We describe the first successful clinical application of a new discovery technology, epitope-mediated antigen prediction (E-MAP), to the investigation of multiple myeloma. Until now, there has been no reliable, systematic method to identify the cognate antigens of paraproteins. E-MAP is a variation of previous efforts to reconstruct the epitopes of paraproteins, with the significant difference that it provides enough epitope sequence data so as to enable successful protein database searches. We first reconstruct the paraprotein's epitope by analyzing the peptides that strongly bind. Then, we compile the data and interrogate the nonredundant protein database, searching for a close match. As a clinical proof-of-concept, we apply this technology to uncovering the protein targets of para-proteins in multiple myeloma (MM). E-MAP analysis of 2 MM paraproteins identified human cytomegalovirus (HCMV) as a target in both. E-MAP sequence analysis determined that one para-protein binds to the AD-2S1 epitope of HCMV glycoprotein B. The other binds to the amino terminus of the HCMV UL-48 gene product. We confirmed these predictions using immunoassays and immunoblot analyses. E-MAP represents a new investigative tool for analyzing the role of chronic antigenic stimulation in B-lymphoproliferative disorders.
Introduction
For the investigation of disease, existing investigative approaches have included genomic and proteomic methods, which explore the universe of expressed genes and proteins in index patients. Rather than focus on the presence or absence of mRNA species or proteins, as in genomics or proteomics, we instead have developed a method to read out the targeting specificity of the humoral immune system. Since the immune system plays a pivotal role in maintaining health and eliminating disease, it will be especially relevant for diseases with an immunologic component. The method, epitope-mediated antigen prediction (E-MAP), comprises 2 steps: (1) reconstruction of the antibody epitope using a peptide combinatorial library, and (2) bioinformatic interrogation of the protein database for matching sequences. To date, this type of protein identification method has not been practical. It has not previously been possible to accurately predict the amino acids comprising the true epitope of the targeted protein. Moreover, the nonredundant protein database is so large that even if an accurate epitope had been identified, too many irrelevant proteins are retrieved in a typical database search. There have been several published attempts to identify antigens for multiple myeloma (MM) paraproteins by probing paraproteins with combinatorial peptide libraries.1,,–4 The investigators tried to link the experimentally determined peptide epitopes with entries in the protein database. However, the published peptide sequences that were identified were insufficiently informative or accurate to yield meaningful database hits.1,,–4 No predictions from the protein database search were experimentally validated.
In our previous work, we described the theoretical reasons why searching the nonredundant protein database in this manner requires an epitope that is at least 7 amino acids long, and with approximately 70% accuracy to the native linear sequence.5 Less than that typically yields too many irrelevant matches, relegating the true match to a position too low in the rank order to be accurately identified. E-MAP attains these thresholds by using high stringency methods of phage panning as well as a postpanning phage clone selection technique. E-MAP combines phage display of a random combinatorial library with a new bioinformatic search method. Briefly, we used a random peptide combinatorial library, expressed in M13 phage, to create a molecular mold that conforms to the patients' paraprotein antigen binding site. By analyzing many different phage peptide inserts, all of which bind to the paraprotein, we can deduce the amino acid sequence of the original epitope. E-MAP thereby identifies the epitope without knowing in advance anything about the paraprotein's true antigen.
Methods
Serum collection
Patient serum discards were collected in accordance with the Declaration of Helsinki and with the approval and under the guidelines of the institutional review board of the Boston University Medical Center (BMC) from BMC's clinical pathology laboratory. For MM sera, the serum protein electrophoresis (SPEP) records were inspected for the identification of samples exhibiting the presence of a potential monoclonal component in the gamma globulin region of the gel. Medical records of patients evidencing a gammopathy on SPEP were retrieved and screened for the clinical diagnosis of multiple myeloma. The serum samples of identified patients with MM were collected, aliquoted and stored at −20°C. Serum samples of 8 individuals (4 male, 4 female) without clinical manifestation of disease were also collected as normal control sera and used for negative depletions during phage library biopanning.
Patient information
Patients 12 and 20 both have a clinical diagnosis of multiple myeloma, with excess plasma cells on bone marrow biopsy and serum and urinary paraproteins. Patient 12, a 60-year-old man, has an IgG-kappa paraprotein. Patient 20, an 84-year-old woman, also has an IgG-kappa paraprotein. At the time of serum collection, patient 12's main clinical problem was renal insufficiency, which had improved from 15 g to 1 g of urinary protein per 24 hours. Patient 20, on the other hand, had been diagnosed 8 months prior and had completed 2 cycles of melphalan and prednisone. She had been diagnosed with cord compression after laminectomy and irradiation to T5-T9.
