Abstract
The Ity/Lsh/Bcg locus encodes the macrophage protein Slc11a1/Nramp1, which protects inbred mice against infection by diverse intracellular pathogens including Leishmania, Mycobacterium, and Salmonella. Human susceptibility to infectious and inflammatory diseases, including rheumatoid arthritis, inflammatory bowel disease, and tuberculosis, shows allelic association with a highly polymorphic regulatory, Z-DNA–forming microsatellite of (GT/AC)n dinucleotides within the proximal SLC11A1 promoter. We surmised that cis-acting allelic polymorphisms may underlie heritable differences in SLC11A1 expression and phenotypic variation in disease risk. However, it is unclear what may underlie such variation in SLC11A1 allele expression. Here we show that hypoxia-inducible Factor 1 (HIF-1) regulates allelic variation in SLC11A1 expression by binding directly to the microsatellite during macrophage activation by infection or inflammation. Targeted Hif-1α ablation in murine macrophages attenuated Slc11a11 expression and responsiveness to S typhimurium infection. Our data also showed that HIF-1 may be functionally linked to complex prototypical inflammatory diseases associated with certain SLC11A1 alleles. As these alleles are highly polymorphic, our finding suggests that HIF-1 may influence heritable variation in SLC11A1-dependent innate resistance to infection and inflammation within and between populations. This report also suggests that microsatellites may play critical roles in the directional evolution of complex heritable traits by regulating gene expression phenotypes.
Introduction
A genetic basis for resistance to infection by intracellular pathogens was proposed several years ago.1-6 This concept was confirmed upon the positional cloning of the dominant autosomal gene Ity/Lsh/Bcg based on its ability to confer on inbred mice an innate resistance to infection by diverse intracellular pathogens.7 These include Salmonella typhimurium,2 Leishmania donovani,3 and some species of Mycobacterium such as M bovis5 and M intracellulare.8 Targeted ablation of Ity/Lsh/Bcg9 or allelic exchange in susceptible mice10 confirmed its requirement for resistance to infection. Ity/Lsh/Bcg encodes the phagocyte-specific solute carrier 11a1 protein Slc11a1 (formerly Nramp1),11-13 which restricts pathogen replication by inducing the expression of major histocompatibility complex (MHC) class II molecules, cytokines (eg, TNFα), and chemokines.14 SLC11A1/Slc11a1 belongs to a family of polytopic membrane proteins whose functions include divalent cation acquisition in mammals (DCT1/DMT1/NRAMP2),15 taste perception in the fruit fly (malvolio),16 and stress signal transduction in plants (EIN2).17 In mice, a single nonsynonymous mutation in Slc11a1 codon 169 determines resistance (Gly169) or susceptibility (Asp169) to intracellular bacterial and leishmanial infections.7 Compared with susceptible mice, macrophages from mice that are resistant to infection also show increased oxidative burst and expression of the MHC class II transactivator CIITA, and I-A antigen.18-21
Human genetic variation and disease association studies have undergone a paradigm shift from mutation detection in susceptibility genes to whole-genome scans for single nucleotide polymorphisms (SNPs)22 and transcriptional analyses of the cognate genes to better understand the evolution of complex traits such as disease risk. Cis and/or trans-acting regulatory variation accounts greatly for quantitative differences in such traits.23-31 Evidence suggests that structural variation in candidate susceptibility genes (eg, due to mutations) cannot wholly account for such differences. In most cases, there are no identifiable mutations within such genes, including SLC11A1,32,33 yet there are demonstrable phenotypic differences (eg, in resistance to infection). No mutations occur in (human) SLC11A1 analogous to the functional coding region mutation in the murine ortholog, despite the apparent association of the gene with a variety of infectious and inflammatory diseases, including tuberculosis and rheumatoid arthritis (reviewed by Blackwell et al14,34 ). Disease association was found between specific (noncoding) polymorphic loci that included introns and microsatellites in both the 5′ and 3′ regions of the gene.32,35 Population-based resequencing studies have not found any deleterious mutations or SLC11A1 isoforms,32,33 ruling out protein structural differences as the basis for phenotypic variation in disease risk.
In this study, we hypothesized that quantitative differences in SLC11A1 transcription may underlie variation in disease susceptibility. We show that a highly polymorphic microsatellite of (GT/CA)n repeats within the proximal SLC11A1 promoter regulates variation in allele expression. We also show that this microsatellite has Z-DNA–forming propensity and binds to the transcriptional regulator hypoxia-inducible factor-1 (HIF-1) both in vitro and in vivo when macrophages are activated by pathogen or proinflammatory signals. Furthermore, HIF-1 accentuated differences in SLC11A1 allele expression. Although HIF-1 has hitherto been associated primarily with oxygen homeostasis and energy metabolism, our data imply that it may regulate interindividual differences in innate immunity. We therefore suggest that regulatory variation in SLC11A1 allele expression may determine heritable differences in immune responses to infection and inflammation.
Materials and methods
Plasmid constructs
The SLC11A1 promoter (385 bp) was amplified from human (placental) genomic DNA with PCR primers (MWG Biotech, Ebersberg, Germany) derived from NCBI GenBank accession no. U57605: sense (NR1pS), TAC GGT ACC GGG GTC TTG GAA CTC CAG ATC AAA G; and antisense (NR1pA), TAC AAG CTT CGA GTG CCC TGC CTC TTA CAT C (KpnI and HindIII restriction sites are underlined). The promoter was subcloned into the KpnI-HindIII (NEB, Hitchin, United Kingdom) sites of pGL3Basic vector (Promega, Southampton, United Kingdom) to generate SLC11A1-luc. Plasmid -137-luc was constructed by subcloning a fragment of the promoter amplified with the primer CAT GGT ACC CAG ATG TGT TGT GGG GCA CAG GGC and NR1pA. To generate enhancer plasmids, phosphorylated complementary microsatellite oligonucleotides (Table S1, available at the Blood website; see the Supplemental Tables link at the top of the online article) were mixed in equimolar amounts in 1 × DNA ligation buffer (NEB), incubated for 5 minutes at 95°C in a heating block, and annealed by slow cooling to less than 37°C. The duplexes were subcloned directionally into the NheI-XhoI sites of pGL3Promoter vector (Promega) and verified by sequencing (MWG Biotech).
Microsatellite interchange
Reconstitution of full-length allele-specific promoters by microsatellite interchange and generation of the hypoxia response element (HRE)–null mutant (GT)22 and the regulatory SNP −209T/C (otherwise denoted −237T/C)36,37 were performed simultaneously by swapping the microsatellite in SLC11A1-luc with those of other naturally occurring alleles (Figure 3A; Table S2) using the QuikChange Multi Site-Directed Mutagenesis System (Strategene, Amsterdam, the Netherlands) as instructed by the manufacturer. Mutagenesis was confirmed by DNA sequencing.
Cell culture, transfection, and reporter assays
Cell-culture media and supplements were obtained from Invitrogen (Paisley, United Kingdom), and cell lines were obtained from the European Collection of Animal Cell Cultures (Porton Down, United Kingdom). Human monocytes (THP-1) and murine macrophages (RAW264.7) were cultured in RPMI 1640 medium supplemented with antibiotics/antimycotics and 10% FBS. Baby hamster kidney cells (BHK) and the C4.5 and Ka13.5 cell lines (kindly provided by Peter Ratcliffe, Oxford University, Oxford, United Kingdom)38 were cultured in Dulbecco modified Eagle medium (DMEM) containing GlutaMAX1 and high glucose (Invitrogen). For transfections, cells were grown in Costar 24-well plates (Corning, Cambridge, MA), and transfected with 100 ng SLC11A1-luc using Lipofectamine 2000 (Invitrogen); 50 ng pSVβgal (Promega) was used as internal control. For transactivation assays, SLC11A1-luc was cotransfected with 100 ng pcDNA3-HIF-1α or pcDNA3-HIF-2α expression plasmids (kindly provided by Ingo Flamme, Bayer Health Care, Wuppertal, Germany). RAW264.7 transfectants were left untreated or treated for 24 hours with 1 μg/mL LPS from Escherichia coli (serotype 055:B6; Sigma, Poole, United Kingdom) or S typhimurium (Sigma), ManLAM and PiLAM (obtained from Germain Puzo, Institut de Pharmacologie et de Biologie Structurale du Centre National de la Recherche Scientifique, Toulouse, France), and 1 μg/mL LPS plus IFNγ (100 U/mL; Roche, Lewes, United Kingom). Luciferase expression was determined with the assay reagent (Promega), while β-galactosidase was assayed with Beta-Glo (Promega) in a microplate luminometer (Tropix TR717; Applied Biosystems, Warrington, United Kingdom); luciferase activities were normalized to β-galactosidase levels.
