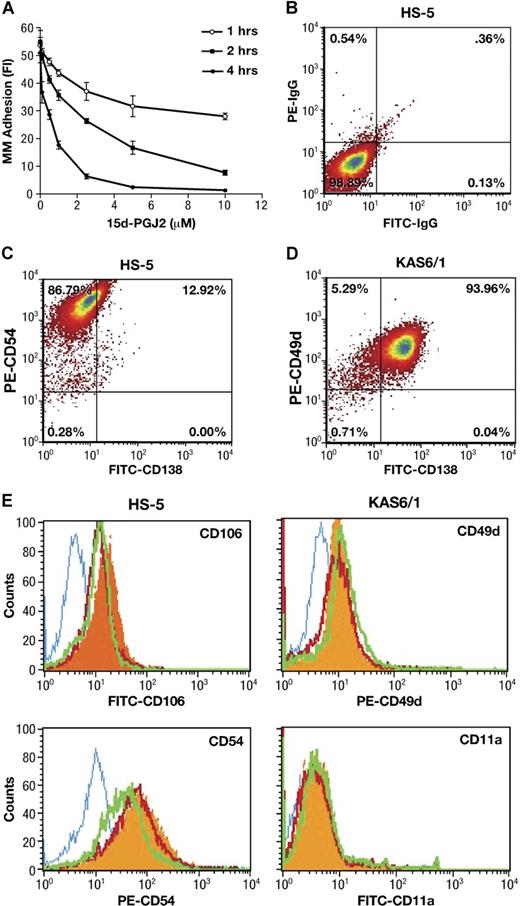
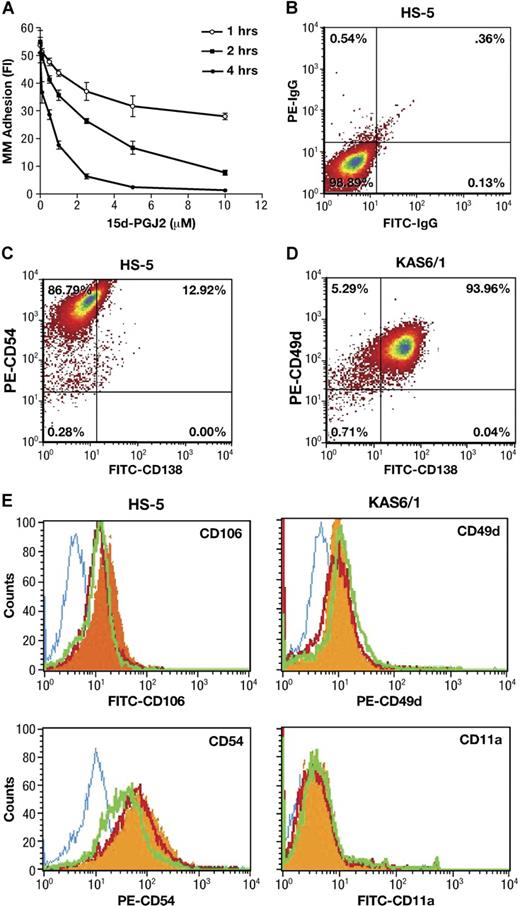
Interactions between malignant plasma cells and stroma may be crucial for the localization as well as the egress of tumor cells from sites of origin. We first used a calcein-AM fluorescence-based adhesion assay25 to explore whether PPARγ directly affects cell-cell adhesion between BMSCs and MM. As shown in Figure 1A, 15-d-PGJ2 significantly reduced KAS6/1 myeloma cells binding to HS-5 stromal cells in a dose- and time-dependent manner. In addition, similar inhibitory effects of PPARγ ligands 15-d-PGJ2 and troglitazone were observed (Table 1) with fluorescence-labeled KAS6/1 cells binding to primary human BMSCs, or mouse AP myeloma cells adhering to primary murine BMSCs, respectively. These data indicate that PPARγ agonists inhibit MM cell adherence to bone marrow stromal cells.
Effect of PPARγ on adherence of MM cells to bone marrow stromal cells. (A) Human KAS6/1 cells were labeled with calcein-AM. After 1 hour, the above fluorescence-labeled KAS6/1 cells were washed and added into HS-5 cell–coated 96-well plates. The cocultured cells were treated with 15-d-PGJ2 and incubated for 1, 2, or 4 hours. After washing, the fluorescence intensity of remaining labeled adhesive MM cells was tested using a fluorescence microplate reader. (B-E) HS-5 cells were cocultured with KAS6/1 cells in the presence or absence of 15-d-PGJ2 or troglitazone for 24 hours. Cells were harvested, washed with PBS, and stained with fluorescence-labeled antibodies. The expression of adhesion molecules was determined using a FACS Calibur flow cytometer. (B) HS-5 cell harvests were stained with FITC-IgG and PE-IgG control; (C) HS-5 cell harvests were stained with FITC-anti-CD138 and PE-anti-CD54; (D) KAS6/1 cell harvests were stained with FITC-labeled anti-CD138 and PE-anti-CD49d; and (E) HS-5 cell harvests were stained with mouse IgG isotype control or fluorescence-conjugated mouse antihuman antibodies against CD106 or CD54. KAS6/1 cell harvests were labeled with fluorescence-conjugated antibodies against CD49d or CD11a, respectively. Solid histogram indicates antigens for untreated cells; open histogram (blue), isotype control; open histogram (red), 15-d-PGJ2–treated cells; open histogram (green), troglitazone-treated cells.
Effect of PPARγ on adherence of MM cells to bone marrow stromal cells. (A) Human KAS6/1 cells were labeled with calcein-AM. After 1 hour, the above fluorescence-labeled KAS6/1 cells were washed and added into HS-5 cell–coated 96-well plates. The cocultured cells were treated with 15-d-PGJ2 and incubated for 1, 2, or 4 hours. After washing, the fluorescence intensity of remaining labeled adhesive MM cells was tested using a fluorescence microplate reader. (B-E) HS-5 cells were cocultured with KAS6/1 cells in the presence or absence of 15-d-PGJ2 or troglitazone for 24 hours. Cells were harvested, washed with PBS, and stained with fluorescence-labeled antibodies. The expression of adhesion molecules was determined using a FACS Calibur flow cytometer. (B) HS-5 cell harvests were stained with FITC-IgG and PE-IgG control; (C) HS-5 cell harvests were stained with FITC-anti-CD138 and PE-anti-CD54; (D) KAS6/1 cell harvests were stained with FITC-labeled anti-CD138 and PE-anti-CD49d; and (E) HS-5 cell harvests were stained with mouse IgG isotype control or fluorescence-conjugated mouse antihuman antibodies against CD106 or CD54. KAS6/1 cell harvests were labeled with fluorescence-conjugated antibodies against CD49d or CD11a, respectively. Solid histogram indicates antigens for untreated cells; open histogram (blue), isotype control; open histogram (red), 15-d-PGJ2–treated cells; open histogram (green), troglitazone-treated cells.
PPARγ agonists decrease cell-cell contact-mediated expression of adhesive molecules
Multiple myeloma cells localize within the bone marrow via an interaction of cell surface adhesion molecules with their respective ligands on bone marrow stromal cells and extracellular matrix proteins.26 We, therefore, examined the expression of integrin ligands (VCAM-1 and ICAM-1) on HS-5 cells and their receptors (VLA-4 and LFA-1) in KAS6/1 cells. Adherent HS-5 cells were stimulated with KAS6/1 cells and treated with 15-d-PGJ2 or troglitazone for 24 hours. At the end of incubation, KAS6/1 cells were washed out and the remaining HS-5 cells were harvested. Cell harvests were washed with PBS and stained with fluorescence-labeled antibodies. The expression of adhesion molecules was determined by FACS.
Since adhering myeloma cells may affect the purity of HS-5 stromal cell harvests even after the washout, we first used anti-CD138 staining (a specific cell surface marker for MM cells) to assay the phenotype of either HS-5 or KAS6/1 cell harvest after washout. The results are shown in Figure 1B-D. Approximately 10% to 15% CD138+ cells were detected in HS-5 harvest after washout, whereas approximately 94% CD138+ cells were detected in KAS6/1 myeloma cell harvest. After treatment with PPARγ agonists, the CD138+ cells in HS-5 harvest were reduced from 12.9% (± 2.6%) for control group to 3.7% (± 0.9%) for the 15-d-PGJ2–treated group, or 0.5% (± 0.1%) for the troglitazone-treated group (P < .05 versus control group). These data suggested that the stromal cell harvest contained a little population of adhering MM cells after washout, although most of the MM cells could be removed from the coculture system. PPARγ agonists decreased adhesion of MM cells to BMSCs, which was consistent with the results (Figure 1A) obtained from the calcein-AM fluorescence-based adhesion assay.
Next, the HS-5 cells were stained with fluorescence-labeled anti-CD106 or anti-CD54; the KAS6/1 cells were stained with fluorescence-labeled anti-CD49d or anti-CD11a. The above-labeled cells were analyzed using FACS (Figure 1E). In the case of HS-5 (CD138−) stromal cells stimulated by KAS6/1 binding, the expression of VCAM-1 (CD106) and ICAM (CD54) was inhibited by treatment of 15-d-PGJ2 and troglitazone. For KAS6/1 (CD138+) myeloma cells, the expression of their respective receptors VLA-4 (CD49d) and LFA-1 (CD11a) was not significantly inhibited by 15-d-PGJ2 and troglitazone (Table 2). This suggests PPARγ activation principally inhibits expression of adhesive molecules (VCAM and ICAM) on bone marrow stromal cells.