E-MAP overview
E-MAP leverages the specificity of patients' immune responses to disease-relevant targets and requires no prior knowledge about the protein. E-MAP links pathologic antibodies of unknown specificity, isolated from patient sera, to their cognate antigens in the protein database. The E-MAP process first involves reconstruction of a predicted epitope using a peptide combinatorial library expressed in M13 phage. We then search the protein database for closely matching amino acid sequences. E-MAP was designed to overcome 2 important problems that were previously encountered with this approach: (1) short predicted epitopes yield too many irrelevant matches from a database search, and (2) the predicted epitopes derived from phage display of peptide combinatorial libraries may not sufficiently accurately represent the epitope in the native antigen. In a previous paper, we found that epitopes generally need to have at least 7 amino acids, with an overall accuracy of more than 70% to the native protein, in order to correctly identify the protein in a nonredundant protein database search.5 Therefore, the E-MAP technique incorporates new innovations to improve the accuracy of the epitope prediction and gain greater information content. For example, since many predicted epitopes often fail to achieve the 7 amino acid threshold, E-MAP incorporates a new concept of paired epitope searches. Whereas a single epitope may yield insufficient information content, 2 epitopes from the same protein satisfy the information requirement for successful database searching. The assumptions and theory behind E-MAP are described elsewhere.5
Peptide combinatorial library biopanning
Biopanning using MM paraproteins was performed essentially as previously described for other monoclonal antibodies,6,–8 with some modifications. Equal amounts of protein A– and protein G–coated paramagnetic beads (Invitrogen, Carlsbad, CA) were mixed and washed twice with IBBT (ImmunoPure [A/G] IgG Binding Buffer + 0.05% Tween-20; Pierce Biotechnology, Rockford, IL). The beads were then incubated with an appropriate dilution (in IBBT) of a patient's serum, or pooled sera from 8 normal controls, containing a 2.5 molar excess of antibodies over the bead-carrying capacity, overnight at 4°C with rocking. The beads were washed 8 times with IBBT to remove other serum proteins. The biopannning scheme entailed tandem positive, negative, positive selections, in each round. These stringently selected phage clones, once neutralized, were allowed to infect Escherichia coli.
Phage amplification and titering
Selection of phage clones
Replicate lifts were created by sequentially laying nitrocellulose membranes (BioTrace NT, Pure Nitrocellulose Transfer Membrane, 0.2-μm pore size/82-mm-diameter discs; Pall Life Sciences, East Hills, NY) onto the phage agar plates at 4°C for 40 minutes. Membranes were marked for orientation, carefully lifted from the agar, and placed at 65°C to dry for 5 minutes. The membranes were blocked with 5% milk in TBST (8 g sodium chloride, 0.2 g potassium chloride, 3 g Trizma base [Sigma Chemicals, St Louis, MO]; 1 L ddH2O, with 0.5% Tween-20) and washed twice with TBST. The selecting patient serum was prepared in TBST (final IgG concentration of 5 μg/mL) and placed on the membrane for 2 hours at room temperature or at 4°C overnight. The membranes were then washed 8 times with TBST, 5 minutes per wash, and incubated with goat anti–human IgG horseradish peroxidase (HRP; Southern Biotech, Birmingham, AL), 1:5000 in TBST, for 1.5 hours. A chemiluminescence protocol was used to visualize patterns of immunoreactivity (ECL Western Blotting Detection Reagents; Amersham Biosciences, Piscataway, NJ). Developed films could be oriented onto the corresponding agar plates. The most immunoreactive spots (representing distinct plaque colonies) were picked and grown individually for further analysis.