Yeast 1-hybrid assay
A yeast 1-hybrid assay was performed using the Matchmaker One-Hybrid System (Clontech, Basingstoke, United Kingdom). The SLC11A1 promoter was subcloned upstream of the lacZ gene at the HindIII-KpnI sites of pLacZi (Clontech) to generate pLacZi-NR1; the latter was linearized with ApaI (NEB) and integrated at the ura3 locus in YM4271 cells. Prototrophs were selected on synthetic dextrose (SD) medium lacking uracil (BIO101; Anachem, Luton, United Kingdom) to generate the reporter yeast strain SLC11A1-LacZiYM. GAL4BD-HIF-1α was constructed by subcloning HIF-1α cDNA from pcDNA3-HIF-1α into the BamHI-SalI sites of pGBT9 (Clontech) in-frame with the GAL4 DNA-binding domain; GAL4AD-ARNT was obtained by subcloning full-length HIF-1β/aryl hydrocarbon receptor nuclear translocator (ARNT) cDNA from pSport-ARNT (kindly provided by Christopher Bradfield, University of Wisconsin, Madison) into the BamHI-SalI sites of pGAD424 (Clontech) in-frame with the GAL4 activation domain. Competent SLC11A1-LacZiYM cells were prepared by the lithium acetate method and transformed with GAL4BD-HIF-1α alone or with GAL4AD-ARNT. Prototrophs were again selected on SD agar plates lacking leucine and uracil (for GAL4AD-ARNT transformants); tryptophan and uracil (for GAL4BD-HIF-1α transformants); and leucine, tryptophan, and uracil (for GAL4AD-ARNT and GAL4BD-HIF-1α cotransformants). After 2 days at 30°C, isolated colonies were picked and restreaked on SD/-Leu-Ura, SD/-Trp-Ura, and SD/-Leu-Trp-Ura agar plates or grown in the respective liquid drop-out medium. LacZ transactivation was determined by a colony-lift filter assay with 334 μg/mL final concentration of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal; Invitrogen) in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 [pH 7.0], and 0.27% β-mercaptoethanol), or by chemiluminescence using the Gal-Screen system (Applied Biosystems).
Conditional HIF-1α knockout mice, Western blotting, and real time RT-PCR
HIF-1a-LysMcre knockout mice have been described previously.39,40 Bone marrow–derived macrophages were isolated from wild-type and conditional knockout mice and cultured for 7 days in DMEM/10% FBS and 30% GM-CSF–conditioned medium. The macrophages were either uninfected or infected for 4 or 24 hours with S typhimurium (ATCC no. 13311; American Type Culture Collection, Manassas, VA), at a multiplicity of infection of 10; extracellular bacteria were killed with 100 μg/mL gentamicin and 5 μg/mL penicillin 1 hour after initiating infection. For Western blotting, macrophages were harvested after 4 hours, washed with PBS, and lysed with RIPA buffer. A total of 20 μg of nuclear extracts were resolved on a 4% to 12% Tris-tricine gel (Invitrogen) in MOPS buffer and transferred onto PVDF membrane. HIF-1α was detected with a 1:1000 dilution of rabbit polyclonal anti–HIF-1α antibody (Novus Biologicals, Littleton, CO). For real-time reverse transcription–polymerase chain reaction (RT-PCR), infected macrophages were incubated for 24 hours; RNA was purified with Trizol (Invitrogen) and reverse transcribed with the Superscript kit (Invitrogen). PCR was performed with the TaqMan SYBR Green Universal Master Mix (Applied Biosystems) using primers for Slc11a1 (forward 5′-GGA CAG TTC GTG ATG GAG GG-3′; reverse 5′-TTG AGT AGA TCG TTG AGG CCG-3′) and Nos2A (forward, 5′-ACC CTA AGA GTC ACA AAA TGG C-3′; reverse, 5′-TTG ATC CTC ACA TAC TGT GGA CG-3′). Quantitative real-time PCR was performed in quadruplicate for both Slc11a1 and Nos2A using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems), and was normalized to 18S rRNA levels.
Indirect immunofluorescence microscopy and immunohistochemistry
To determine HIF-1α activation in response to proinflammatory stimuli, RAW264.7 cells were grown on chamber slides (Nunc, Paisley, United Kingdom) and transfected with 200 ng pcDNA3/HIF-1α-FLAG. Approximately 12 hours after transfection, the cells were either untreated or treated with 1 μg/mL E coli LPS and 100 U/mL IFNγ. After another 12 hours, the cells were washed with PBS, fixed with 3.7% paraformaldehyde, and permeabilized with PBS, 0.05% saponin, and 5% nonfat milk. FLAG-tagged HIF-1α was detected with FITC-conjugated anti-FLAG antibody (Sigma); cells were counterstained for nuclei with DAPI (Vector Laboratories, Peterborough, United Kingdom). Images were captured with a Leica DC200 digital camera (Leica, Milton Keynes, United Kingdom) linked to a Zeiss Axioplan fluorescence microscope (Carl Zeiss, Welwyn Garden City, United Kingdom). Images were subsequently processed using Adobe Photoshop version 7 (Adobe Systems, Uxbridge, United Kingdom).
To determine HIF-1α and SLC11A1 expression in inflammatory disease, formalin-fixed, paraffin-embedded human synovial tissue sections with active rheumatoid arthritis (RA) were stained for HIF-1α using a 1:20 dilution of the monoclonal antibody ESEE 12241 (kindly provided by K. C. Gatter, Oxford University, United Kingdom); SLC11A1 was detected with a 1:200 dilution of anti-SLC11A1 antibody11 (kindly provided by Philip Gros, McGill University, Montréal, Canada). Tissue sections (3 μm) were deparaffinized, and endogenous peroxidase activity was quenched with methanol and 3% H2O2 for 15 minutes. Thereafter, slides were placed in antigen-unmasking buffer (pH 6.0; ILEM, Cortemaggiore, Italy) and microwaved (3 × 4 minutes) for antigen retrieval. A breast cancer tissue section with strong diffuse HIF-1α expression and uninvolved normal tissue were used as positive and negative controls, respectively. Primary antibodies were applied overnight at 4°C; normal rabbit IgG was used as negative control. Following washing with TBS, sections were incubated with Kwik mouse anti-rabbit biotinylated secondary antibody (Shandon-Upshaw, Pittsburgh, PA) for 15 minutes and washed in TBS. Kwik streptavidin peroxidase reagent (Shandon-Upshaw) was applied for 15 minutes. After washing in TBS, sections were treated for 15 minutes with 3,3′-diaminobenzidine, and briefly counterstained with hematoxylin. Images were acquired with a Nikon Eclipse E400 optical microscope, captured and processed with the Nikon Automatic Camera Tamer-1 (ACT-1) version 2 software (Nikon Instruments, Kingston, United Kingdom).
EMSA
Electrophoretic mobility shift assay (EMSA) was performed as previously described42 using nuclear extracts from untreated THP-1 cells (control) or from cells treated for 6 hours with 1 μg/mL LPS or 100 nM of the bacterial chemotactic peptide N-formyl-methionine-leucine-phenylalanine (fMLP). Double-stranded allele-specific microsatellite oligonucleotides (Table S3) were end-labeled with T4 polynucleotide kinase (NEB) and γ32P-ATP (Amersham Biosciences, Amersham, United Kingdom); 1 pmol of each oligonucleotide was incubated for 20 minutes in binding buffer with approximately 10 μg nuclear extracts. Cold competitor oligonucleotide (100 pmol) was added for 20 minutes at room temperature where necessary. Binding reactions were incubated at room temperature for 30 minutes and then resolved on 6% nondenaturing gels.
Full-length HIF-1α and ARNT were expressed in vitro using the T7 and Sp6 TNT Quick Coupled transcription and translation systems, respectively (Promega). Equal amounts of synthetic HIF-1α and ARNT were mixed in binding buffer and incubated at room temperature for 30 minutes to form heterodimers. HIF-1α or ARNT alone, unprocessed reticulocyte lysate (negative control), and the heterodimers were incubated with 1 pmol radiolabeled microsatellite DNA (of allele 3) or (GT)22; 100 pmol cold microsatellite DNA was added as competitor. Binding reactions were incubated for a further 20 minutes before electrophoresis.
ChIP assays
THP-1 cells were grown to mid-log phase and either untreated or treated for 6 hours with 1 μg/mL E coli LPS/100 U/mL IFNγ or 100 μg/mL zymosan A (Sigma). Chromatin immunoprecipitation (ChIP) assays were performed as previously described42 using an assay kit (Upstate Biotechnology, Lake Placid, NY). Samples were incubated overnight at 4°C with 5 μg anti–HIF-1α polyclonal antibody to the human protein (H-206/sc-10790; Santa Cruz Biotechnology, Santa Cruz, CA); control immunoprecipitation was performed with 5 μg of a nonspecific IgG (Sigma). DNA was purified by phenol-chloroform extraction and ethanol precipitation; 2 μL of the DNA was used for PCR with primers NR1pS and NR1pA as follows: 95°C (5 minutes), then 95°C (30 seconds), 60°C (1 minute), and 72°C (1 minute) for 35 cycles with a final extension at 72°C (10 minutes).
Detection of Z-DNA formation in vitro and in vivo
To determine microsatellite Z-DNA-forming propensity, radiolabeled allele 3 microsatellite was incubated for 2.5 hours at 37°C in EMSA buffer and 100 μM hexamine CoCl2 to induce Z-DNA formation. Aliquots (1 pmol) were incubated in EMSA buffer with various dilutions or 1 μL of undiluted anti–Z-DNA antibody Z2243 (kindly provided by B. David Stollar, Tufts University, Boston, MA), in a final volume of 10 μL for 2 hours at room temperature; 1 μL of undiluted c-Myc antibody (Cell Signaling Technology/NEB, Hitchin, United Kingdom) was used as control. Samples were resolved on 6% gels.