PPARγ agonists reduced IL-6 secretion triggered by adhesion of MM cells to BMSCs
Stimulation of myeloma cells via these cell surface molecules, either directly or via tumor cell adhesion to stromal cells, can induce autocrine or paracrine production of IL-6.27 We, therefore, tested whether the PPARγ agonists affected the levels of soluble IL-6 in cocultured MM and BMSCs systems. As shown in Table 3, binding of KAS6/1 myeloma cells, like TNFα stimulation, induced strong increases in IL-6 secretion from human primary BMSCs. In contrast, IL-6 secretion by MM cells alone was negligible, which was consistent with the previous reports by Chauhan et al27 Troglitazone and 15-d-PGJ2 significantly reduced IL-6 release from fresh primary human BMSCs induced by KAS6/1 cell binding or TNFα stimulation, whereas these ligands had little effect on the basal level of IL-6 secretion by BMSCs themselves. Similar results on IL-6 secretion were obtained from stromal cell line HS-5 cells triggered by binding of KAS6/1 or ANBL-6 myeloma cells (Table 4). Troglitazone and 15-d-PGJ2, but not the PPARα agonist WY14643, reduced IL-6 release from HS-5 cells induced by adhesion of human MM cells.
Effect of PPARγ ligands on interaction between stromal cells with drug-resistant MM cells
Since the effectiveness of hormonal therapy to multiple myeloma is limited by the development of resistance, it is interesting to determine whether PPARγ ligands impact interaction between stromal cells and drug-resistant MM cells. We used MM.1R, a drug-resistant multiple myeloma cell line, and its parent sensitive cell line, MM.1S, to test PPARγ expression and its ligands' effects. Like KAS6/1 cells, both MM.1S and MM.1R expressed PPARγ (Figure 2A) determined by Western blotting. As expected, dexamethasone decreased cell growth of MM.1S, but not of MM.1R, which was consistent with the previous report.28,29 Troglitazone and 15-d-PGJ2 remarkably inhibited cell growth of both MM.1S and MM.1R cells in a dose-dependent manner (Figure 2B,C). There was no obvious difference in IC50 values between MM.1R and MM.1S for both PPARγ ligands. Moreover, the effects of PPARγ ligands on drug-resistant MM cell–induced IL-6 release from stromal cells were examined. MM.1S or MM.1R cells were cocultured with HS-5 stromal cells and treated with PPARγ ligands. Troglitazone and 15-d-PGJ2 could reduce IL-6 production by HS-5 cells triggered by adhesion of both MM.1R and MM.1S cells (Figure 2D). These data indicate that PPARγ ligands overcome dexamethasone-resistant MM cell adhesion to BMSCs through inhibiting IL-6 secretion.
Effect of PPARγ ligands on Dex-resistant MM cells. (A) Expression of PPARγ on MM1S and MM.1R cell lines detected by Western blot. (B,C) Dose-dependent inhibition of PPARγ ligands on cell growth of both MM.1S and MM.1R cells. Proliferation of quiescent MM.1R (B) or MM.1S cells (C) in culture was examined following treatment with 10 ng/M IL-6 and increasing concentrations (abscissa) of 15-d-PGJ2 (○), troglitazone (□), or dexamethasone (▵) for 16 hours at 37°C. [3H]-thymidine incorporation was analyzed. (D) Effects of PPARγ ligands on drug-resistant MM cell–induced IL-6 release from stromal cells. HS-5 cells were cocultured with MM.1S or MM.1R cells and treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone for 48 hours. IL-6 concentration was determined in supernatants. Error bars represent the mean (± SE) from 3 independent experiments.
Effect of PPARγ ligands on Dex-resistant MM cells. (A) Expression of PPARγ on MM1S and MM.1R cell lines detected by Western blot. (B,C) Dose-dependent inhibition of PPARγ ligands on cell growth of both MM.1S and MM.1R cells. Proliferation of quiescent MM.1R (B) or MM.1S cells (C) in culture was examined following treatment with 10 ng/M IL-6 and increasing concentrations (abscissa) of 15-d-PGJ2 (○), troglitazone (□), or dexamethasone (▵) for 16 hours at 37°C. [3H]-thymidine incorporation was analyzed. (D) Effects of PPARγ ligands on drug-resistant MM cell–induced IL-6 release from stromal cells. HS-5 cells were cocultured with MM.1S or MM.1R cells and treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone for 48 hours. IL-6 concentration was determined in supernatants. Error bars represent the mean (± SE) from 3 independent experiments.
Inhibition of IL-6 by PPARγ agonists is relative to PPARγ expression
We further analyzed whether PPARγ ligands affect MM-induced IL-6 transcription of BMSCs cells using ribonuclease protection assays. As demonstrated in Figure 3A, KAS6/1 cells themselves showed a very low level of IL-6 mRNA expression. However, adhesion of KAS6/1 cells increased IL-6 expression by HS-5 cells. PPARγ ligands 15-d-PGJ2 or troglitazone, but not PPARα ligand WY14643, markedly inhibited IL-6 mRNA expression in HS-5 cells induced by KAS6/1 cell binding. These observations suggest that PPARγ ligands inhibit IL-6 secretion and mRNA expression of BMSCs induced by MM cell adhesion.
PPARγ ligands decreased IL-6 transcription and secretion largely by a PPARγ-dependent mechanism. (A) Adherent HS-5 cells were stimulated with KAS6/1 cells and treated with PPAR ligands 15-d-PGJ2, troglitazone, or WY14643 for 6 hours. KAS6/1 cells were washed out and collected. The remaining HS-5 cells were then harvested. The RNA samples, prepared from the above-harvested KAS6/1 cells or HS-5 cells, were hybridized with [33P]-labeled RNA probes corresponding to transcripts for individual human IL-6 gene, according to the BD PharMingen protocol. RNase protected fragments were separated on 5% polyacrylamide gel electrophoresis (PAGE). (B) SiRNA-mediated knockdown of PPARγ alters its ligands' inhibition of IL-6 production. HS-5 cells were transfected with SiRNA-PPARγ or control vector and cultured for 96 hours. The above HS-5 cells were then treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone and incubated with or without KAS6/1 cells for an additional 48 hours. IL-6 concentration was determined in supernatants. Insert figure shows knockdown of PPARγ expression in HS-5 cells transfected with SiRNA-PPARγ. HS-5 cells were transduced with SiRNA-PPARγ or control vector and incubated for 96 hours. Proteins were then extracted from the cells, and the levels of PPARγ expression were examined by Western blotting. The β-actin bands served as an internal control for equal total protein loading. (C) HS-5 cells were cotransfected with a PPARγ wild-type expression vector (pSG5-hPPARγ2) or control pSG5 plasmid, an IL-6 promoter luciferase reporter gene construct, and a pRL-TK control vector for 24 hours. Cells were then treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone and incubated with or without KAS6/1 cells for an additional 48 hours. Luciferase activity of lysed cells was measured. The level of firefly luciferase activity was normalized to that of the Renilla luciferase activity. Insert figure shows the level of PPARγ expression in HS-5 cells transfected with PPARγ wild-type expression vector. (D) Effect of PPARγ ligands on transactivation of IL-6 promoter. HS-5 cells were transfected with the indicated IL-6 promoter luciferase reporter gene constructs. Twenty-four hours after transfection, KAS6/1 cells were added directly to the transfected confluent HS-5 BMSCs pretreated with 15-d-PGJ2 or troglitazone for 2 hours. The transfected HS-5 cells were also cultured alone as a control. The cocultured cells were incubated for an additional 24 hours. After washing out KAS6/1 cells, luciferase activity of adherent HS-5 cells was measured. Error bars represent the mean (± SE) from 3 independent experiments.
