The presence of valine (V) at position 158 of FcγRllla (CD16) is known to improve clinical response to rituximab in indolent non-Hodgkin lymphoma (NHL). Little is known about the basic mechanisms for this observation. We examined natural killer (NK) cells from healthy donors representing the FcγRIIIa-158 polymorphic subgroups (V/V, V/F, and F/F) for gene transcript and cell surface CD16 expression, rituximab binding, and rituximab-dependent NK cell-mediated cytotoxicity. We observed higher levels of FcγRIIIa transcripts among individuals with the FcγRIIIa-158 V/V versus V/F or F/F genotype (P < .001); increased cell surface CD16 expression by quantitative flow cytometry on NK cells from individuals expressing at least one valine at FcγRIIIa-158 versus F/F (P = .029); as well as augmented rituximab binding and rituximab-mediated, antibody-dependent cellular cytotoxicity (ADCC). These results suggest that individuals expressing at least one valine at FcγRIIIa-158 might, in part, have better clinical outcomes due to increased CD16 expression, rituximab binding, and rituximab-mediated ADCC.
Introduction
Rituximab is a CD20-directed, IgG1-chimeric monoclonal antibody (mAb) used to treat patients with B-cell lymphomas and various autoimmune disorders. Both quantitative as well as qualitative differences in natural killer (NK) cell function may explain rituximab clinical activity. Higher circulating NK cell levels and responses to rituximab have been reported in patients with indolent non-Hodgkin lymphoma (NHL), suggesting that antibody-dependent cellular cytotoxicity (ADCC) enacted by NK cells may be a primary mechanism by which rituximab functions.1,2 Moreover, responses to rituximab may depend upon polymorphisms present in the FcγRIIIa (CD16) receptor, a receptor mainly expressed on NK cells.3,–5
Polymorphisms in position 48 and 158 of the FcγRIIIa receptor expression have been reported to influence human IgG1 binding and ADCC activity.6,,–9 Polymorphisms at position 158 result in either valine (V) or phenylalanine (F) expression,6,8,9 the former of which is associated with increased depletion of peripheral blood B cells10 and response to rituximab in patients with indolent NHL3,–5 but not chronic lymphocytic leukemia (CLL).11 At position 48, polymorphisms of the FcγRIIIa receptor result in expression of either leucine, arginine, or histidine, the first of which is linked to FcγRIIIa-158F and the latter 2 with the FcγRIIIa-158V polymorphisms.5,8,9 However, the binding of IgG1 to FcγRIIIa appears to occur independently of position 48 polymorphisms most likely on the basis of tight genetic linkage to FcγRIIIa-158 polymorphisms.5,8 Genetic linkage between polymorphisms in FcγRIIa (CD32), a receptor also implicated in predicting rituximab clinical response, and FcγRIIIa has recently been demonstrated by us and points to the primacy of FcγRIIIa-158 polymorphisms in predicting rituximab response.12
While these studies suggest that variable responses to rituximab among FcγRIIIa-158 polymorphic groups are likely the result of qualitative (ie, antibody affinity) differences, the possibility that quantitative differences in cell surface CD16 expression, rituximab binding, and ADCC activity have not been addressed. As such, we sought to delineate differences in FcγRIIIa gene expression, cell surface CD16 expression, rituximab binding, and rituximab-dependent ADCC activity in NK cells isolated from healthy individuals representing the 3 FcγRIIIa-158 polymorphic subgroups (V/V, V/F, and F/F).
Materials and methods
FcγRIIIa-158 genotype analysis
We analyzed the genotype of 52 unrelated healthy individuals by sequencing exon 4 of the FcγRIIIa gene. FcγRIIIa-158 polymorphisms were determined by allele-specific reverse transcription polymerase chain reaction (RT-PCR) and direct sequencing of genomic DNA, as we previously described.5 Genomic DNA was extracted from peripheral blood using a DNA isolation kit (Qiagen, Valencia, CA). The study was approved by the Dana Farber Cancer Institute's Institutional Review Board, and written consent was obtained from each donor in accordance with the Declaration of Helsinki.
Cell isolation and culture
Peripheral blood mononuclear cells (PBMNCs) were isolated using Ficoll-Paque (Amersham, Uppsala, Sweden). NK cells were selected from PBMNCs using the NK-cell isolation kit II (Miltenyi, Auburn, CA) resulting in more than 95% purity (CD3−/CD56+). ARH-77 and Daudi cells were cultured as previously described.
RT-PCR analysis
FcγRIIIa gene expression was determined by quantitative real-time RT-PCR (Applied Biosystems, Foster City, CA). RNA was extracted from NK cells. Primer sequences were as follows: FcγRllla sense (5′-CCAAAAGCCACACTCAAAGAC-3′) and antisense (5′-ACCCAGGTGGAAAGAATGATG-3′); TaqMan probe (5′-AACATCACCATCACTCAAGGTTTGG-3′). The quantity of FcγRIIIa mRNA in each sample was normalized to the relative quantity of HR-18S.