To determine Z-DNA formation in vivo, BHK cells were transfected with 10 μg SLC11A1-luc or the microsatellite construct NR1HREab-luc; pGL3Basic was used as negative control. After 24 hours, the cells were cross-linked with formaldehyde; chromatin was extracted and (without sonication) aliquots were taken for input controls, or treated with and without approximately 20 μg Z22, and with preimmune rabbit serum. After immunoprecipitation, plasmid DNA was purified on Nucleospin miniprep columns (Macherey-Nagel, Düren, Germany) and transformed into XL10 Gold ultracompetent cells (Stratagene Europe, Amsterdam, The Netherlands). Transformants were enumerated for each construct in triplicate after incubating at 37°C overnight on Luria-Bertani broth (LB)/ampicillin (100 μg/mL) agar plates.
Statistical analysis
Data were analyzed with GraphPad Prism (GraphPad Software, San Diego, CA). Where appropriate, data sets were analyzed by the Mann-Whitney U test or the 1-way analysis of variance, Student-Newman-Keuls multiple-comparisons test. P values less than .05 were accepted as significant. Results were expressed as means plus or minus SEM.
Results
The SLC11A1 promoter contains a regulatory microsatellite polymorphism
To determine transcriptional regulation of SLC11A1, we amplified 385 bp of its promoter from human genomic DNA and subcloned it upstream of the luciferase gene to produce SLC11A1-luc; this construct included a highly polymorphic microsatellite of (GT/CA)n dinucleotides. DNA sequencing confirmed that we had cloned allele 3 (Figure 1).34,35 Deletion mapping, transfection of both sense and antisense promoter constructs into BHK cells, and luciferase assays showed an orientation-independent enhancer function restricted to the microsatellite region of the promoter (Figure 2A). This suggested that the promoter may be bidirectional in nature; such promoters abound in the human genome.49
Nucleotide sequence of the SLC11A1 (allele 3) promoter. Double-underlined nucleotide (G) is the putative transcription start site.44 The polypurine/polypyrimidine tract [(GT/CA)n] and Z-DNA–forming microsatellite sequence is single-underlined. The microsatellite has a z score (Z-DNA–forming potential) of 12 598.14 (z score minimum is 700), as determined with the ZHunt algorithm.45,46 HREs47,48 are in boldface. Shaded thymidine is a regulatory SNP; it was subsequently mutated to the more common cytosine SNP (T → C) simultaneously with microsatellite interchange.
Nucleotide sequence of the SLC11A1 (allele 3) promoter. Double-underlined nucleotide (G) is the putative transcription start site.44 The polypurine/polypyrimidine tract [(GT/CA)n] and Z-DNA–forming microsatellite sequence is single-underlined. The microsatellite has a z score (Z-DNA–forming potential) of 12 598.14 (z score minimum is 700), as determined with the ZHunt algorithm.45,46 HREs47,48 are in boldface. Shaded thymidine is a regulatory SNP; it was subsequently mutated to the more common cytosine SNP (T → C) simultaneously with microsatellite interchange.
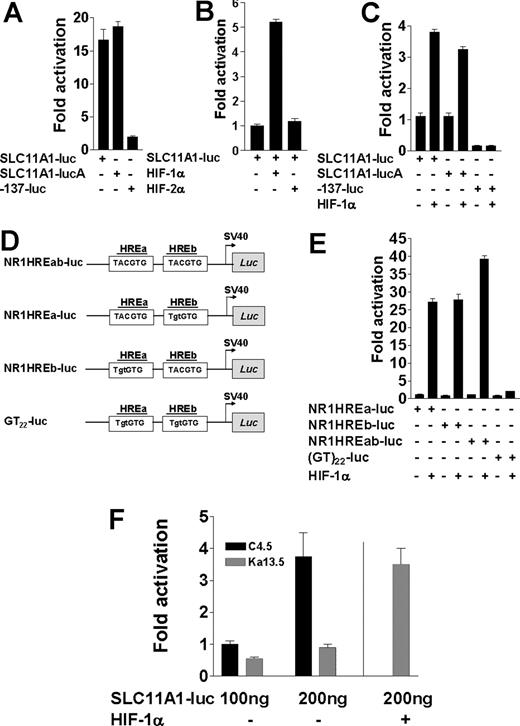
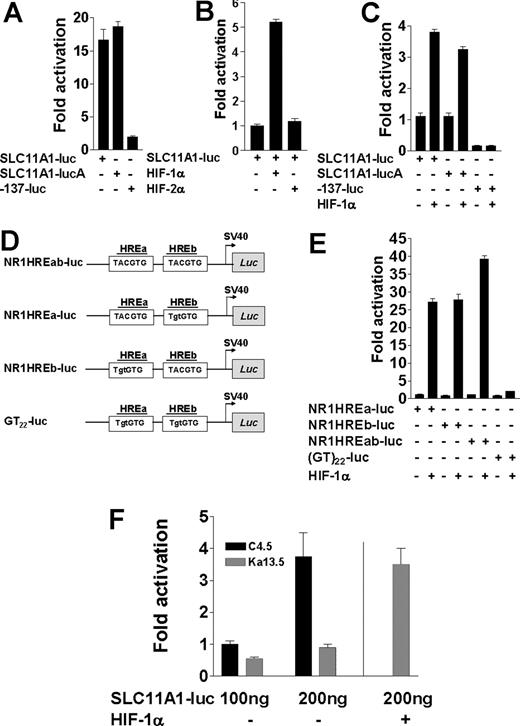
The polymorphic microsatellite in the proximal SLC11A1 promoter directs transcriptional regulation by HIF-1α. (A) Luciferase activities from 100 ng SLC11A1-luc constructs in both sense and antisense (SLC11A1-lucA) orientations compared with the construct -137-luc, which lacks the polymorphic microsatellite. Fold activation was with respect to pGL3Basic activity. (B) Comparison of SLC11A1-luc transcriptional activation by HIF-1α and HIF-2α. (C) HIF-1α regulation of SLC11A1 promoter activity is orientation independent and requires the polymorphic microsatellite. Luciferase expression was determined for promoter constructs transfected without or with 100 ng HIF-1α; fold activation is a ratio of transactivation and basal luciferase activities. Empty pcDNA3.1 vector was added where necessary to ensure equivalent amounts of plasmids. Activity of -137-luc is in comparison with SLC11A1-luc. For A-C, data are means (±SEM). (D) Duplex microsatellite oligonucleotide constructs with intact HREa and HREb (NR1HREab-luc) or where either or both HREs were mutated (mutations are shown in lowercase) to generate NR1HREa-luc, NR1HREb-luc, and GT22-luc, respectively; duplexes were subcloned into pGL3Promoter vector (see Table S1). (E) Transactivation of HRE-luciferase constructs by HIF-1α. The constructs (100 ng each) from panel D were transfected alone or with 100 ng HIF-1α plasmid into BHK cells. Luciferase activities were normalized to β-galactosidase internal control. Data are representative of 5 experiments (means ± SEM). (F) HIF-1α–deficient cells cannot support SLC11A1 expression. SLC11A1-luc (100 ng) was transfected alone or with 100 ng HIF-1α plasmid into wild-type (C4.5) and HIF-1α–deficient (Ka13.5) cells; luciferase activity was normalized to β-galactosidase levels. A vertical line separates Ka13.5 cells trans-complemented with HIF-1α by cotransfection. Data are representative of 5 independent experiments (means ± SEM).
The polymorphic microsatellite in the proximal SLC11A1 promoter directs transcriptional regulation by HIF-1α. (A) Luciferase activities from 100 ng SLC11A1-luc constructs in both sense and antisense (SLC11A1-lucA) orientations compared with the construct -137-luc, which lacks the polymorphic microsatellite. Fold activation was with respect to pGL3Basic activity. (B) Comparison of SLC11A1-luc transcriptional activation by HIF-1α and HIF-2α. (C) HIF-1α regulation of SLC11A1 promoter activity is orientation independent and requires the polymorphic microsatellite. Luciferase expression was determined for promoter constructs transfected without or with 100 ng HIF-1α; fold activation is a ratio of transactivation and basal luciferase activities. Empty pcDNA3.1 vector was added where necessary to ensure equivalent amounts of plasmids. Activity of -137-luc is in comparison with SLC11A1-luc. For A-C, data are means (±SEM). (D) Duplex microsatellite oligonucleotide constructs with intact HREa and HREb (NR1HREab-luc) or where either or both HREs were mutated (mutations are shown in lowercase) to generate NR1HREa-luc, NR1HREb-luc, and GT22-luc, respectively; duplexes were subcloned into pGL3Promoter vector (see Table S1). (E) Transactivation of HRE-luciferase constructs by HIF-1α. The constructs (100 ng each) from panel D were transfected alone or with 100 ng HIF-1α plasmid into BHK cells. Luciferase activities were normalized to β-galactosidase internal control. Data are representative of 5 experiments (means ± SEM). (F) HIF-1α–deficient cells cannot support SLC11A1 expression. SLC11A1-luc (100 ng) was transfected alone or with 100 ng HIF-1α plasmid into wild-type (C4.5) and HIF-1α–deficient (Ka13.5) cells; luciferase activity was normalized to β-galactosidase levels. A vertical line separates Ka13.5 cells trans-complemented with HIF-1α by cotransfection. Data are representative of 5 independent experiments (means ± SEM).