PPARγ ligands decreased IL-6 transcription and secretion largely by a PPARγ-dependent mechanism. (A) Adherent HS-5 cells were stimulated with KAS6/1 cells and treated with PPAR ligands 15-d-PGJ2, troglitazone, or WY14643 for 6 hours. KAS6/1 cells were washed out and collected. The remaining HS-5 cells were then harvested. The RNA samples, prepared from the above-harvested KAS6/1 cells or HS-5 cells, were hybridized with [33P]-labeled RNA probes corresponding to transcripts for individual human IL-6 gene, according to the BD PharMingen protocol. RNase protected fragments were separated on 5% polyacrylamide gel electrophoresis (PAGE). (B) SiRNA-mediated knockdown of PPARγ alters its ligands' inhibition of IL-6 production. HS-5 cells were transfected with SiRNA-PPARγ or control vector and cultured for 96 hours. The above HS-5 cells were then treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone and incubated with or without KAS6/1 cells for an additional 48 hours. IL-6 concentration was determined in supernatants. Insert figure shows knockdown of PPARγ expression in HS-5 cells transfected with SiRNA-PPARγ. HS-5 cells were transduced with SiRNA-PPARγ or control vector and incubated for 96 hours. Proteins were then extracted from the cells, and the levels of PPARγ expression were examined by Western blotting. The β-actin bands served as an internal control for equal total protein loading. (C) HS-5 cells were cotransfected with a PPARγ wild-type expression vector (pSG5-hPPARγ2) or control pSG5 plasmid, an IL-6 promoter luciferase reporter gene construct, and a pRL-TK control vector for 24 hours. Cells were then treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone and incubated with or without KAS6/1 cells for an additional 48 hours. Luciferase activity of lysed cells was measured. The level of firefly luciferase activity was normalized to that of the Renilla luciferase activity. Insert figure shows the level of PPARγ expression in HS-5 cells transfected with PPARγ wild-type expression vector. (D) Effect of PPARγ ligands on transactivation of IL-6 promoter. HS-5 cells were transfected with the indicated IL-6 promoter luciferase reporter gene constructs. Twenty-four hours after transfection, KAS6/1 cells were added directly to the transfected confluent HS-5 BMSCs pretreated with 15-d-PGJ2 or troglitazone for 2 hours. The transfected HS-5 cells were also cultured alone as a control. The cocultured cells were incubated for an additional 24 hours. After washing out KAS6/1 cells, luciferase activity of adherent HS-5 cells was measured. Error bars represent the mean (± SE) from 3 independent experiments.
To determine whether the PPARγ ligand inhibition of IL-6 transcription and secretion is relative to PPARγ expression, we used a SiRNA for PPARγ to test whether interfering with PPARγ expression would lead to reduction of PPARγ ligand inhibition on IL-6 secretion induced by MM cell adhesion. HS-5 cells were transfected with a SiRNA-PPARγ or control vector. After transduction of SiRNA-PPARγ into HS-5 cells for 96 hours, more than 90% reduction of PPARγ expression analyzed by immunoblotting was observed (Figure 3B insert). Although treatment of PPARγ ligands results in a significant decrease of the KAS6/1 cell binding–inducible IL-6 production in the control SiRNA vector–transfected HS-5 cells, such inhibition of these PPARγ ligands 15-d-PGJ2 and troglitazone was considerably abolished in SiRNA-PPARγ–transfected HS-5 cells (Figure 3B). These results suggest transcriptional inhibition of IL-6 by PPARγ agonists is largely dependent on the expression and activation of PPARγ in the BMSCs.
We also examined whether overexpression of PPARγ affected its ligands inhibiting IL-6 transcription of BMSCs. The HS-5 cells were cotransfected with control pSG5 plasmid, a PPARγ expression plasmid, and an IL-6 promoter luciferase reporter gene construct. In the presence of 15-d-PGJ2 and troglitazone, KAS6/1 cell–inducible IL-6 promoter transactivation in PPARγ-overexpressing HS-5 cells (Figure 3C insert) was less than that in control pSG5-transfected HS-5 cells (Figure 3C), suggesting that overexpression of PPARγ enhances its ligands inhibiting IL-6 transcription by BMSCs. These results support the notion that the transcriptional inhibition of IL-6 by PPARγ agonists is relative to expression of PPARγ.
Activated PPARγ suppresses synergistic activation of IL-6 promoter by C/EBPβ and NF-κB
Since transcription factors NF-κB and C/EBPβ synergistically activate transcription of IL-6,30 we used a wild-type and a series of mutant IL-6 promoter luciferase constructs to assess whether PPARγ ligands affect C/EBPβ/NF-κB–mediated IL-6 promoter activation. HS-5 cells were transiently transfected with −224-bp IL-6 promoter construct and several mutation constructs containing a single C/EBP site mutation (−158 to −145 or −87 to −76), a single NF-κB site mutation (−75 to −63), or triple (2 C/EBP sites and 1 NF-κB site) mutations22,–24 for 24 hours; KAS6/1 cells were added to the confluent transfected HS-5 cells pretreated with 15-d-PGJ2 or troglitazone for 1 hour. The cocultured cells were incubated for another 24 hours. After KAS6/1 cells were washed out, the remaining adherent HS-5 cells were then harvested for analysis of luciferase activity (Figure 3D). Compared with transfected HS-5 cells alone with the above IL-6 promoter constructs, adherence of KAS6/1 cells resulted in a 7-fold increase in luciferase activity of transfected HS-5 cells with −224-bp wild-type construct and also a 7-fold increase in that with a single C/EBP site mutant (−158 to −145 or −87 to −76) constructs, but not that with triple (2 C/EBP and 1 NF-κB sites) mutant constructs. Both PPARγ ligands showed a significant inhibition of IL-6 promoter transactivation. In the case of the transfected HS-5 cells with the construct with single mutation of the NF-κB site, KAS6/1 binding to HS-5 cells induced a lower level of IL-6 promoter luciferase activity than that with the wild-type construct. Moreover, 15-d-PGJ2 lost its inhibition of transactivation of IL-6 promoter containing a NF-κB–binding site mutation, indicating that the NF-κB site is the most important positive regulatory element and the target site for 15-d-PGJ2 inhibition of IL-6 promoter transactivation in BMSCs. In contrast, no detectable luciferase activity was observed in transfected KAS6/1 cells themselves with an IL-6 promoter luciferase construct.
Furthermore, we assayed the effect of the different C/EBP-binding site mutations on PPARγ ligand inhibition of IL-6 promoter transactivation. Compared with inhibitory effects of PPARγ ligand on the wild-type IL-6 promoter luciferase activity stimulated by KAS6/1 cell adhesion, the extent of this suppression was markedly reduced in HS-5 cells transfected with a construct containing a mutation of the 5′ C/EBP site (−158 to −145) with intact 3′ C/EBP (−87 to −76) and NF-κB sites, whereas such PPARγ ligand inhibition was not significantly affected by the mutation of the 3′ C/EBP site with intact 5′ C/EBP and NF-κB. These data suggest that the 5′ C/EBP, but not the 3′ C/EBP site, is involved in PPARγ ligand down-regulating IL-6 promoter activity.
PPARγ cross talks with NF-κB mediated by PGC-1
Since specific targeting of NF-κB can overcome the growth and survival advantage conferred both by tumor cell binding to BMSCs and cytokine secretion in the BM milieu,9,31,–33 we performed an EMSA analysis to evaluate the effect of PPARγ in activation of NF-κB by cell-cell contact or TNF-α (5 ng/mL) stimulation. As shown in Figure 4A, the NF-κB DNA-binding activity of HS-5 cells was enhanced by stimulation of TNF-α or binding of KAS6/1 cells and could be supershifted by p50 or p65, and also attenuated by antibody (αPGC-1) to the NF-κB coactivator PGC-1. Such enhanced NF-κB DNA-binding activity was suppressed by PPARγ agonists 15-d-PGJ2 or troglitazone, but not by PPARα agonist WY14643. GW9662, a pure antagonist of PPARγ,34 could overcome this effect. Moreover, the extent of the above inhibitory effect induced by 15-d-PGJ2 is much larger than that by troglitazone. In addition, similar results on transactivation of NF-κB were obtained from HS-5 cells transfected with the 3 × NF-κB–binding sites–luciferase reporter construct (Figure 4B). The transactivation of NF-κB induced by binding of KAS6/1 or ANBL-6 cells was reduced by PPARγ agonists 15-d-PGJ2 and troglitazone, but not by PPARγ antagonist GW9662. These data suggest that PPARγ ligand inactivation of IL-6 transcription correlates to the inhibition on NF-κB binding and transactivation.
Effect of PPARγ ligands on activation of NF-κB. (A) Nuclear extracts corresponding to 5 μg protein were incubated in the absence of antibody anti-p50, anti-p65, anti–PGC-1, or normal rabbit serum (NRS) in combination with a [32P]-labeled NF-κB oligonucleotide probe. Arrows indicate migrational location of each nonsupershifted NF-κB–DNA complex. (B) HS-5 cells were transfected with a 3 × NF-κB–binding element–pGL3 promoter luciferase construct. Cells were then pretreated with 1 μM 15-d-PGJ2, 10 μM troglitazone, or 50 μM GW9662 for 2 hours, and then incubated with or without KAS6/1 or ANBL-6 cells for 24 hours. Error bars represent the mean (± SE) from 3 independent experiments. (C) The coactivator PGC-1 coimmunoprecipitates with p50 or p65 in KAS6/1 cell adhesion–induced HS-5 cells. HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then added with KAS6/1 cells for 24 hours before lyses. Western blotting analysis with anti-p65 (top panel), or p50 (middle panel), or anti–PGC-1 (bottom panel) was performed on anti–PGC-1 immunoprecipitates. (D) The coactivator PGC-1 coimmunoprecipitates with PPARγ in HS-5 cells. HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then added with KAS6/1 cells for 24 hours before lyses. Western blotting analysis with either anti-PPARγ (top panel) or anti–PGC-1 (bottom panel) was performed on anti–PGC-1 immunoprecipitates. (E) The above cell extracts were immunoprecipitated with anti-PPARγ. Western blotting analysis was performed with anti-p50 and anti-p65. (F) HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then simulated with KAS6/1 for 1 hour before lyses. Western blotting analysis was performed with anti–phospho-IKKα, anti-IKKα, anti–phospho-IκBα, or anti-IκBα.