DNA insert sequencing
Phage clones that had high specific immunoreactivity for the selecting antibody by the enzyme-linked immunosorbent assay (ELISA) as well as by the immunoblot, were submitted for further analysis, by sequencing the nucleotide inserts coding for the combinatorial peptides, as previously described.6,–8
Consensus motif elicitation
Motifs are identified as previously described.5 Briefly, the nucleic acid sequences are translated in silico using the Translate tool (ExPASy Proteomics Server, Swiss Institute of Bioinformatics [SIB], http://ca.expasy.org). The translated protein sequences are verified to be in frame by identification of invariant elements of the cpIII protein. A list of the primary amino acid sequences of the variable inserts is then compiled in the FASTA format and submitted to a motif-elucidation bioinformatics algorithm called MEME. The MEME [multiple expectation-maximization for motif elicitation]9 web utility software tool is available on the University of California, San Diego server (http://meme.sdsc.edu/meme/intro.html). MEME is used to aid the objective and standardized determination of any motifs present in the sequenced peptide inserts. The output contains the submitted oligopeptides rank-ordered for the relative presence of any dominant motif determinants. The output also affords position-specific scoring matrices (PSSMs) characterizing and capturing the sequence variation seen at each of the positions of the motif, and these were stored as separate files. As the size of the motif cannot be predicted ahead of time, a patient's specific set of peptides are submitted several times with different constraints. For instance, setting a minimum motif expectation length of 4 amino acids and then sequentially increasing the maximum length to 5, 6, 7, etc, amino acids, there comes a point where the determined motif length plateaus regardless of increasing expectation length. This exercise also reveals which span of the determined motif is most preserved, as at suboptimal lengths MEME will choose the motif span containing the most conserved anchor residues.
Serum protein electrophoresis
Patient sera are applied to precast protein β1/β2 agarose gels in a Hydrasys electrophoresis instrument (SEBIA-USA, Norcross, GA) according to the manufacturer's instructions.10
Serum protein electrophoresis immunoblots for IgG and phage
This method is used in Figure 4. Patient sera were diluted in phosphate-buffered saline (PBS) and 10-μL aliquots were loaded and run on a precast protein β1/β2 agarose gel in a Hydrasys instrument (SEBIA-USA, Norcross, GA) according to the manufacturer's instructions. The automated program was stopped after phoresis (40 Vh, ∼ 5 min) and not allowed to proceed to the gel-drying step. The gel was removed from the instrument and contact blotted onto a nitrocellulose membrane (Protran BA83 0.2 μm nitrocellulose membrane; Whatman, Florham Park, NJ, or NitroBind Cast pure nitrocellulose 0.45 μm; General Electric Water & Process Technologies, Minnetonka, MN) under 100 g of weight for 30 minutes at room temperature. Placement of the gel relative to the membrane was noted with ink, marking sample lanes and other features of interest. The gel was then blocked with 2% milk PBST for 1 hour at room temperature. The membrane was rinsed twice with PBST and specific phage, prepared in 1% milk PBST, and incubated overnight at 4°C with rocking. The membrane was then washed 3 times, 10 minutes each, with PBST. Mouse anti–M13-HRP conjugate was added, prepared as 1:5000 in 1% milk PBST, for 1.5 hours at room temperature. The membrane was washed twice with PBST, once with PBS, and any retained phage were visualized using a standard chemiluminescence protocol. Also, serum protein electrophoresis (SPEP) blots were undertaken with patient sera diluted 1:1000 in PBS and these blots were developed with goat anti–human IgG HRP to reveal the location of the paraprotein, as an internal control for each run.
Agarose gel affinity-transfer immunoblot
This method is used in Figure 6. For this assay, proteins are electrophoretically separated in an agarose gel. The proteins are then contact blotted onto an antigen-coated nitrocellulose membrane. Protein transfer requires that serum antibodies in the gel bind to antigen on the nitrocellulose membrane. Only immunoglobulins capable of binding to the antigen adhere. The nitrocellulose membrane is otherwise saturated with irrelevant proteins, largely preventing nonspecific protein transfer. Immunoglobulins that are bound to the nitrocellulose sheet are then visualized with a human IgG–specific antibody-enzyme conjugate.
Nitrocellulose membranes were incubated with specific phage prepared in 0.5 M bicarbonate buffer (pH 8.0), overnight, at 4°C with rocking. The membranes were then rinsed with PBST and blocked for 1 hour with 2% milk PBST. In this variation of the immunoblot, the gels are allowed to contact the antigen-coated nitrocellulose membranes for 30 minutes at room temperature, sandwiched between 2 glass plates. The relative position of the gels to the membranes is marked in ink, and the gels are removed. The membranes are thoroughly washed 3 times in PSBT for a total of 30 minutes. Membranes are then incubated with goat anti–human IgG HRP conjugate prepared as 1:5000 in 1% milk PBST for 1.5 hours at room temperature or overnight at 4°C, with rocking. Membranes were washed twice with PBST and once with PBS before development by chemiluminescence.