Sequence analysis revealed 2 tandem putative cryptic HREs—TACGTG47,48 (Figure 1)—within the microsatellite. Cotransfection of BHK cells with SLC11A1-luc and either HIF-1α or HIF-2α expression vectors revealed that while HIF-2α could not transactivate the promoter, HIF-1α induced up to 4-fold increase in luciferase expression under normoxic conditions (P = .002; Figure 2B,C). As SLC11A1 is almost exclusively expressed in myeloid cells, this finding suggests that target gene transactivation by either HIF-1α or HIF-2α may be context dependent, and that the former may be more important for macrophage activation. Microsatellite constructs with 1 or both putative HREs (hereafter designated NR1HREa and NR1HREb; Figure 2D and Table S1) were furthermore strongly transactivated by HIF-1α (P < .001), whereas luciferase expression was abrogated with the microsatellite construct (GT)22-luc in which both HREs were mutated (Figure 2E). These findings were corroborated by transfecting SLC11A1-luc into HIF-1α wild-type (C4.5) and HIF-1α–null (Ka13.5) CHO cell lines.38 While we could detect only scant activity in the latter, luciferase expression in C4.5 increased in a dose-dependent manner (P = .02). We observed comparable luciferase activity in Ka13.5 cells only upon cotransfection of both SLC11A1-luc and HIF-1α (Figure 2F). The microsatellite therefore appears necessary and sufficient to direct SLC11A1 regulation by HIF-1α. This could explain its high conservation in humans as a consequence of strong positive directional selection; it thus appears to be a true regulatory polymorphism.
HIF-1α regulates SLC11A1 allele expression phenotypes from the cognate microsatellites
A total of 9 SLC11A1 alleles have been identified in different populations across the world; these alleles differ not only in microsatellite length (Figure 3A), but also in prevalence, as they are distributed along ethnic and racial lines.14,32,36 These distributions may account for regional geographic variations in disease-type susceptibility. Certain polymorphisms of the microsatellite are associated with susceptibility to infectious and inflammatory diseases.14 To assess whether the microsatellite could determine differences in allele expression, we performed microsatellite interchange by site-directed mutagenesis, replacing the microsatellite in SLC11A1-luc with other naturally occurring alleles (Figure 3A; Table S2). We found that NR1HREb was deleted in allele 4, while in allele 7, it was mutated to TATGTG. The presence of only NR1HREa in these alleles corroborates data in Figure 2E, indicating that both HREs were independent and functionally nonadditive. We also mutated both HREs within SLC11A1-luc to generate a3*GT22 and created a single HRE deletion, a3delHREa, to simulate alleles 4 and 7. As the SLC11A1 microsatellite is often inherited along with unique SNPs, we also generated and compared alleles 2 and 3 variants in which regulatory SNPs have been reported.36,37 Using these constructs in transfections, we found allelic variation in luciferase expression that was accentuated approximately 3.4- to 11.3-fold by HIF-1α (P = .04 to < .001), with allele 3 and its SNP variants driving the highest luciferase expression. By contrast, luciferase expression was diminished with a3*GT22 (Figure 3B). These findings suggest a strong association between HIF-1α and microsatellite-dependent SLC11A1 allele expression phenotypes. Although these data were obtained in a surrogate system, they are consistent with expression phenotypes underlying stochastic, interindividual variation in gene expression.23,24,26-28,30 We speculate that these differences in SLC11A1 allele expression may account for heritable variation in susceptibility to infection and/or inflammation within and between populations. Furthermore, these experiments provide strong evidence that microsatellites may determine the directional evolution of complex traits by regulating gene expression phenotypes.
HIF-1α regulates SLC11A1 allele expression phenotypes from the cognate microsatellites. (A) Alignment of microsatellites of SLC11A1 alleles identified in different (human) individuals and populations. HIF-1α response elements (TACGTG) are underlined. Note HREb deletion or mutation in alleles 4 and 7, respectively; the control (nonnatural) mutant microsatellite (GT)22 is also shown for comparison. Nucleotides (not shown) juxtaposing either side of each microsatellite are identical. Using allele 1 as reference, dashes indicate deletions, red-coded nucleotides are identical residues, green-coded residues are conserved in at least 5 alleles, and black prints indicate nonconserved residues. (B) One hundred nanograms each of allele-specific promoter constructs derived by microsatellite interchange were transfected alone or with 100 ng HIF-1α expression vector. Allele 3 microsatellite (a3) was interchanged (*) with microsatellites of alleles 1 (a1), 2 (a2), 6 (a6), and 7 (a7). Regulatory SNPs in allele 2 (a3*a2TG and a3*a2CG) and allele 3 (a3TG*a3CG) were also generated. The mutant HRE-null microsatellite a3*GT22 carried the a3CG SNP, and a3delHREa is allele 3 with a mutated NR1HREa. Fold activation is with respect to pGL3Basic. Results are representative of 5 experiments (means ± SEM).
HIF-1α regulates SLC11A1 allele expression phenotypes from the cognate microsatellites. (A) Alignment of microsatellites of SLC11A1 alleles identified in different (human) individuals and populations. HIF-1α response elements (TACGTG) are underlined. Note HREb deletion or mutation in alleles 4 and 7, respectively; the control (nonnatural) mutant microsatellite (GT)22 is also shown for comparison. Nucleotides (not shown) juxtaposing either side of each microsatellite are identical. Using allele 1 as reference, dashes indicate deletions, red-coded nucleotides are identical residues, green-coded residues are conserved in at least 5 alleles, and black prints indicate nonconserved residues. (B) One hundred nanograms each of allele-specific promoter constructs derived by microsatellite interchange were transfected alone or with 100 ng HIF-1α expression vector. Allele 3 microsatellite (a3) was interchanged (*) with microsatellites of alleles 1 (a1), 2 (a2), 6 (a6), and 7 (a7). Regulatory SNPs in allele 2 (a3*a2TG and a3*a2CG) and allele 3 (a3TG*a3CG) were also generated. The mutant HRE-null microsatellite a3*GT22 carried the a3CG SNP, and a3delHREa is allele 3 with a mutated NR1HREa. Fold activation is with respect to pGL3Basic. Results are representative of 5 experiments (means ± SEM).
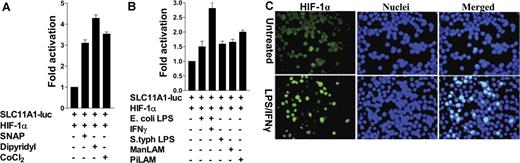
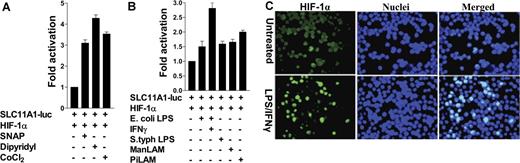
HIF-1α inducers and/or stabilizers directly affect SLC11A1 regulation
From first principles, agents and/or pathways that induce and stabilize HIF-1α could also influence SLC11A1 expression. To examine this further, we found that the pharmacologic hypoxia mimetics and HIF-1α stabilizers cobalt chloride (CoCl2) and dipyridyl (an iron chelator), and the nitric oxide (NO) donor S-nitroso-N-acetyl penicillamine, enhanced luciferase expression in cotransfections of SLC11A1-luc and HIF-1α (P = .002; Figure 4A). These results are consistent with observations that changes in macrophage-labile iron pool and redox status affect HIF-1α expression and SLC11A1 activation of phagocyte defenses.21,50 Next, as surrogate SLC11A1-innate immune stimuli, we used E coli and S typhimurium LPS, and mannose-capped lipoarabinomannan (ManLAM) from the slow-growing mycobacterium M bovis BCG, and phosphoinositide-capped LAM (PiLAM) from the fast-growing nonpathogenic M smegmatis. Both LPS and lipoarabinomannan species enhanced SLC11A1-luc transactivation by HIF-1α in RAW264.7 cells (P = .02 to .002). Costimulation with LPS and IFNγ induced the highest level of luciferase expression (Figure 4B). HIF-1α activation was further confirmed by indirect immunofluorescence of RAW 264.7 cells transfected with FLAG-tagged HIF-1α; LPS/IFNγ treatment of the cells caused HIF-1α nuclear translocation (Figure 4C). These results indicate that microbe-associated molecular patterns can stimulate the HIF-1α–SLC11A1 axis, and are consistent with evidence of normoxic induction of HIF-1α by LPS, proinflammatory cytokines, and whole live bacteria.39,40,50-52
HIF-1α inducers and/or stabilizers directly influence SLC11A1 regulation. (A) Representative data from cells cotransfected with SLC11A1-luc and HIF-1α, and untreated or treated with 100 μM CoCl2, 100 μM dipyridyl, and 200 μM of the nitrosative stressor S-nitroso-N-acetypenicillamine. (B) Pathogen-associated molecular patterns differentially activate SLC11A1 through HIF-1α. RAW264.7 cells cotransfected with SLC11A1-luc and HIF-1α were treated with lipoarabinomannans (ManLAM or PiLAM) and E. coli or S. typhimurium LPS, with or without IFNγ. Transactivation data are representative of 3 independent experiments (means ± SEM). (C) Activation of HIF-1α by proinflammatory stimuli causes its translocation to the nucleus. Immunofluorescence of RAW264.7 cells transfected with HIF-1α cDNA and left untreated or treated with LPS (1 μg/mL) and IFNγ (100 U/mL). HIF-1α was detected with FITC-labeled anti-FLAG antibody; nuclei were counterstained with DAPI. Image was captured with a Leica DC200 digital camera and software (version 2.51), using a 40×/0.70 numeric aperture (NA) oil objective lens, and processed with Adobe Photoshop 7. Magnification is ×40.