Effect of PPARγ ligands on activation of NF-κB. (A) Nuclear extracts corresponding to 5 μg protein were incubated in the absence of antibody anti-p50, anti-p65, anti–PGC-1, or normal rabbit serum (NRS) in combination with a [32P]-labeled NF-κB oligonucleotide probe. Arrows indicate migrational location of each nonsupershifted NF-κB–DNA complex. (B) HS-5 cells were transfected with a 3 × NF-κB–binding element–pGL3 promoter luciferase construct. Cells were then pretreated with 1 μM 15-d-PGJ2, 10 μM troglitazone, or 50 μM GW9662 for 2 hours, and then incubated with or without KAS6/1 or ANBL-6 cells for 24 hours. Error bars represent the mean (± SE) from 3 independent experiments. (C) The coactivator PGC-1 coimmunoprecipitates with p50 or p65 in KAS6/1 cell adhesion–induced HS-5 cells. HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then added with KAS6/1 cells for 24 hours before lyses. Western blotting analysis with anti-p65 (top panel), or p50 (middle panel), or anti–PGC-1 (bottom panel) was performed on anti–PGC-1 immunoprecipitates. (D) The coactivator PGC-1 coimmunoprecipitates with PPARγ in HS-5 cells. HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then added with KAS6/1 cells for 24 hours before lyses. Western blotting analysis with either anti-PPARγ (top panel) or anti–PGC-1 (bottom panel) was performed on anti–PGC-1 immunoprecipitates. (E) The above cell extracts were immunoprecipitated with anti-PPARγ. Western blotting analysis was performed with anti-p50 and anti-p65. (F) HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then simulated with KAS6/1 for 1 hour before lyses. Western blotting analysis was performed with anti–phospho-IKKα, anti-IKKα, anti–phospho-IκBα, or anti-IκBα.
We further studied the molecular mechanism on PPARγ ligands inhibiting NF-κB DNA binding by testing for a physical interaction among NF-κB, PPARγ, and its coactivator PGC-1. Adherent HS-5 cells were stimulated with KAS6/1 cells and treated with 15-d-PGJ2 or troglitazone. After washing out KAS6/1 cells, the remaining bound HS-5 cells were lysed and immunoprecipitated (IP) with a PGC-1–specific antibody; immunoprecipitates were developed on Western blots (WBs) with a p50- or p65-specific antibody. NF-κB (p50 or p65) can be coprecipitated with PGC-1 in BMSCs (Figure 4C), whether KAS6/1 MM cell adhesion occurs or not, indicating that PGC-1 is constitutively associated with NF-κB, although PGC-1 was originally identified as a transcriptional coactivator of the nuclear receptor PPARγ.35,36 Notably, the complex between PGC-1 and p65 NF-κB was markedly decreased in KAS6/1 binding to HS-5 cells treated with 15-d-PGJ2 and troglitazone, although expression of the PGC-1 determined by anti–PGC-1 was unchanged by any treatment. Furthermore, the above HS-5 cell extracts were immunoprecipitated with a PGC-1–specific antibody; immunoprecipitates were developed on Western blots with a PPARγ-specific antibody. The complex between PGC-1 and PPARγ was increased upon KAS6/1 cells binding to HS-5 cells in the presence of either 15-d-PGJ2 or troglitazone (Figure 4D), suggesting that PGC-1 indeed interacts with liganded PPARγ. Moreover, such PPARγ interaction with PGC-1 induced by 15-d-PGJ2 was significantly stronger than that induced by troglitazone. However, the NF-κB (p65 or p50) could not be detected in PPARγ immunoprecipitates under the same condition (Figure 4E). Therefore, PGC-1 may function as a bridge partner to mediate NF-κB interaction with ligand-bound PPARγ, although NF-κB did not associate directly with PPARγ. The data indicate that activated PPARγ may inhibit NF-κB DNA binding through competition for limiting amounts of a cofactor PGC-1 docking with NF-κB.
15-d-PGJ2 directly inactivates IκB and IκB kinase
In addition to PPARγ-dependent functions, 15-d-PGJ2, the physiological ligand for PPARγ, has also been shown to have complex PPARγ-independent activities. Several lines of evidence suggested 15-d-PGJ2 is a cell type–specific regulator of intracellular kinases, including IκB kinase (IKK).37,–39 We, therefore, analyzed whether PPARγ ligands' inhibition of NF-κB activation in BMSCs was associated with phosphorylation of IκB and IκB kinase. The HS-5 cells lysates were immunoprecipitated with a specific antibody against IKKα or IκBα and blotted with a specific antibody against phospho-IKKα or phospho-IκBα. As Figure 4F shows, 15-d-PGJ2, but not troglitazone, could inhibit the phosphorylation of IKKα and IκBα (top panel). The blot was stripped and total levels of IKKα and IκBα were detected, which displayed the same loading levels (Figure 4F lower panel). Densitometric analysis demonstrated that 15-d-PGJ2 resulted in approximately 50% loss of phosphorylation/nonphosphorylation protein ratio for both IKKα and IκBα. These data indicated that 15-d-PGJ2, compared with a synthetic ligand troglitazone, may have additional PPARγ-independent effects on the NF-κB pathway involving specific inactivation of IKKα and IκBα, thus explaining the more potent effects of 15-d-PGJ2 on NF-κB activation, as noted in Figure 4A.
PPARγ interactions with C/EBPβ
Because the IL-6 promoter requires participation of both NF-κB and C/EBPβ,24 we also explored how C/EBPβ was involved in PPARγ ligand inhibition of IL-6 by BMSCs. HS-5 cells were stimulated with KAS6/1 cells and treated with 15-d-PGJ2 or troglitazone. After washing out KAS6/1 cells, adherent HS-5 cells were lysed; the effect on C/EBP-binding activity was detected by EMSA. The C/EBPβ DNA-binding activity of HS-5 cells was enhanced by stimulation of KAS6/1 cells and could be supershifted by anti-C/EBPβ. Such enhanced C/EBPβ DNA-binding activity was suppressed by troglitazone and 15-d-PGJ2 (Figure 5A).
PPARγ ligand inhibition of C/EBPβ mediated by PPARγ direct association with C/EBPβ. (A) HS-5 cells were pretreated with 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then incubated with KAS6/1 cells for 2 hours. After washing out KAS6/1 cells, adherent HS-5 cells were lysated. Nuclear extracts corresponding to 5 μg protein were incubated in the absence of antibody C/EBPβ or normal rabbit serum (NRS) in combination with a [32P]-labeled C/EBP oligonucleotide probe. Arrows indicate migration location of each nonsupershifted C/EBPβ-DNA complex. (B) HS-5 cells were transfected with a 3 × C/EBPβ-binding element–pGL3 promoter luciferase construct. Cells were then pretreated with 1 μM 15-d-PGJ2, 10 μM troglitazone, or 50 μM GW9662 for 2 hours, and then incubated with or without KAS6/1 or ANBL-6 myeloma cells for 24 hours. Error bars represent the mean (± SE) from 3 independent experiments. (C) The PPARγ coimmunoprecipitates with C/EBPβ in KAS6/1 adhesion-induced HS-5 cells. HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then added with KAS6/1 cells for 24 hours before lyses. Western blotting analysis with anti-C/EBPβ (top panel) or anti-PPARγ (bottom panel) was performed on anti-PPARγ immunoprecipitates. The densitometric relative units in each lane were indicated beneath each blot. (D) No significant changes in C/EBPβ expression in HS-5 cells. The HS-5 cells were treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then simulated with KAS6/1 for 12 hours before lyses. Western blotting analysis was performed with anti-C/EBPβ. The densitometric relative units in each lane were indicated beneath each blot.
PPARγ ligand inhibition of C/EBPβ mediated by PPARγ direct association with C/EBPβ. (A) HS-5 cells were pretreated with 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then incubated with KAS6/1 cells for 2 hours. After washing out KAS6/1 cells, adherent HS-5 cells were lysated. Nuclear extracts corresponding to 5 μg protein were incubated in the absence of antibody C/EBPβ or normal rabbit serum (NRS) in combination with a [32P]-labeled C/EBP oligonucleotide probe. Arrows indicate migration location of each nonsupershifted C/EBPβ-DNA complex. (B) HS-5 cells were transfected with a 3 × C/EBPβ-binding element–pGL3 promoter luciferase construct. Cells were then pretreated with 1 μM 15-d-PGJ2, 10 μM troglitazone, or 50 μM GW9662 for 2 hours, and then incubated with or without KAS6/1 or ANBL-6 myeloma cells for 24 hours. Error bars represent the mean (± SE) from 3 independent experiments. (C) The PPARγ coimmunoprecipitates with C/EBPβ in KAS6/1 adhesion-induced HS-5 cells. HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then added with KAS6/1 cells for 24 hours before lyses. Western blotting analysis with anti-C/EBPβ (top panel) or anti-PPARγ (bottom panel) was performed on anti-PPARγ immunoprecipitates. The densitometric relative units in each lane were indicated beneath each blot. (D) No significant changes in C/EBPβ expression in HS-5 cells. The HS-5 cells were treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then simulated with KAS6/1 for 12 hours before lyses. Western blotting analysis was performed with anti-C/EBPβ. The densitometric relative units in each lane were indicated beneath each blot.