Quantitative flow cytometry
CD16 receptors were quantified using the QuantiBRITE system. NK cells (2 × 105) were stained with 5 μL (0.287 mg/mL) of anti-CD16 PE bead-conjugated mAb for 20 minutes at 4°C (BD Biosciences, San Jose, CA). After incubation, NK cells were washed twice and resuspended in 1× PBS. Prior to each analysis, the flow cytometer was calibrated by QuantiBRITE PE calibration beads. CD16 receptors were assessed by gating 104 (CD3−CD56+) cells. Samples were analyzed using CellQuest software (BD Biosciences).
Rituximab binding to NK cells
Rituximab (Genentech BioOncology, San Francisco, CA) binding was determined using an indirect method as previously described,7 using an anti-CD16 (3G8 clone) mAb. NK cells (2 × 105) were incubated with rituximab at concentrations of 10, 50, 100, and 200 μg/mL for 30 minutes at 4°C, followed by incubation with anti-CD16 FITC and anti-CD56 PE mAbs at 4°C for 20 minutes. After incubation, NK cells were washed with PBS and analyzed by flow cytometry. CD16 median fluorescence intensity (MFI) was determined by gating on CD3-CD56+ lymphocytes. Rituximab binding was defined as percentage of inhibition for binding of anti-CD16 mAb and calculated as follows: [(MFI without rituximab) − (MFI with rituximab)] × 100/(MFI without rituximab).
ADCC assays
ADCC experiments were performed using NK cells as effectors cells. To avoid killer cell immunoglobulin-like receptor (KIR) dependent ADCC, HLA class I expressing (ARH-77) and nonexpressing (Daudi) cell lines (both of which are CD20+) were used as a target cells. Cells were incubated with 10 μg/mL rituximab and human IgG1 (control) for 1 hour, washed twice, and cocultured (5 × 103 per well) in varying ratios with effector cells for 4 hours at 37°C with 5% CO2. A colorimetric-based lactate dehydrogenase (LDH) assay (CytoTox 96; Promega, Madison, WI) was used and cytotoxicity calculated according to manufacturer's instructions.
Statistical analysis
Differences among polymorphic groups were compared by the Kruskal-Wallis and Mann-Whitney tests. The correlations between gene expression and CD16 receptors were assessed using the Pearson correlation coefficient.
Results and discussion
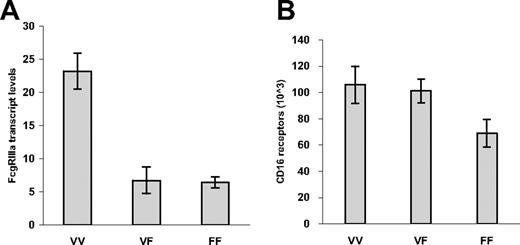
We first analyzed the gene expression of FcγRIIIa by performing real-time RT-PCR analysis for 13 donors whose genotyping demonstrated FcγRIIIa-158 V/V (n = 4), V/F (n = 4), and F/F (n = 5). Individuals with the FcγRIIIa-158 V/V genotype expressed higher FcγRIIIa transcript levels versus those individuals with the FcγRIIIa V/F and F/F genotype (P < .001). However, no significant difference in FcγRIIIa transcripts was observed between individuals with the FcγRIIIa V/F and F/F genotypes (Figure 1A). This observation is particularly intriguing in view of the fact that the absolute number of CD16 receptors per NK cell was significantly higher in donors who expressed at least one valine at FcγRIIIa-158 (ie, were either V/V or V/F) versus F/F (P = .029; Figure 1B). The basis for these discordant findings between FcγRIIIa transcript expression and cell surface protein levels among individuals with FcγRIIIa-V/F remains to be clarified but may reflect relative differences in transcript or protein stability and/or recycling of CD16 at the cell surface imposed by the expression of valine.
FcγRIIIa transcript expression on NK cells for 13 donors. Transcript expression was assayed by real-time RT-PCR analysis (A), and cell surface CD16 expression by quantitative flow cytometry (B), on NK cells for donors whose genotyping demonstrated FcγRIIIa-158 V/V (n = 4), V/F (n = 4), and F/F (n = 5). P is less than .001 for transcript levels for individuals with FcγRIIIa-158 V/V versus V/F or F/F genotype. P equals .029 for CD16 expression for individuals expressing at least one valine at FcγRIIIa-158 versus F/F. Values represent means plus or minus SE.
FcγRIIIa transcript expression on NK cells for 13 donors. Transcript expression was assayed by real-time RT-PCR analysis (A), and cell surface CD16 expression by quantitative flow cytometry (B), on NK cells for donors whose genotyping demonstrated FcγRIIIa-158 V/V (n = 4), V/F (n = 4), and F/F (n = 5). P is less than .001 for transcript levels for individuals with FcγRIIIa-158 V/V versus V/F or F/F genotype. P equals .029 for CD16 expression for individuals expressing at least one valine at FcγRIIIa-158 versus F/F. Values represent means plus or minus SE.