HIF-1α inducers and/or stabilizers directly influence SLC11A1 regulation. (A) Representative data from cells cotransfected with SLC11A1-luc and HIF-1α, and untreated or treated with 100 μM CoCl2, 100 μM dipyridyl, and 200 μM of the nitrosative stressor S-nitroso-N-acetypenicillamine. (B) Pathogen-associated molecular patterns differentially activate SLC11A1 through HIF-1α. RAW264.7 cells cotransfected with SLC11A1-luc and HIF-1α were treated with lipoarabinomannans (ManLAM or PiLAM) and E. coli or S. typhimurium LPS, with or without IFNγ. Transactivation data are representative of 3 independent experiments (means ± SEM). (C) Activation of HIF-1α by proinflammatory stimuli causes its translocation to the nucleus. Immunofluorescence of RAW264.7 cells transfected with HIF-1α cDNA and left untreated or treated with LPS (1 μg/mL) and IFNγ (100 U/mL). HIF-1α was detected with FITC-labeled anti-FLAG antibody; nuclei were counterstained with DAPI. Image was captured with a Leica DC200 digital camera and software (version 2.51), using a 40×/0.70 numeric aperture (NA) oil objective lens, and processed with Adobe Photoshop 7. Magnification is ×40.
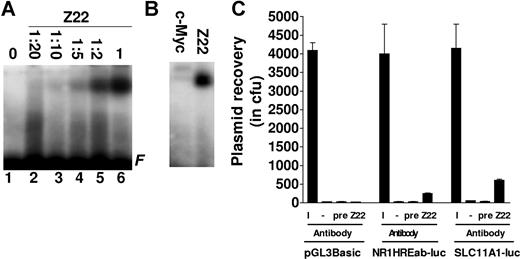
The SLC11A1 microsatellite has Z-DNA–forming propensity in vitro and in vivo
As microsatellites can adopt a non-B or Z-DNA conformation,53-55 we asked whether the SLC11A1 promoter microsatellite could form Z-DNA. In EMSAs with hexamine CoCl2–treated microsatellite DNA, we observed that the Z-DNA–specific antibody Z2243 bound to the microsatellite in a concentration-dependent manner, while a control antibody (against c-Myc) did not (Figure 5A,B). To determine microsatellite propensity to form Z-DNA in vivo, we transfected pGL3Basic, SLC11A1-luc, and the microsatellite construct NR1HREab-luc separately into BHK cells. Chromatin was immunoprecipitated without or with Z22 and preimmune serum; immunoprecipitated chromatin was used to transform competent E. coli cells. Enumerating ampicillin-resistant colonies, we recovered more plasmids from cells transfected with SLC11A1-luc and NR1HREab-luc using Z22 (P = .002) compared with immunoprecipitation from cells transfected with pGL3Basic, nonimmunoprecipitated chromatin, and ChIP with preimmune serum (Figure 5C). These data suggest that the microsatellite has Z-DNA–forming propensity both in vitro and in vivo. Due to differences in microsatellite length, alleles may also differ in Z-DNA–forming potential, and we surmise that this could represent another facet of SLC11A1 transcriptional regulation.
The SLC11A1 promoter microsatellite has Z-DNA-forming propensity. (A) Z-DNA binding assay. Allele 3 microsatellite (a3) was treated with 100 μM hexamine CoCl2 and incubated without (lane 1) or with various dilutions of anti–Z-DNA antibody Z22 (lanes 2-5) and undiluted Z22 (lane 6). F indicates free probe. (B) Radiolabeled a3 was incubated with undiluted c-Myc and Z-DNA antibodies. (C) Z-DNA ChIP. SLC11A1-luc, NR1HREab-luc, and pGL3Basic were transfected into BHK cells, and plasmids were recovered from chromatin with Z22; negative controls were without antibody (−) or treated with preimmune serum (pre). Plasmids were also recovered from 10% of input chromatin (I). Data are presented as averages of colony forming units from 3 replicates. Error bars denote SEM.
The SLC11A1 promoter microsatellite has Z-DNA-forming propensity. (A) Z-DNA binding assay. Allele 3 microsatellite (a3) was treated with 100 μM hexamine CoCl2 and incubated without (lane 1) or with various dilutions of anti–Z-DNA antibody Z22 (lanes 2-5) and undiluted Z22 (lane 6). F indicates free probe. (B) Radiolabeled a3 was incubated with undiluted c-Myc and Z-DNA antibodies. (C) Z-DNA ChIP. SLC11A1-luc, NR1HREab-luc, and pGL3Basic were transfected into BHK cells, and plasmids were recovered from chromatin with Z22; negative controls were without antibody (−) or treated with preimmune serum (pre). Plasmids were also recovered from 10% of input chromatin (I). Data are presented as averages of colony forming units from 3 replicates. Error bars denote SEM.
HIF-1 binds directly to the SLC11A1 microsatellite in vitro and in vivo
Next, we performed EMSA to determine HIF-1 binding to allele-specific microsatellites (Table S3) using nuclear extracts from THP-1 monocytes with or without LPS treatment. We found that LPS stimulated differential HIF-1 binding to the microsatellites, while binding to the mutant microsatellite (GT)22 was poor (Figure 6A). Similar results were obtained with monocytes treated with the proinflammatory bacterial chemotactic tripeptide fMLP (Figure 6B). Microsatellite HIF-1 binding specificity was confirmed in EMSA using full-length synthetic HIF-1α and ARNT (Figure 6C) as well as glutathione S-transferase fusion proteins of their respective bHLH/PAS domains (data not shown). In both cases, HIF-1α/ARNT heterodimers bound to the wild-type but not to the mutant (GT)22 microsatellites; binding could be completely suppressed with a 100-fold molar excess of the cold microsatellite probe.
HIF-1α/ARNT heterodimers bind directly to the microsatellite in vitro and in vivo. EMSA was performed with nuclear extracts from THP-1 cells either untreated or treated for 6 hours with (A) 1 μg/mL E. coli LPS and (B) 100 nM fMLP. Alleles 1 to 7 microsatellites or the mutant (GT)22 were used for binding assays in panel A; shifts for only alleles 1 to 6 are shown in panel B. In both cases, lane 1 contains no nuclear extract (ie, a3 oligonucleotide only [−] and lane 2 (Un) contains untreated control nuclear extract; arrow shows specific HIF-1 binding. (C) In vitro–translated HIF-1α (lane 3), ARNT (lane 4) or HIF-1α/ARNT heterodimers (lanes 5 and 7) were incubated with untreated (lanes 3-6 and 8) or hexamine CoCl2-treated allele 3 microsatellite (lane 7). HIF-1α/ARNT heterodimers bound (arrows) to wild-type but not to the mutant microsatellite (GT)22 (lane 8). Binding was competitively inhibited by excess cold microsatellite DNA (lane 6). Lane 1 shows oligonucleotide only; lane 2, negative control with untranslated reticulocyte lysate; NS, nonspecific binding component(s); F, free probe in panels A-C. (D) Chromatin immunoprecipitation of the SLC11A1 promoter from THP-1 cells treated with LPS/IFNγ or zymosan was performed using an anti–HIF-1α antibody; a nonspecific antibody (NS) was used as negative control, and 10% input chromatin was used as positive control for PCR. (E) Yeast 1-hybrid assay for HIF-1α/ARNT occupancy and transactivation of the SLC11A1 promoter. A total of 4 independent GAL4BD-HIF-1α/GAL4AD-ARNT yeast cotransformants were tested for Leu-Trp-Ura prototrophy and for qualitative β-galactosidase expression (blue coloration) by colony-lift filter assay, after exposing the prototrophs to the β-galactosidase substrate X-gal for 4 hours. (F) Quantitative β-galactosidase assay. Yeast cells with a chromosomally-integrated copy of the promoter were either untransformed or transformed with GAL4BD-HIF-1α, GAL4AD-ARNT, or both, and β-galactosidase expression was determined by chemiluminescence. SD/Leu-Trp-Ura medium was used as background control. Error bars denote SEM.