HS-5 cells were also transfected with a 3 × C/EBPβ-binding sites–luciferase reporter construct. Transfected HS-5 cells were then stimulated with KAS6/1 or ANBL-6 myeloma cells and treated with 15-d-PGJ2, troglitazone, or GW9662. As shown in Figure 5B, the transactivation of C/EBPβ in transfected HS-5 cells was stimulated by adhesion of KAS6/1 or ANBL-6 cells. Both PPARγ ligands, 15-d-PGJ2 and troglitazone, inhibited activity of the C/EBP-luciferase gene reporter in HS-5 cells mediated by myeloma cell binding. We further used a coimmunoprecipitation experiment to test for a complex formation between PPARγ and C/EBPβ. HS-5 cells were stimulated with KAS6/1 cells and treated with 15-d-PGJ2 or troglitazone. Adherent HS-5 cell extracts were prepared and immunoprecipitated with a PPARγ-specific antibody; immunoprecipitates were developed on Western blots with a C/EBPβ-specific antibody. As shown in Figure 5C, the C/EBPβ can be coprecipitated with PPARγ in cells treated with 15-d-PGJ2 and troglitazone. These data indicate that a direct physical protein-protein interaction occurs between nuclear receptor PPARγ and C/EBPβ. In contrast, no significant changes in C/EBPβ expression (Figure 5D) were observed in control HS-5 cells or in KAS6/1 cell–stimulated HS-5 cells in the presence or absence of all of the above PPARγ ligands, which was confirmed by densitometric analysis. These data suggest that a direct physical complex of PPARγ with C/EBPβ, but not expression of C/EBPβ protein, may be involved in PPARγ ligands' inactivation of IL-6.
Discussion
Adhesion of MM cells to BMSCs is mediated via adhesion molecules.40,–42 Recent studies suggest that the VCAM-1 that is constitutively expressed in bone marrow stromal cells plays an important role in regulating the behavior of neighboring myeloma cells.43 A PPARγ ligand pioglitazone has been demonstrated to inhibit the expression of VCAM-1 on activated human umbilical vein endothelial cells after IL-1β stimulation.44 In the present study, we have found that PPARγ agonists effectively inhibit MM cell adhesion to BMSCs (both primary cells and cell line) (Tables 1,2; Figure 1A) and expression of adhesion molecules such as ICAM-1 and VCAM-1 by BMSCs (Figure 1E). These observations suggest that PPARγ ligands may function as a class of inhibitor of MM through preventing cell-cell interactions between myeloma cells and marrow stromal cells mediated principally via ICAM-1 and VCAM-1.
Multiple myeloma cell adherence to BMSCs induces up-regulation of IL-6 transcription and secretion in BMSCs.45,–47 We demonstrated that MM cell binding increased markedly IL-6 mRNA expression and secretion by BMSCs. PPARγ agonists interfere with IL-6 transcription (Figure 3A) and production (Tables 3,4) triggered by direct contact between MM cells and BMSCs. SiRNA-mediated knockdown of PPARγ in BMSCs attenuates its agonist inhibition of IL-6 (Figure 3B), whereas overexpression of PPARγ enhances such inhibition (Figure 3C). Thus, these findings indicate that PPARγ ligands' suppression of IL-6 gene expression and secretion is largely through PPARγ functionally down-regulating IL-6 promoter transactivation. More importantly, we have demonstrated that both MM.1S and MM.1R cell lines expressed PPARγ (Figure 2A). PPARγ agonists could inhibit cell growth of MM.1R (Figure 2B) and reduce IL-6 production of stromal cells triggered by adhesion of MM.1R cells (Figure 2D). These data suggested that PPARγ ligands would be able to overcome resistance, indicating the clinically applicable possibility of PPARγ ligands in multiple myeloma.
The increase in adhesion molecules facilitates activation of transcriptional factors, and in turn, activation of IL-6 transcription. Adhesion-mediated activation of NF-κB up-regulates adhesion molecules and further enhances adhesion of MM cells to BMSCs.48 This binding confers resistance to apoptosis and also triggers NF-κB–dependent secretion of IL-6. Transcriptional regulation of the IL-6 gene is complex and involves multiple transcriptional factors. Both C/EBPβ and NF-κB play a central role in the transcription of IL-6 gene expression. However, the relationship between PPARγ and NF-κB or C/EBPβ in BMSCs/MM remains to be clarified. Using a series of IL-6 promoter luciferase constructs containing wild-type and binding site mutations, we have found that inhibitory effects of PPARγ agonists in IL-6 transcription resulting from MM adhesion-induced BMSCs were due to disruption of the cooperation between NF-κB and C/EBPβ. Moreover, NF-κB– and 5′-C/EBP–binding sites are involved in PPARγ ligands' down-regulation of the IL-6 promoter activity in BMSCs in response to MM cell adhesion (Figure 3D).
Although several lines of evidence indicated PPARγ directly associated with NF-κB in different cell lines,49 our results delineate mechanisms on PPARγ interfering with NF-κB and C/EBPβ in MM cell–adhesive BMSCs through direct or coactivator PGC-1–mediated protein-protein interaction. Cofactors have been described to bridge the gap between transcription factors and components of basal transcription machinery. The coactivator CREB-binding protein (CBP) and its homologue p300 can enhance the transcriptional activity of p65.50 Similarly, a potentiating effect on NF-κB and AP-1–driven transactivation was reported for SRC-1.51 Competition between glucocorticoid receptor (GR) and the driving transcriptional factors for a limited amount of coactivator CBP/p300 or SRC-1 in the cell would account for the repressive action by GRs.52,,–55 Suzawa et al56 showed NF-κB seems to be recruited preferentially to PPARγ, at least in bone marrow stem cells, by its physical interaction with PGC-2, a PPARγ AF-1–specific coactivator. In the case of bone marrow stromal cells, we observed that ligand-bound PPARγ might use its coactivator PGC-1 as a bridging protein to associate with NF-κB (predominantly p65), although NF-κB did not associate directly with PPARγ. PGC-1 was originally identified as a transcriptional coactivator of the nuclear receptor PPARγ.35,36 The addition of PPARγ-specific ligands could induce tethering of PPARγ binding to PGC-1 (Figure 4D). First we found that PGC-1 is also a cofactor shared by NF-κB and PPARγ (Figure 4C). The data suggested that ligand-bound PPARγ competes for the limiting amounts of PGC-1, and that PGC-1 dissociates with NF-κB and decreases its DNA binding, and transactivation; while under these conditions, PGC-1 increased association with PPARγ. Taken together, it therefore seems that ligand-bound PPARγ competes for PGC-1 docking to NF-κB (Figure 4C,D).
It is worthwhile to note that 15-d-PGJ2, but not troglitazone, may have additional inhibitory activities by direct inactivation of IKK and IκB. Our data showed the 15-d-PGJ2 also directly inhibits IKKα and IκBα phosphorylation (Figure 4F), which is consistent with the previous observation35 that specific inhibition of NF-κB by IκB kinase (IKK) inhibitor down-regulates both constitutive and induced IL-6 secretion. In macrophages, 15-d-PGJ2 was shown to block IκB kinase (IKK) activity, possibly through covalent modifications of critical cysteine residues in IKKβ.39 Even though 15-d-PGJ2 was found to have blocked aspects of NF-κB activation by direct inhibition of IκB and its kinase IKK, the majority of the effects of 15-d-PGJ2 were apparently dependent on PPARγ. PPARγ-SiRNA was able to abolish 15-d-PGJ2 inhibition of IL-6 production (Figure 3B).