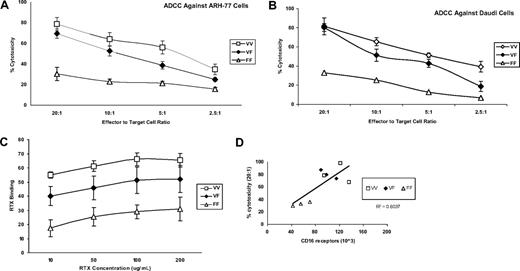
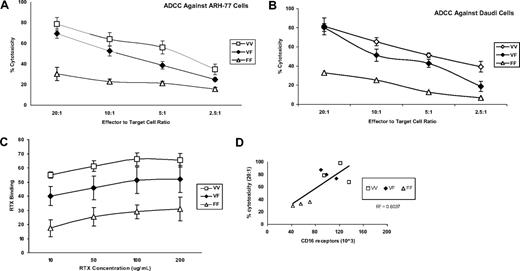
We next evaluated the functional implications of FcγRIIIa-158 polymorphisms by studying rituximab binding and rituximab-dependent NK cell-mediated cytotoxicity. Both rituximab binding (Figure 2A) as well as rituximab-mediated ADCC activity by NK cells (Figure 2B,C) increased with the presence of at least one valine, in comparison with the donors' homozygous for phenylalanine (V/V> V/F> F/F), at all concentrations (10 to 200 μg/mL) of rituximab studied. The cytotoxicity induced by the control (10 μg huIgG1) was less than 10% (data not shown). These results are unlikely to be explained by KIR mismatching because HLA class I expressing (ARH-77) and nonexpressing (Daudi) cell lines were used as target cells. Independent of the FcγRIIIa-158 polymorphic subgroup, rituximab-mediated ADCC activity was observed to correlate with the number of cell surface CD16 receptors (Figure 2D). These results are in agreement with previous studies demonstrating increased binding of IgG1-class antibodies, including rituximab among individuals expressing valine at FcγRIIIa-158,6,,–9,13 and suggest that the expression level of cell surface CD16 may also contribute to augmented rituximab binding and ADCC activity in addition to possible differences in binding affinity.
Characterization of NK-cell RTX-binding and RTX-mediated ADCC activity by NK cells against Daudi and ARH-77 CD20+ B cells. RTX-mediated ADCC activity by NK cells against (A) ARH-77 and (B) Daudi CD20+ B cells was assayed for 9 donors whose genotyping demonstrated FcγRIIIa-158 V/V (n = 3), V/F (n = 3), and F/F (n = 3) and (C) NK-cell RTX binding. (D) The correlation between cell-surface CD16 receptors and RTX-dependent ADCC activity for these individuals. Values represent means plus or minus SE.
Characterization of NK-cell RTX-binding and RTX-mediated ADCC activity by NK cells against Daudi and ARH-77 CD20+ B cells. RTX-mediated ADCC activity by NK cells against (A) ARH-77 and (B) Daudi CD20+ B cells was assayed for 9 donors whose genotyping demonstrated FcγRIIIa-158 V/V (n = 3), V/F (n = 3), and F/F (n = 3) and (C) NK-cell RTX binding. (D) The correlation between cell-surface CD16 receptors and RTX-dependent ADCC activity for these individuals. Values represent means plus or minus SE.
The results of these studies may help to explain augmented responses to rituximab observed with the addition of certain agents known to up-regulate CD16, including one study wherein the addition of IL-2 to rituximab appeared to selectively result in clinical responses among rituximab-refractory patients expressing FcγRIIIa-158 F/F.14 Further exploration of agents aimed at augmenting CD16 expression, particularly in context with newer CD20-directed mAb-bearing enhanced Fc binding and ADCC activity,15,16 may lead to improved responses among patients with indolent NHL, including for those individuals expressing FcγRIIIa-158 F/F.
In summary, the results of these studies suggest that individuals expressing at least one valine at FcγRIIIa-158 might in part have better clinical outcomes due to increased CD16 expression.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was funded by the Peter and Helen Bing Fund for Waldenstrom macroglobulinemia, the Bailey Family Fund at the Dana-Farber Cancer Institute, and a National Institutes of Health Career Development Award (K23CA087977-03) (S.P.T.).
National Institutes of Health
Authorship
Contribution: E.H. designed the study, performed research, analyzed the data, and wrote the first draft of the manuscript; L.X., designed, performed, and analyzed experiments in molecular biology; D.D.S, performed various pertinent research; Z.R.H., collected samples and analyzed the data; B.T.C. performed various pertinent research; S.V. and M.M. designed, performed, and analyzed experiments in molecular biology; Y.C. performed various pertinent research; R.J.M. collected samples and analyed the data; X.L. performed various pertinent research; E.A.D. collected samples and analyzed the data; A.K., C.M., and K.C.A. contributed advice to the design and interpretation of the study; E.A.F. designed, performed, and analyzed experiments in molecular biology; and S.P.T. designed the study, oversaw the experiments, and wrote the final draft of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steven P. Treon, Bing Center for Waldenstrom's Macroglobulinemia, Dana-Farber Cancer Institute, M548, 44 Binney St, Boston, MA 02115; e-mail:steven_treon@dfci.harvard.edu.