HIF-1α/ARNT heterodimers bind directly to the microsatellite in vitro and in vivo. EMSA was performed with nuclear extracts from THP-1 cells either untreated or treated for 6 hours with (A) 1 μg/mL E. coli LPS and (B) 100 nM fMLP. Alleles 1 to 7 microsatellites or the mutant (GT)22 were used for binding assays in panel A; shifts for only alleles 1 to 6 are shown in panel B. In both cases, lane 1 contains no nuclear extract (ie, a3 oligonucleotide only [−] and lane 2 (Un) contains untreated control nuclear extract; arrow shows specific HIF-1 binding. (C) In vitro–translated HIF-1α (lane 3), ARNT (lane 4) or HIF-1α/ARNT heterodimers (lanes 5 and 7) were incubated with untreated (lanes 3-6 and 8) or hexamine CoCl2-treated allele 3 microsatellite (lane 7). HIF-1α/ARNT heterodimers bound (arrows) to wild-type but not to the mutant microsatellite (GT)22 (lane 8). Binding was competitively inhibited by excess cold microsatellite DNA (lane 6). Lane 1 shows oligonucleotide only; lane 2, negative control with untranslated reticulocyte lysate; NS, nonspecific binding component(s); F, free probe in panels A-C. (D) Chromatin immunoprecipitation of the SLC11A1 promoter from THP-1 cells treated with LPS/IFNγ or zymosan was performed using an anti–HIF-1α antibody; a nonspecific antibody (NS) was used as negative control, and 10% input chromatin was used as positive control for PCR. (E) Yeast 1-hybrid assay for HIF-1α/ARNT occupancy and transactivation of the SLC11A1 promoter. A total of 4 independent GAL4BD-HIF-1α/GAL4AD-ARNT yeast cotransformants were tested for Leu-Trp-Ura prototrophy and for qualitative β-galactosidase expression (blue coloration) by colony-lift filter assay, after exposing the prototrophs to the β-galactosidase substrate X-gal for 4 hours. (F) Quantitative β-galactosidase assay. Yeast cells with a chromosomally-integrated copy of the promoter were either untransformed or transformed with GAL4BD-HIF-1α, GAL4AD-ARNT, or both, and β-galactosidase expression was determined by chemiluminescence. SD/Leu-Trp-Ura medium was used as background control. Error bars denote SEM.
To provide evidence of HIF-1 recruitment to the promoter in vivo and in response to proinflammatory stimuli, we performed ChIP assays with THP-1 cells treated with LPS/IFNγ or zymosan. The promoter could be immunoprecipitated with an anti–HIF-1α antibody, but not with a control antibody of the same isotype. This assay also showed that LPS/IFNγ increased promoter occupancy by HIF-1 compared with zymosan, as indicated by a greater amount of immunoprecipitated chromatin (Figure 6D). Finally promoter occupancy by HIF-1 could be reconstituted in vivo using the yeast 1-hybrid assay. Yeast cells with a chromosomally integrated copy of the SLC11A1 promoter fused to lacZ and cotransformed with GAL4BD–HIF-1α and GAL4AD-ARNT were robustly prototrophic on selection medium and produced very high levels of β-galactosidase (Figure 6E,F), while control transformations did not. Taken together, the data strongly suggest allelic variation in HIF-1 recruitment to the SLC11A1 promoter. Furthermore, HIF-1 binding may be enhanced by macrophage stimulation with pathogen or proinflammatory signals.
HIF-1α is required for SLC11A1 activation during infection
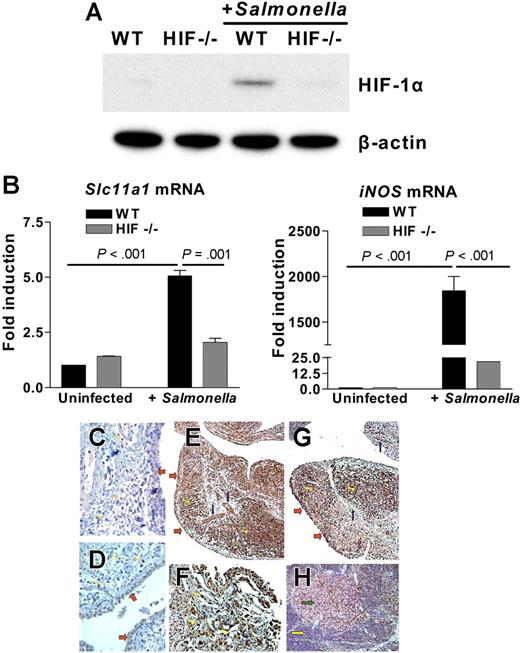
To test for a functional link between SLC11A1/Slc11a1 and HIF-1α during infection by the prototypical intracellular pathogen S. typhimurium, we used conditional HIF-1a-LysMcre knockout mice in which HIF-1α expression has been selectively ablated in myeloid cells by Cre-mediated recombination driven by the Lysozyme M (LysM) promoter.39,40 We challenged bone marrow–derived macrophages from wild-type and HIF-1a-LysMcre mice with S. typhimurium and found increased HIF-1α expression in macrophages from wild-type (compared with uninfected controls) mice, but not in HIF-1a-LysMcre mice (Figure 7A). In parallel, we detected a 5-fold increase in Slc11a1 mRNA in wild-type macrophages compared with uninfected controls or HIF-1a-LysMcre mice (P = .001; Figure 7B left panel). To rule out differences in Slc11a1 allele type as a confounding variable, we genotyped both wild-type and HIF-1a mutant mice and found that they both carried the Asp169 allele (data not shown). We also found distinct Slc11a1 microsatellite-length polymorphisms between mouse strains, each with 1 or 2 consensus HIF-1 binding site(s), TGCGTG37,38 (H.K.B. et al, unpublished data, June 2005). The observations therefore suggest that as with SLC11A1, HIF-1α may regulate Slc11a1 responses to infection independently of the codon 169 mutation.7 In parallel with Slc11a1 activation, inducible NO synthase gene (iNOS) was also very strongly activated by S. typhimurium infection in wild-type compared with HIF-1a-LysMcre macrophages (P < .001; Figure 7B right panel). This result may reflect synergy or epistatic interactions between Slc11a1 and iNOS, a known stabilizer of HIF-1α,56 in macrophage priming to fight infection, and that HIF-1α insufficiency could affect SLC11A1 expression and susceptibility to infection.
HIF-1α is functionally linked to prototypical SLC11A1-associated diseases. (A) S typhimurium infection up-regulates HIF-1α. Bone marrow–derived macrophages from wild-type (WT) or conditional HIF-1a-LysMcre (HIF−/−) mice were infected with Salmonella (multiplicity of infection = 10). HIF-1α expression was detected by Western blotting using β-actin expression as loading control. (B) Slc11a1 and iNOS are activated in Salmonella infection. Slc11a1 and iNOS expression was determined and compared in WT and HIF-1a-LysMcre mice by quantitative real-time RT-PCR; expression levels were normalized to 18S ribosomal RNA levels. Fold induction is with respect to uninfected controls; data represent averages of 4 replications (means ± SEM). (C-H) Association between HIF-1α and SLC11A1 in inflammatory disease. Immunohistochemical staining of control, nonarthritic tissue with antibodies to (C) HIF-1α or (D) SLC11A1 shows that neither protein is induced or expressed in the synovium, vessels, or lymphocytes. (E) RA tissue sections were immunolabeled with anti-SLC11A1 antibody; SLC11A1 expression was detected in synovial cells (red arrows), vessels (blue arrows), and infiltrating lymphocytes (yellow arrows). (F) A × 200 magnification of panel E to show infiltrating lymphocytes stained for SLC11A1. (G) HIF-1α shows a pattern of expression that is similar to SLC11A1 expression in RA tissue sections. (H) SLC11A1 reactivity was also detected in lymphocytes in reactive sinus histiocytosis (green arrow shows reactive lymph node) but not in a control lymph node (yellow arrow). All sections were counterstained with hematoxylin. Images were acquired with a Nikon Eclipse E400 microscope using 20×/0.75 NA (panels C, D, F, and H; magnification ×200) or 10×/0.45 NA objective lenses (panels E and G; magnification ×100); They were processed with Nikon ACT-1 software version 2 and assembled with Adobe Photoshop version 7.
HIF-1α is functionally linked to prototypical SLC11A1-associated diseases. (A) S typhimurium infection up-regulates HIF-1α. Bone marrow–derived macrophages from wild-type (WT) or conditional HIF-1a-LysMcre (HIF−/−) mice were infected with Salmonella (multiplicity of infection = 10). HIF-1α expression was detected by Western blotting using β-actin expression as loading control. (B) Slc11a1 and iNOS are activated in Salmonella infection. Slc11a1 and iNOS expression was determined and compared in WT and HIF-1a-LysMcre mice by quantitative real-time RT-PCR; expression levels were normalized to 18S ribosomal RNA levels. Fold induction is with respect to uninfected controls; data represent averages of 4 replications (means ± SEM). (C-H) Association between HIF-1α and SLC11A1 in inflammatory disease. Immunohistochemical staining of control, nonarthritic tissue with antibodies to (C) HIF-1α or (D) SLC11A1 shows that neither protein is induced or expressed in the synovium, vessels, or lymphocytes. (E) RA tissue sections were immunolabeled with anti-SLC11A1 antibody; SLC11A1 expression was detected in synovial cells (red arrows), vessels (blue arrows), and infiltrating lymphocytes (yellow arrows). (F) A × 200 magnification of panel E to show infiltrating lymphocytes stained for SLC11A1. (G) HIF-1α shows a pattern of expression that is similar to SLC11A1 expression in RA tissue sections. (H) SLC11A1 reactivity was also detected in lymphocytes in reactive sinus histiocytosis (green arrow shows reactive lymph node) but not in a control lymph node (yellow arrow). All sections were counterstained with hematoxylin. Images were acquired with a Nikon Eclipse E400 microscope using 20×/0.75 NA (panels C, D, F, and H; magnification ×200) or 10×/0.45 NA objective lenses (panels E and G; magnification ×100); They were processed with Nikon ACT-1 software version 2 and assembled with Adobe Photoshop version 7.