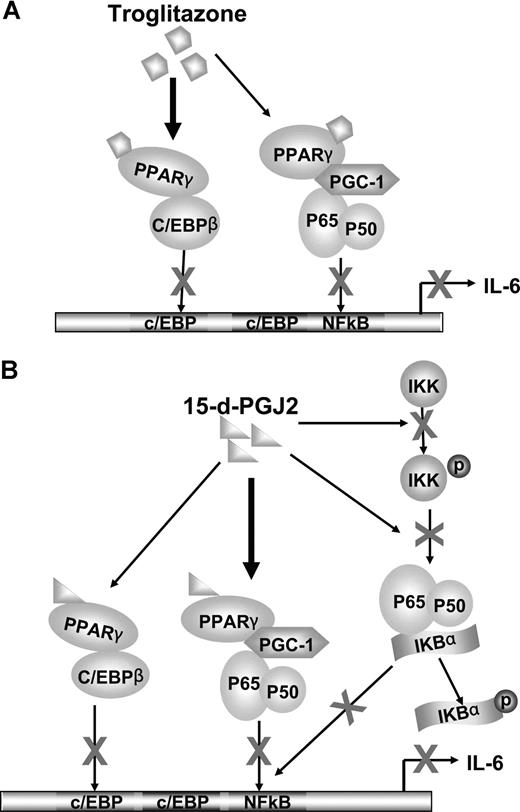
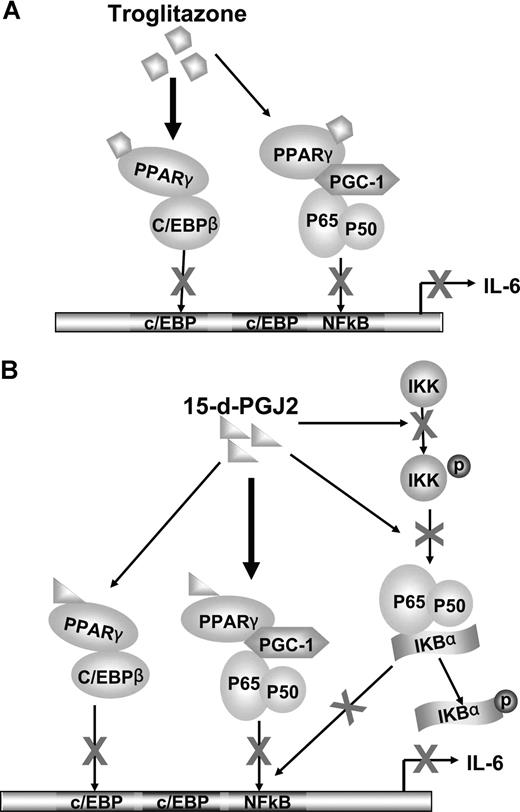
We also observed the effects of 15-d-PGJ2 and troglitazone on another important IL-6 promoter transcription factor, C/EBPβ. Both ligands appeared to stimulate a complex formation between PPARγ and C/EBPβ, blocking transcriptional promoter activity (Figure 5A-D). The complexity of gene promoter regulation such as IL-6 is convoluted by the different mechanisms of actions of the PPARγ nuclear receptor agonists that have ligand-dependent/independent functions. In addition, cross talk between transcriptional systems may be involved in direct transcriptional factor interactions or modifying activities contributed by coregulatory proteins. With this in mind, we have observed that the synthetic ligand troglitazone appears to require PPARγ for inhibition of both NF-κB and C/EBPβ. Troglitazone-PPARγ activation stimulates direct PPARγ association with c/EBPβ blocking transcriptional activity. Troglitazone-PPARγ, however, did not directly associate with p65/p50 subunits, but rather, recruited the coactivator PGC-1 to PPARγ, thus making it less available to the NF-κB complex (Figure 6A). The natural ligand, 15-d-PGJ2, showed both PPARγ-dependent and -independent effects. With regard to the regulation of NF-κB, 15-d-PGJ2 directly inhibited phosphorylation of IKKα and IκBα. Not only did 15-d-PGJ2 stimulate association of the coactivator PGC-1 to PPARγ concomitant with decreased association with p65/p50, but also 15-d-PGJ2 stimulated direct PPARγ complex association with C/EBPβ, in turn blocking C/EBPβ transcriptional activity (Figure 6B).
Cross talk between PPARγ and NF-κB or between PPARγ and C/EBPβ. (A) Troglitazone inhibition of IL-6 transcription. C/EBPβ and NF-κB bind to the promoter region of the IL-6 gene and their cooperation is needed to activate IL-6 transcription. The nuclear receptor PPARγ can be activated by troglitazone. Predominately, the complex between C/EBPβ and troglitazone-bound PPARγ leads to decreased DNA binding and transactivation of C/EBPβ, in turn inhibiting gene expression of IL-6. In addition, PGC-1, a coactivator, is shared by both PPARγ and NF-κB. After activation by ligands, ligand-bound PPARγ competes for the limited amounts of PGC-1. Therefore, NF-κB dissociates with PGC-1 and decreases NF-κB DNA-binding and transactivation, leading to blocked IL-6 transcription. (B) 15-d-PGJ2 inhibition of IL-6 transcription. Although 15-d-PGJ2 also shares the above ligand-bound PPARγ down-regulation mechanisms on C/EBPβ and NF-κB, 15-d-PGJ2, compared with troglitazone, prefers to use PGC-1 as a bridging protein to associate with NF-κB. In addition, 15-d-PGJ2 inactivates NF-κB through decreasing phosphorylation of IKK and IκB.
Cross talk between PPARγ and NF-κB or between PPARγ and C/EBPβ. (A) Troglitazone inhibition of IL-6 transcription. C/EBPβ and NF-κB bind to the promoter region of the IL-6 gene and their cooperation is needed to activate IL-6 transcription. The nuclear receptor PPARγ can be activated by troglitazone. Predominately, the complex between C/EBPβ and troglitazone-bound PPARγ leads to decreased DNA binding and transactivation of C/EBPβ, in turn inhibiting gene expression of IL-6. In addition, PGC-1, a coactivator, is shared by both PPARγ and NF-κB. After activation by ligands, ligand-bound PPARγ competes for the limited amounts of PGC-1. Therefore, NF-κB dissociates with PGC-1 and decreases NF-κB DNA-binding and transactivation, leading to blocked IL-6 transcription. (B) 15-d-PGJ2 inhibition of IL-6 transcription. Although 15-d-PGJ2 also shares the above ligand-bound PPARγ down-regulation mechanisms on C/EBPβ and NF-κB, 15-d-PGJ2, compared with troglitazone, prefers to use PGC-1 as a bridging protein to associate with NF-κB. In addition, 15-d-PGJ2 inactivates NF-κB through decreasing phosphorylation of IKK and IκB.
In conclusion, PPARγ and its ligands may block IL-6 secretory pathways by inhibiting MM-BMSC adhesion and also independently block IL-6 transcription by BMSCs through cross talk with NF-κB and C/EBPβ. The molecular mechanisms of PPARγ ligands on the regulation of multiple transcription factors have proven, not surprisingly, complex. Given that IL-6 is the key growth and survival factor of multiple myeloma cells, and is particularly involved in the origin of all benign and malignant plasma cell expansions as well as MM cell resistance, the effects and targets of the PPARγ ligands on aspects of multiple myeloma biology and bone marrow stromal cells may be clinically relevant.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute (NCI), Center for Cancer Research, and also funded in part with federal funds from the NCI, NIH, under contract no. NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
We are very grateful to Dr Elaine Hurt and Dr Adrian Wiestner in the Metabolism Branch of NCI for providing us with the human bone marrow aspirate, and Drs Xin Chen and Huifang Dong for their help in mouse bone marrow harvest.
National Institutes of Health
Authorship
Contribution: L.H.W. designed the research, analyzed data, and wrote the paper; X.Y.Y. performed key experiments; X.Z. performed some experiments; W.L.F. is the laboratory chief and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Li Hua Wang and William L. Farrar, PO Box B, National Cancer Institute-Frederick, National Institutes of Health, Frederick, MD 21702; e-mail:lhwang@ncifcrf.gov and farrar@ncifcrf.gov.