Glycoprotein b, AD-2S1 epitope, and UL-48 gene product ELISA
This method is used in Figure 5. A peptide corresponding to the sequence of the AD-2S1 epitope of glycoprotein B, HCMV, was synthesized (AnaSpec, San Jose, CA). The peptide sequence was Acetyl-KKSHRANETIYNTTLKYGDVTGTNTTK-Biotin CONH2. Also, a peptide corresponding to the sequence of the UL-48 gene product was synthesized (AnaSpec). The peptide sequence was Biotin-PEG2-MSNTAPGPTVANKRDEKHRH-CONH2. The biotin is separated from the peptide by 2 repeating units of polyethylene glycol as an extension. The carboxy terminus was amidated. The peptides were dissolved in PBST, 0.1% milk (100 μL/well; 20 μg/mL). Immulon 4HBX ELISA plate wells were coated with 100 μL of 10 μg/mL streptavidin (ImmunoPure Streptavidin; Product no. 21125; Pierce Biotechnology) in carbonate-bicarbonate buffer (0.05 M, pH 9.6), overnight at 4°C. The microtiter plate wells were rinsed and blocked with 5% milk in PBST for 2 hours at room temperature. After rinsing the wells 8 times with PBST, the UL-48 or gpB peptides were added to different plates and incubated for 2 hours at 37°C. During this incubation, the UL-48 or gpB peptide attached to the streptavidin coating via its biotin moiety. The microtiter wells were then rinsed 10 times with PBST. Patient sera were added at a 1:1250 dilution, in PBST/0.1% nonfat dry milk, and incubated overnight at 4°C. Wells were then rinsed and goat anti–human IgG HRP conjugate (Southern Biotech, Birmingham, AL; 1:5000 dilution in PBST, 0.1% milk) was added and incubated for 2 hours at 37°C. Finally, the wells were rinsed with PBST prior to adding the ABTS (2,2′-Azino-bis(3-Ethylbenzthazoline-6-sulfonic acid); Sigma Chemicals) substrate (1 mg/mL), dissolved in 0.05 M phosphate, 0.05M citric acid, and 1 μL/mL of 3% H2O2 pH 5. We read absorbance at 405 nm.
Results
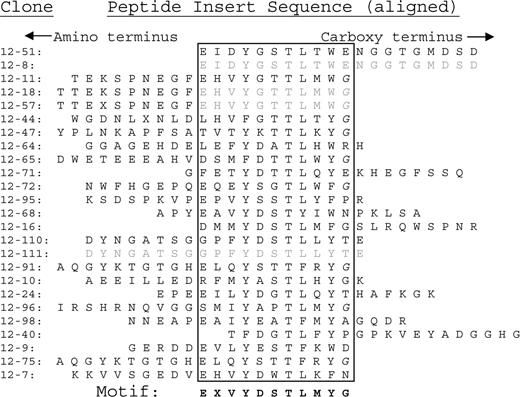
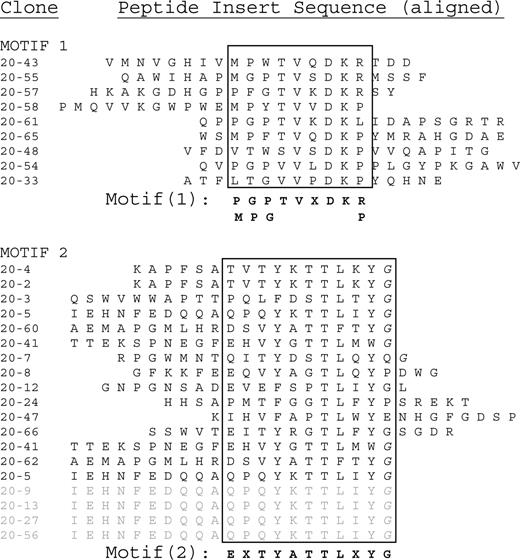
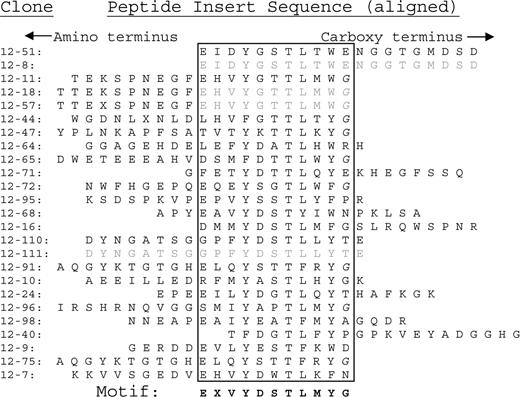
In order to validate the predictive accuracy of E-MAP for the investigation of human disease, we randomly selected a group of 9 sera from patients with MM. Patients 12 and 20 were among this first group to be analyzed. The patient characteristics are described in “Patient information.” We analyzed the 9 patient paraproteins and determined the consensus sequences of the epitopes to which each paraprotein binds (see “Consensus motif elicitation”). Sequence data from phage inserts that bind to paraproteins from patients 12 and 20 are shown in Figures 1 and 2, respectively. Each sequence is positioned so as to align areas of homology (within the box).