HIF-1α and SLC11A1 are overexpressed in inflammatory disease
Finally, we asked whether HIF-1α may be involved in prototypical human inflammatory diseases associated with SLC11A1 allelic variation. For example, association was observed between the high expressor allele 3 and susceptibility to RA.14,57 Furthermore, overexpression of HIF-1α in myeloid cells by conditional ablation of its negative regulator, the von Hippel-Lindau protein (pVHL), caused protracted inflammatory responses in mice, while HIF-1a deletion prevented inflammation.39 We performed (parallel) immunohistochemistry on uninvolved tissue sections from control patients without RA and on pathologic RA tissue sections for both SLC11A1 and HIF-1α expression. Although neither HIF-1α (Figure 7C) nor SLC11A1 (Figure 7D) was expressed in normal uninvolved tissue, we found pronounced SLC11A1 immunoreactivity in synovial, endothelial, and inflammatory cells in RA samples (Figure 7E-F). Based on cell morphology, CD3+ T cells, and CD20+ B cells, these inflammatory cells were predominantly lymphocytes. SLC11A1 expression coincided with the up-regulation of HIF-1α in inflammatory cells within RA sections (Figure 7G). We also found that lymph nodes with reactive sinus histiocytosis characteristic of RA stained for SLC11A1, while no reactivity was detected in normal lymphoid follicles (Figure 7H). These results suggest that HIF-1α may be an important regulatory quantitative trait locus linked to inflammatory diseases associated with SLC11A1. However, the myriad interacting pathways in autoimmune disease susceptibility58 preclude our assertion of synergy between HIF-1α and SLC11A1 in RA susceptibility and pathogenesis. In other words, we do not discount contribution from other components of the inflammatory response.
Discussion
Innate immunity is orchestrated by a highly complex interplay of several factors and interwoven pathways. For example, the Toll-like receptors (TLRs), signaling through NFκB and promoting proinflammatory cytokine gene expression, directly affect innate immune responses to a variety of pathogen-associated molecular patterns.59 Defects in TLR4 signaling are associated with blunted immune responses to LPS.60,61 iNOS also contributes to macrophage antimicrobial capacity,62 and iNOS-mutant mice are susceptible to M tuberculosis infection.63 These observations suggest that resistance to infection may be a multigenic trait determined by epistatic interactions between several genes. Recent evidence however suggests that HIF-1α may be a critical nexus integrating innate immune responses to infection and inflammation. For example, HIF-1α has been shown to control myeloid cell activation, incipient inflammation, and bacterial killing by regulating the expression of neutrophil proteases, antimicrobial peptides, iNOS, and TNF-α.39,40 HIF-1α is also induced by a range of bacteria and viruses and, as with bacteria, its stabilization through loss of pVHL confers resistance to viral infection.64-67 Taken together, these data strongly suggest that HIF-1α may have a role far beyond its more traditional functions in oxygen homeostasis, energy metabolism, and angiogenesis.68 In particular, it may confer immunity to a broad range of infections by regulating specific immune responses, including those that are SLC11A1 dependent.
Cis-acting regulatory variation contributes to heritable differences in gene expression, phenotypic diversity, and the directional evolution of complex quantitative traits. Microsatellites constitute a potentially large source of such variation because they abound in and punctuate eukaryotic genomes, occurring at more than 100 000 copies per mammalian genome.53 Microsatellites such as the SLC11A1 (GT/CA)n dinucleotide repeat have a propensity to form Z-DNA, an unstable left-handed form of DNA transiently induced during gene transcription by a moving RNA polymerase, and stabilized by negative supercoiling.54,55 Although generally considered “junk” DNA, useful only as neutral phylogenetic markers, microsatellites are nonrandomly distributed, occurring proximally to the transcription initiation sites of actively transcribed genes.45 They have been shown to either activate or repress gene transcription in a context-dependent manner.69,70 For example, the SW1/SNF-like BAF chromatin remodeling complex is recruited to and induces Z-DNA in the colony-stimulating factor 1 gene promoter to activate the gene.71 A number of other Z-DNA–binding proteins are now known; these include the poxvirus virulence factor E3L, the interferon-inducible protein DLM-1, and the RNA editing enzyme adenosine deaminase, ADAR1.72-76 However, no transcription factor has been identified that regulates gene expression from Z-DNA,54,55 thus making HIF-1α the only transcription factor to be shown to bind and affect gene expression from Z-DNA–forming microsatellites.
The singular contribution of genetic loci to (non-Mendelian) quantitative traits is immeasurably small due to confounding epistatic interactions between genes.77 We have shown here that a polymorphic microsatellite, acting in synergy with regulatory SNPs in the SLC11A1 promoter, can direct phenotypic variation in allele expression. We found no other polymorphisms within 1 kb of the promoter from sequencing the genes of 30 to 35 individuals from several geographical populations (J.M.B. et al, manuscript in preparation). Although distal SNPs may be involved,22 our data strongly suggest that the microsatellite may sufficiently direct variation in allele expression, and thus determine interindividual differences in handling SLC11A1-related diseases. Due to their inherent instability,78 the SLC11A1 microsatellites may also be hotspots of rapid change in allele type and frequency within populations (Figure 3A), thus helping to establish a broad spectrum of phenotypically different innate immune responses. We also showed that HIF-1α acts in trans to accentuate differences in allele expression from the cognate microsatellites. As the SLC11A1 alleles are highly polymorphic and are distributed according to race and ethnicity, HIF-1α may therefore determine within- and between-population differences in susceptibility to infection and inflammation; this may enable our understanding of such differences in innate immunity. This model is conceptually exquisite in its simplicity and specificity in that while the microsatellite may provide directional selection (because it is heritable), it is also a molecular switch for precise, pulsatile control of SLC11A1 expression by HIF-1α, which is induced by infection and inflammation.39,40 Our report thus suggests HIF-1α as a “master regulator” of innate immunity, with the implication that quantitative or qualitative differences in its expression due to regulatory SNPs or structural variation, respectively, could directly affect SLC11A1-disease association. Future genetic and functional studies at both SLC11A1 and HIF-1a loci will address this proposition. However, since disease susceptibility is complex, such studies could face major challenges, as they would require large population cohorts and statistical power. They may also be confounded by epistasis,77 gene-environment interactions,79 and gene dosage effects or copy-number polymorphisms.80-82 Nonetheless, our finding may provide an important framework to re-examine the contribution of HIF-1α to innate immunity in general.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Ingo Flamme for FLAG-tagged pcDNA3-HIF-1α and HIF-2α plasmids, P. Gros for an anti-SLC11A1 antibody, Christopher Bradfield for pSport-ARNT/HIF-1β plasmid, B. David Stollar for anti–Z-DNA antibody, Peter Ratcliffe for C4.5 and Ka13.5 cell lines, K. C. Gatter for anti–HIF-1α antibody (ESEE), and Germain Puzo for lipoarabinomannans. We also thank Willie Russell for advice on Z-DNA binding assays, and Tony Segal and Jill Norman for critically reading the manuscript and for helpful suggestions.
W.W.A.-S. and S.G. were supported by Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior, Conselho Nacional de Desenvolvimento Cientifico e Tecnológico and Fundação de Amparo à Pesquisa do Estado de São Paulo, Brazil. H.K.B. and S.K.S.S. are grateful to the Charles Wolfson Trust and the Biotechnology and Biological Sciences Research Council for grant support.
Authorship
Contribution: H.K.B. designed and performed research and wrote the paper; C.P. and A.G. designed and performed research; W.W.A.-S. performed research; H.S.M and H.C. contributed vital materials; S.G and M.K designed and performed research; R.S.J. designed research and contributed vital materials; and J.M.B, V.N., and S.K.S.S. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Henry K. Bayele, Department of Biochemistry & Molecular Biology, University College London, NW3 2PF, United Kingdom; e-mail: h.bayele@medsch.ucl.ac.uk.