![Figure 2. Effect of PPARγ ligands on Dex-resistant MM cells. (A) Expression of PPARγ on MM1S and MM.1R cell lines detected by Western blot. (B,C) Dose-dependent inhibition of PPARγ ligands on cell growth of both MM.1S and MM.1R cells. Proliferation of quiescent MM.1R (B) or MM.1S cells (C) in culture was examined following treatment with 10 ng/M IL-6 and increasing concentrations (abscissa) of 15-d-PGJ2 (○), troglitazone (□), or dexamethasone (▵) for 16 hours at 37°C. [3H]-thymidine incorporation was analyzed. (D) Effects of PPARγ ligands on drug-resistant MM cell–induced IL-6 release from stromal cells. HS-5 cells were cocultured with MM.1S or MM.1R cells and treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone for 48 hours. IL-6 concentration was determined in supernatants. Error bars represent the mean (± SE) from 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/13/10.1182_blood-2006-07-038026/3/m_zh80010810620002.jpeg?Expires=1769232777&Signature=rrsFCInzltVfUY6qh6IdwoEFTn5M6uum8N3zXGsxOh47zCRYFcdBmTxqmEq9Eqv-ypFin1LcbQzidcgRceO9-GeMmVzw9iDkzwC8jA2IxWJVatz7XH9qXVxPfURsbjg9rhsf0DPUzTOqJFZ1Uw9L-owXK8~Pmgl-Lhs8fjJnQK2r9M1YmXgLI1Ks6Yr61bWltlzlk9di2kLi~~nyNQ1apEFDGePZ8nRaIQ5hPBsmI6hE-F0fZvJgqj8X0GZBlNj19WEqv4CJCjPAMVyhf4IqrA9N74MwsdFgPjUSKfFjD2YAUse-9fgHJsmCIa5T1jaEGXGp-U5o0t3ijN-uT4FHqA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. PPARγ ligands decreased IL-6 transcription and secretion largely by a PPARγ-dependent mechanism. (A) Adherent HS-5 cells were stimulated with KAS6/1 cells and treated with PPAR ligands 15-d-PGJ2, troglitazone, or WY14643 for 6 hours. KAS6/1 cells were washed out and collected. The remaining HS-5 cells were then harvested. The RNA samples, prepared from the above-harvested KAS6/1 cells or HS-5 cells, were hybridized with [33P]-labeled RNA probes corresponding to transcripts for individual human IL-6 gene, according to the BD PharMingen protocol. RNase protected fragments were separated on 5% polyacrylamide gel electrophoresis (PAGE). (B) SiRNA-mediated knockdown of PPARγ alters its ligands' inhibition of IL-6 production. HS-5 cells were transfected with SiRNA-PPARγ or control vector and cultured for 96 hours. The above HS-5 cells were then treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone and incubated with or without KAS6/1 cells for an additional 48 hours. IL-6 concentration was determined in supernatants. Insert figure shows knockdown of PPARγ expression in HS-5 cells transfected with SiRNA-PPARγ. HS-5 cells were transduced with SiRNA-PPARγ or control vector and incubated for 96 hours. Proteins were then extracted from the cells, and the levels of PPARγ expression were examined by Western blotting. The β-actin bands served as an internal control for equal total protein loading. (C) HS-5 cells were cotransfected with a PPARγ wild-type expression vector (pSG5-hPPARγ2) or control pSG5 plasmid, an IL-6 promoter luciferase reporter gene construct, and a pRL-TK control vector for 24 hours. Cells were then treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone and incubated with or without KAS6/1 cells for an additional 48 hours. Luciferase activity of lysed cells was measured. The level of firefly luciferase activity was normalized to that of the Renilla luciferase activity. Insert figure shows the level of PPARγ expression in HS-5 cells transfected with PPARγ wild-type expression vector. (D) Effect of PPARγ ligands on transactivation of IL-6 promoter. HS-5 cells were transfected with the indicated IL-6 promoter luciferase reporter gene constructs. Twenty-four hours after transfection, KAS6/1 cells were added directly to the transfected confluent HS-5 BMSCs pretreated with 15-d-PGJ2 or troglitazone for 2 hours. The transfected HS-5 cells were also cultured alone as a control. The cocultured cells were incubated for an additional 24 hours. After washing out KAS6/1 cells, luciferase activity of adherent HS-5 cells was measured. Error bars represent the mean (± SE) from 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/13/10.1182_blood-2006-07-038026/3/m_zh80010810620003.jpeg?Expires=1769232777&Signature=kPk10SvsaJxfZGhHkQa4HHHNF0esdzfyrHhumDTyUe1gcClBZw2OrGG2J9bIC-hJYe4h28Emk7z09t70tka5mZDzMZ7XlLkkrL~MAqZQ1JGxT~PhgfbIpezb5cG~QgJ8mpQmI1cnVdBGSHSjR8J2kiDehlTqcbLEMk6AlPbi8Gx4pcfvwvBCaLUeprH6S7RpLcuHT5~-LsD3Z7nfoFiVJA-fXWVKG8pO7HQEllkc1Jd7lgbE2qFnFBv3gIbBdjuX0f1H-Xv02R8YyeMxrhnppBejOD1bfhSFhHEMquVEzLn1fVTfb9OV9uY80eJjL~4Ab7vI4DlCdGfA23gu1CJ-UA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Effect of PPARγ ligands on activation of NF-κB. (A) Nuclear extracts corresponding to 5 μg protein were incubated in the absence of antibody anti-p50, anti-p65, anti–PGC-1, or normal rabbit serum (NRS) in combination with a [32P]-labeled NF-κB oligonucleotide probe. Arrows indicate migrational location of each nonsupershifted NF-κB–DNA complex. (B) HS-5 cells were transfected with a 3 × NF-κB–binding element–pGL3 promoter luciferase construct. Cells were then pretreated with 1 μM 15-d-PGJ2, 10 μM troglitazone, or 50 μM GW9662 for 2 hours, and then incubated with or without KAS6/1 or ANBL-6 cells for 24 hours. Error bars represent the mean (± SE) from 3 independent experiments. (C) The coactivator PGC-1 coimmunoprecipitates with p50 or p65 in KAS6/1 cell adhesion–induced HS-5 cells. HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then added with KAS6/1 cells for 24 hours before lyses. Western blotting analysis with anti-p65 (top panel), or p50 (middle panel), or anti–PGC-1 (bottom panel) was performed on anti–PGC-1 immunoprecipitates. (D) The coactivator PGC-1 coimmunoprecipitates with PPARγ in HS-5 cells. HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then added with KAS6/1 cells for 24 hours before lyses. Western blotting analysis with either anti-PPARγ (top panel) or anti–PGC-1 (bottom panel) was performed on anti–PGC-1 immunoprecipitates. (E) The above cell extracts were immunoprecipitated with anti-PPARγ. Western blotting analysis was performed with anti-p50 and anti-p65. (F) HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then simulated with KAS6/1 for 1 hour before lyses. Western blotting analysis was performed with anti–phospho-IKKα, anti-IKKα, anti–phospho-IκBα, or anti-IκBα.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/13/10.1182_blood-2006-07-038026/3/m_zh80010810620004.jpeg?Expires=1769232777&Signature=EwsAYWxbBB1do38N0o8mGyz7s3q5DP9fNYpKZ3PwrcCiR-qF8uLdDDqSf6EkBDI~UVqWnqLLorUsrU7dXQSed6CR4gwLIJxmOLLDskMXm6gsyJn2U~73FpLDVPP-HxMgsAl5plev5kSsHREH25noIkSuRtMxE~ijeRLCsErM1-lcHbSjPWtEoDAslG0OzdrCGhf8CGoQq8wjrnSNYf2ofMaI6StaiLX-mnnhG8PbOfNpDl77N3K8~3d~gVv~Dd3pYm921qYf~uFYHKiBmlFILoQF4VKYvx7KRBa-X2zXI8RKz3Cvq8395ZjnZ-aMMBidytqByoe8Wtxul0thgW-HOQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. PPARγ ligand inhibition of C/EBPβ mediated by PPARγ direct association with C/EBPβ. (A) HS-5 cells were pretreated with 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then incubated with KAS6/1 cells for 2 hours. After washing out KAS6/1 cells, adherent HS-5 cells were lysated. Nuclear extracts corresponding to 5 μg protein were incubated in the absence of antibody C/EBPβ or normal rabbit serum (NRS) in combination with a [32P]-labeled C/EBP oligonucleotide probe. Arrows indicate migration location of each nonsupershifted C/EBPβ-DNA complex. (B) HS-5 cells were transfected with a 3 × C/EBPβ-binding element–pGL3 promoter luciferase construct. Cells were then pretreated with 1 μM 15-d-PGJ2, 10 μM troglitazone, or 50 μM GW9662 for 2 hours, and then incubated with or without KAS6/1 or ANBL-6 myeloma cells for 24 hours. Error bars represent the mean (± SE) from 3 independent experiments. (C) The PPARγ coimmunoprecipitates with C/EBPβ in KAS6/1 adhesion-induced HS-5 cells. HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then added with KAS6/1 cells for 24 hours before lyses. Western blotting analysis with anti-C/EBPβ (top panel) or anti-PPARγ (bottom panel) was performed on anti-PPARγ immunoprecipitates. The densitometric relative units in each lane were indicated beneath each blot. (D) No significant changes in C/EBPβ expression in HS-5 cells. The HS-5 cells were treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then simulated with KAS6/1 for 12 hours before lyses. Western blotting analysis was performed with anti-C/EBPβ. The densitometric relative units in each lane were indicated beneath each blot.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/13/10.1182_blood-2006-07-038026/3/m_zh80010810620005.jpeg?Expires=1769232777&Signature=BLy2iue3nkKtO8i1H1tsqDKaGba1MUEPJ5SYMKjH028y8NmtgRWVWzJVMK4yS9QQl7kHu9yoD2C1AK5PRl6M5GZ-6yOseNZ6VaD-SWLsBe4JVcq9naElyExoobn-5ulUcyZNLG-4LuYXEmXEYUTofR~wjMSUjcBIBGk0g~JiDzcWPat-hoC0sm9ryZUhzYRBPlD3f8QmsqqXaagkJuERs5WgohV5rtFPvimOunXigEgZb~PMAQLCXtPw5EBkJEvDXBBYfBMKPVlb6pJw5VGsEOxbxfUFepZKpf2CliR9IbJIuwjkcXBOZ-fy-nckciAPTkheyfjTpdZsnwNP08vkfQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 2. Effect of PPARγ ligands on Dex-resistant MM cells. (A) Expression of PPARγ on MM1S and MM.1R cell lines detected by Western blot. (B,C) Dose-dependent inhibition of PPARγ ligands on cell growth of both MM.1S and MM.1R cells. Proliferation of quiescent MM.1R (B) or MM.1S cells (C) in culture was examined following treatment with 10 ng/M IL-6 and increasing concentrations (abscissa) of 15-d-PGJ2 (○), troglitazone (□), or dexamethasone (▵) for 16 hours at 37°C. [3H]-thymidine incorporation was analyzed. (D) Effects of PPARγ ligands on drug-resistant MM cell–induced IL-6 release from stromal cells. HS-5 cells were cocultured with MM.1S or MM.1R cells and treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone for 48 hours. IL-6 concentration was determined in supernatants. Error bars represent the mean (± SE) from 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/13/10.1182_blood-2006-07-038026/3/m_zh80010810620002.jpeg?Expires=1769232778&Signature=fPstMXv9vxmgRdAU0kAyvGrbLKAN6vsj64GMpyAsShu5HvXzu8c~YYCGsdKn-ADjCh57TkAVq8eTToIVHUn2DQ8bwCUdPuJvEXeIhP8mo4LhCW--fqh~kJqrG3fhlawfbLLw3SvNQifF4Z9nUSN6r2BcBd-VDMYd2jRZ2rIJ1FFrYqfHu9iGpEOoFDWyXWwtcnprXhoDrB1EJ5fX70lIkQKw~wA1PbHQtRZ~WDhSJuU2rLyOhfyH1JkQamR7rrVmPoJDP7FnqdnAsHyN8zQBJTjLAwRt-2QNq6hSMR-DRkwCS9gWzB2URiMhYnlAmGZH9GWXVBrgbJA8cb3D1InSMQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. PPARγ ligands decreased IL-6 transcription and secretion largely by a PPARγ-dependent mechanism. (A) Adherent HS-5 cells were stimulated with KAS6/1 cells and treated with PPAR ligands 15-d-PGJ2, troglitazone, or WY14643 for 6 hours. KAS6/1 cells were washed out and collected. The remaining HS-5 cells were then harvested. The RNA samples, prepared from the above-harvested KAS6/1 cells or HS-5 cells, were hybridized with [33P]-labeled RNA probes corresponding to transcripts for individual human IL-6 gene, according to the BD PharMingen protocol. RNase protected fragments were separated on 5% polyacrylamide gel electrophoresis (PAGE). (B) SiRNA-mediated knockdown of PPARγ alters its ligands' inhibition of IL-6 production. HS-5 cells were transfected with SiRNA-PPARγ or control vector and cultured for 96 hours. The above HS-5 cells were then treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone and incubated with or without KAS6/1 cells for an additional 48 hours. IL-6 concentration was determined in supernatants. Insert figure shows knockdown of PPARγ expression in HS-5 cells transfected with SiRNA-PPARγ. HS-5 cells were transduced with SiRNA-PPARγ or control vector and incubated for 96 hours. Proteins were then extracted from the cells, and the levels of PPARγ expression were examined by Western blotting. The β-actin bands served as an internal control for equal total protein loading. (C) HS-5 cells were cotransfected with a PPARγ wild-type expression vector (pSG5-hPPARγ2) or control pSG5 plasmid, an IL-6 promoter luciferase reporter gene construct, and a pRL-TK control vector for 24 hours. Cells were then treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone and incubated with or without KAS6/1 cells for an additional 48 hours. Luciferase activity of lysed cells was measured. The level of firefly luciferase activity was normalized to that of the Renilla luciferase activity. Insert figure shows the level of PPARγ expression in HS-5 cells transfected with PPARγ wild-type expression vector. (D) Effect of PPARγ ligands on transactivation of IL-6 promoter. HS-5 cells were transfected with the indicated IL-6 promoter luciferase reporter gene constructs. Twenty-four hours after transfection, KAS6/1 cells were added directly to the transfected confluent HS-5 BMSCs pretreated with 15-d-PGJ2 or troglitazone for 2 hours. The transfected HS-5 cells were also cultured alone as a control. The cocultured cells were incubated for an additional 24 hours. After washing out KAS6/1 cells, luciferase activity of adherent HS-5 cells was measured. Error bars represent the mean (± SE) from 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/13/10.1182_blood-2006-07-038026/3/m_zh80010810620003.jpeg?Expires=1769232778&Signature=D-Nej0VlJ2IE9T8U1c9lFpW3A9Z0PXf~YQgVSrJhFYzyf0Tc-5ix0hlPOHz4ivWQLPTcMtd2h4m4RJ9Yiv7Akhgv740ze7CZHoVQlJP0tSHd9SaNGHovUZkW-jISGI29LOkRD2CCr6iv0rhwKGJ4Tk6NJ05PHESqgMfNCK2~dpZdha7i53tE9ZS18FMLRKHXrlmtqRP6jv0duNxWcO-cFMn49gARMhrP7tjSERVRytmZMUCZQwTmvYyHpQDKGK7HcObOIE1doDKgPBU-0j4gOBO~zOSGA-n9GqlylyNsnR07DFdnj-9o440SUVSXEJBcdErqfRAjnphVPIR89FAy~w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Effect of PPARγ ligands on activation of NF-κB. (A) Nuclear extracts corresponding to 5 μg protein were incubated in the absence of antibody anti-p50, anti-p65, anti–PGC-1, or normal rabbit serum (NRS) in combination with a [32P]-labeled NF-κB oligonucleotide probe. Arrows indicate migrational location of each nonsupershifted NF-κB–DNA complex. (B) HS-5 cells were transfected with a 3 × NF-κB–binding element–pGL3 promoter luciferase construct. Cells were then pretreated with 1 μM 15-d-PGJ2, 10 μM troglitazone, or 50 μM GW9662 for 2 hours, and then incubated with or without KAS6/1 or ANBL-6 cells for 24 hours. Error bars represent the mean (± SE) from 3 independent experiments. (C) The coactivator PGC-1 coimmunoprecipitates with p50 or p65 in KAS6/1 cell adhesion–induced HS-5 cells. HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then added with KAS6/1 cells for 24 hours before lyses. Western blotting analysis with anti-p65 (top panel), or p50 (middle panel), or anti–PGC-1 (bottom panel) was performed on anti–PGC-1 immunoprecipitates. (D) The coactivator PGC-1 coimmunoprecipitates with PPARγ in HS-5 cells. HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then added with KAS6/1 cells for 24 hours before lyses. Western blotting analysis with either anti-PPARγ (top panel) or anti–PGC-1 (bottom panel) was performed on anti–PGC-1 immunoprecipitates. (E) The above cell extracts were immunoprecipitated with anti-PPARγ. Western blotting analysis was performed with anti-p50 and anti-p65. (F) HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then simulated with KAS6/1 for 1 hour before lyses. Western blotting analysis was performed with anti–phospho-IKKα, anti-IKKα, anti–phospho-IκBα, or anti-IκBα.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/13/10.1182_blood-2006-07-038026/3/m_zh80010810620004.jpeg?Expires=1769232778&Signature=ALC6Yo~mLdOL2SpqcGHDmEMpOgTxMJgm9jqS7uc9FgNa9grX-kbCZrxdjXt0VvKkKQhFuhZFFfzd04PzBVA5Jrwtcp~lYn6TjkFZa3iBArCME7xYYhkozA~9uDlPXqmP05bXM9qSJtYpSSEtouNitM5907wsvnxw5NUgFKEXsgCirZlmDfXAfBBnsdHAGAO-xFMFWnWKTeV36EaQ3d~Bo5U7b6E82uvq3SAQ-bw4~FsVdfLTg-uJHbYPaPyXCUnvegQnNz4X64UzO9qUO15MMrO7qTzb6LMGWTkKWbe6Xe35gL5OSbzSJ8xI~8sKjmdTAJRZFfo4IDZs~dOKuRjbiQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. PPARγ ligand inhibition of C/EBPβ mediated by PPARγ direct association with C/EBPβ. (A) HS-5 cells were pretreated with 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then incubated with KAS6/1 cells for 2 hours. After washing out KAS6/1 cells, adherent HS-5 cells were lysated. Nuclear extracts corresponding to 5 μg protein were incubated in the absence of antibody C/EBPβ or normal rabbit serum (NRS) in combination with a [32P]-labeled C/EBP oligonucleotide probe. Arrows indicate migration location of each nonsupershifted C/EBPβ-DNA complex. (B) HS-5 cells were transfected with a 3 × C/EBPβ-binding element–pGL3 promoter luciferase construct. Cells were then pretreated with 1 μM 15-d-PGJ2, 10 μM troglitazone, or 50 μM GW9662 for 2 hours, and then incubated with or without KAS6/1 or ANBL-6 myeloma cells for 24 hours. Error bars represent the mean (± SE) from 3 independent experiments. (C) The PPARγ coimmunoprecipitates with C/EBPβ in KAS6/1 adhesion-induced HS-5 cells. HS-5 cells were treated with or without 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then added with KAS6/1 cells for 24 hours before lyses. Western blotting analysis with anti-C/EBPβ (top panel) or anti-PPARγ (bottom panel) was performed on anti-PPARγ immunoprecipitates. The densitometric relative units in each lane were indicated beneath each blot. (D) No significant changes in C/EBPβ expression in HS-5 cells. The HS-5 cells were treated with 1 μM 15-d-PGJ2 or 10 μM troglitazone for 2 hours and then simulated with KAS6/1 for 12 hours before lyses. Western blotting analysis was performed with anti-C/EBPβ. The densitometric relative units in each lane were indicated beneath each blot.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/13/10.1182_blood-2006-07-038026/3/m_zh80010810620005.jpeg?Expires=1769232778&Signature=NMZ1UDMv1zLAJH9BQTTcYYJXEy6tZnunSLHVPUmoSkY70mrJ7AAArNBNb6jZNyekemRnIlaJWsam1w5rq5hWm~ccbWzm2BbwAokr2Bvv4AaXUvf7FS5jNQIbW7dqjYIrhFERfzoWw6BKmlz~u1bX9AGMcRcFDV98j~HfMCCMrjYxHnL9XM6Cfj5J03tTJ6UyuOsDahgoYG0n-1zGVH99IxBLpdrFJnjGDUX5Udn8JB5gnQtpWvcqqJVRUH7VNIEdyQw8BFHvQ-yMvHn3zQDN3CTVMtUroTMuF6goWJN~Mm65KHX~r7Ze8gw7aafA0fhF1MQyCWprnEsKBdqPvABtRw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)