![Figure 1. Nucleotide sequence of the SLC11A1 (allele 3) promoter. Double-underlined nucleotide (G) is the putative transcription start site.44 The polypurine/polypyrimidine tract [(GT/CA)n] and Z-DNA–forming microsatellite sequence is single-underlined. The microsatellite has a z score (Z-DNA–forming potential) of 12 598.14 (z score minimum is 700), as determined with the ZHunt algorithm.45,46 HREs47,48 are in boldface. Shaded thymidine is a regulatory SNP; it was subsequently mutated to the more common cytosine SNP (T → C) simultaneously with microsatellite interchange.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/8/10.1182_blood-2006-12-063289/4/m_zh80210708440001.jpeg?Expires=1765088017&Signature=1sdb2dZaf0LOPh-MfFK37X3MVcWZIRXErH5NFr0-NtLx3uhCYHU32oeB9QT~jybiHltNGVG9jVxdgmgYr3phqCWZGqiUPTg~wuzuJTVvsgJ0A53AkI~tR9Br-GtJD1VdYCOlUcGiLg5G-QmZgPAQv4TWQ247Le9FOJoyRMEBtJ0fq8JW227c8SMmyVchu-gYNb2xwKfyWigv9cmz2hp0bZSp4RLNT5CogyUNNovq3AABUYdeAUIipYT1wtXGALnRcO~httU8DvmZzZfR5KKtfwWePD01pTakO6V9f5cWsJDdhZ1rFmgWtd1lriTYECFekvVLAyc~D4iTU1WZBVkQPg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 6. HIF-1α/ARNT heterodimers bind directly to the microsatellite in vitro and in vivo. EMSA was performed with nuclear extracts from THP-1 cells either untreated or treated for 6 hours with (A) 1 μg/mL E. coli LPS and (B) 100 nM fMLP. Alleles 1 to 7 microsatellites or the mutant (GT)22 were used for binding assays in panel A; shifts for only alleles 1 to 6 are shown in panel B. In both cases, lane 1 contains no nuclear extract (ie, a3 oligonucleotide only [−] and lane 2 (Un) contains untreated control nuclear extract; arrow shows specific HIF-1 binding. (C) In vitro–translated HIF-1α (lane 3), ARNT (lane 4) or HIF-1α/ARNT heterodimers (lanes 5 and 7) were incubated with untreated (lanes 3-6 and 8) or hexamine CoCl2-treated allele 3 microsatellite (lane 7). HIF-1α/ARNT heterodimers bound (arrows) to wild-type but not to the mutant microsatellite (GT)22 (lane 8). Binding was competitively inhibited by excess cold microsatellite DNA (lane 6). Lane 1 shows oligonucleotide only; lane 2, negative control with untranslated reticulocyte lysate; NS, nonspecific binding component(s); F, free probe in panels A-C. (D) Chromatin immunoprecipitation of the SLC11A1 promoter from THP-1 cells treated with LPS/IFNγ or zymosan was performed using an anti–HIF-1α antibody; a nonspecific antibody (NS) was used as negative control, and 10% input chromatin was used as positive control for PCR. (E) Yeast 1-hybrid assay for HIF-1α/ARNT occupancy and transactivation of the SLC11A1 promoter. A total of 4 independent GAL4BD-HIF-1α/GAL4AD-ARNT yeast cotransformants were tested for Leu-Trp-Ura prototrophy and for qualitative β-galactosidase expression (blue coloration) by colony-lift filter assay, after exposing the prototrophs to the β-galactosidase substrate X-gal for 4 hours. (F) Quantitative β-galactosidase assay. Yeast cells with a chromosomally-integrated copy of the promoter were either untransformed or transformed with GAL4BD-HIF-1α, GAL4AD-ARNT, or both, and β-galactosidase expression was determined by chemiluminescence. SD/Leu-Trp-Ura medium was used as background control. Error bars denote SEM.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/8/10.1182_blood-2006-12-063289/4/m_zh80210708440006.jpeg?Expires=1765088017&Signature=newO5gIgBD7FjuNt3ezBLbkK14kO44XzxiWXfNY8yDUzJZvGp4zt31i6iNYyvZ4Am0WqJcgDTk3gVR4ub1GykLXKodLo8zp6lg7Q1xiXybnwATBbu4MkmHn5~YClKEOWVaRYkNoYssNNKQbFCWGRBq5r8ORQnoSoDS~DjQCdnzHotqYJgjhUuXMl7MY7czAGbhI8dImh0TyM4vw5Ulb89KrTmlpKQmXC4KfjtGdS7czyDaiQGMRnXwKOeGXntDoc46w0a8jnXs~83RsAesScnseWQmT41nct~Cu88tGEJVm3KOXRsux9kMJjqC~EWWCl~-tDzEjaFubT9~ISAo4MZg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 1. Nucleotide sequence of the SLC11A1 (allele 3) promoter. Double-underlined nucleotide (G) is the putative transcription start site.44 The polypurine/polypyrimidine tract [(GT/CA)n] and Z-DNA–forming microsatellite sequence is single-underlined. The microsatellite has a z score (Z-DNA–forming potential) of 12 598.14 (z score minimum is 700), as determined with the ZHunt algorithm.45,46 HREs47,48 are in boldface. Shaded thymidine is a regulatory SNP; it was subsequently mutated to the more common cytosine SNP (T → C) simultaneously with microsatellite interchange.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/8/10.1182_blood-2006-12-063289/4/m_zh80210708440001.jpeg?Expires=1765199857&Signature=O5mtAFDok-MG4UWNdBC7Opf~2RSbc-TIg4-8aqco9E8WD72fT~gKbFcqd70-i8Mb6BfqUa9AixOgj7ZblJD7p-Qe-sl0XFR~vjIBEKz0blo1BJqwj0ekK7jbAF0C22P1msun~6CC8niQt4QZkxF1CXDRRfvsmYXUGIitLpKvOkMsj3qd9hsrkIT4H9vaLwWMfmYWuM5XcauFK-dwAENtkiCLeDt~OIkfXYoaEOq22HpS~N20sYjiMRNQP-x-jcl6~O3OnbZzNaIkacvw1YcqOirtaozFlec8NobyhW45Gim8Yg1Dtqpgb54keZvnmR9a-Dkafgk7mASCCuDLOX3JAw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 6. HIF-1α/ARNT heterodimers bind directly to the microsatellite in vitro and in vivo. EMSA was performed with nuclear extracts from THP-1 cells either untreated or treated for 6 hours with (A) 1 μg/mL E. coli LPS and (B) 100 nM fMLP. Alleles 1 to 7 microsatellites or the mutant (GT)22 were used for binding assays in panel A; shifts for only alleles 1 to 6 are shown in panel B. In both cases, lane 1 contains no nuclear extract (ie, a3 oligonucleotide only [−] and lane 2 (Un) contains untreated control nuclear extract; arrow shows specific HIF-1 binding. (C) In vitro–translated HIF-1α (lane 3), ARNT (lane 4) or HIF-1α/ARNT heterodimers (lanes 5 and 7) were incubated with untreated (lanes 3-6 and 8) or hexamine CoCl2-treated allele 3 microsatellite (lane 7). HIF-1α/ARNT heterodimers bound (arrows) to wild-type but not to the mutant microsatellite (GT)22 (lane 8). Binding was competitively inhibited by excess cold microsatellite DNA (lane 6). Lane 1 shows oligonucleotide only; lane 2, negative control with untranslated reticulocyte lysate; NS, nonspecific binding component(s); F, free probe in panels A-C. (D) Chromatin immunoprecipitation of the SLC11A1 promoter from THP-1 cells treated with LPS/IFNγ or zymosan was performed using an anti–HIF-1α antibody; a nonspecific antibody (NS) was used as negative control, and 10% input chromatin was used as positive control for PCR. (E) Yeast 1-hybrid assay for HIF-1α/ARNT occupancy and transactivation of the SLC11A1 promoter. A total of 4 independent GAL4BD-HIF-1α/GAL4AD-ARNT yeast cotransformants were tested for Leu-Trp-Ura prototrophy and for qualitative β-galactosidase expression (blue coloration) by colony-lift filter assay, after exposing the prototrophs to the β-galactosidase substrate X-gal for 4 hours. (F) Quantitative β-galactosidase assay. Yeast cells with a chromosomally-integrated copy of the promoter were either untransformed or transformed with GAL4BD-HIF-1α, GAL4AD-ARNT, or both, and β-galactosidase expression was determined by chemiluminescence. SD/Leu-Trp-Ura medium was used as background control. Error bars denote SEM.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/8/10.1182_blood-2006-12-063289/4/m_zh80210708440006.jpeg?Expires=1765199857&Signature=0vMb8fEsNdC-VvyL2VaZWjUJEkIHEm9~3rwZAFkao2OBxajprj8kJ1gZ2tjy2sOBKxG7VEtW0HpjygE0EK0nn1gCyqmLzUPn3U7RxgtJU~~7sy9eUNBPvQj3xsmxWJ4GjjbfmJ2PA5ZBf6leNBG~tovFCfhFOmeWqnGzlTmgrYeWcqoWzzkyQcBw3mWDDfbPjjsYD~bqYStQK27vfpU074UWjsDAtWRyzE~zJiDR9Wm7vAKSreTu5KVrv8rB-szGAwEv13qGv1qKRGBDxWYP504uDf52VvOHXhO9FCSMRUksgVFwJl1q1HyM8bxXQ8cd0kilDxZtPA08rKNDyQqkvw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)