Peptide sequences binding to patient 12's paraprotein. The peptide inserts in the phage library are 20 amino acids long. The sequences are aligned by their consensus residues, which are enclosed within the boxes. The fact that so many consensus sequences are immediately adjacent to the carboxy terminus (right-hand side) of the peptide insert indicates that the next (invariant) amino acid is likely identical to the native sequence. Otherwise, the consensus sequences should have been randomly positioned within the peptide insert. For that reason, we included the next amino acid on the carboxyl side (glycine, G) as part of the consensus sequence. Glycines that are part of the invariant carboxy terminus are italicized (G).
Peptide sequences binding to patient 12's paraprotein. The peptide inserts in the phage library are 20 amino acids long. The sequences are aligned by their consensus residues, which are enclosed within the boxes. The fact that so many consensus sequences are immediately adjacent to the carboxy terminus (right-hand side) of the peptide insert indicates that the next (invariant) amino acid is likely identical to the native sequence. Otherwise, the consensus sequences should have been randomly positioned within the peptide insert. For that reason, we included the next amino acid on the carboxyl side (glycine, G) as part of the consensus sequence. Glycines that are part of the invariant carboxy terminus are italicized (G).
Peptide insert sequences binding to patient 20's paraprotein. The sequences at the bottom of motif 2, from phage clones 20-5 until 20-56, are independent clones having the exact same sequence. These redundant sequences are shaded in gray. This sequence was weighted as only one entry when calculating the dominant motif in MEME.
Peptide insert sequences binding to patient 20's paraprotein. The sequences at the bottom of motif 2, from phage clones 20-5 until 20-56, are independent clones having the exact same sequence. These redundant sequences are shaded in gray. This sequence was weighted as only one entry when calculating the dominant motif in MEME.
A single amino acid consensus sequence emerged from patient 12 (Figure 1). In contrast, 2 distinct motifs (designated motifs 1 and 2) emerged from patient 20 (Figure 2). The consensus sequences for patient 12 and motif 2 of patient 20 both share the following amino acid sequence: E - - Y - - T L - Y G. This finding suggests that the paraproteins from patients 12 and 20 bind to the same exact epitope. We analyzed the amino acid sequences with the MEME software utility.11 The resulting epitope motif from the MEME software utility for the combined data sets is EXVYDTTLXYG. We then submitted 2 different types of database queries, employing the BLAST and MAST search algorithms. We submitted the dominant motif string (EXVYDTTLXYG) to the National Center for Biotechnology Information's “search for short, nearly exact matches” protein-protein BLAST utility. We searched against the nonredundant database, using default settings (PAM 30 matrix, word size 2 and expectation value 20 000), requesting the top 100 hits. Glycoprotein B, human herpes virus 5 (human cytomegalovirus), populated positions 2 through 66 of the search.
We also obtained the same result with an alternative search method that uses the MAST software utility.12 Instead of a single dominant motif (EXVYDTTLXYG), MAST can accept the MEME analysis motif output in the form of a 2-dimensional numeric matrix, the position-specific scoring matrix (PSSM). The PSSM is not simply a dominant motif string, but contains all of the phage clones' peptide insert information, preserving the experimentally observed positional variation within the motif. We submitted the combined PSSM of patients 12 and 20 (motif 2) to MAST, searching against the nonredundant protein database, having set a threshold expectation (E) value of 50. We retrieved 61 hits, 41 of which were entries for glycoprotein B of human cytomegalovirus (HCMV).
Figure 3A compares the predicted epitope (derived from the phage clone sequences shown in Figures 1 and 2) with the relevant portion of the glycoprotein B native sequence. Glycoprotein B closely aligns with the predicted epitope, with 7 exact matches and 2 conserved substitutions. No other glycoprotein B proteins, such as from other herpesviruses, have homologous sequences. We also performed a similar analysis of the second motif (motif 1) from patient 20. We submitted the PSSM to MAST, searching the nonredundant protein database, and retrieved the UL-48 gene product. A comparison of the motif to the UL-48 gene product is shown in Figure 3B. For both motifs shown in Figure 3A and B, our epitope reconstruction yielded a prediction that exceeded the minimum threshold of 7 amino acids, with more than 70% sequence accuracy to the native proteins.
Sequence comparisons. (A) A short segment from the native sequence of glycoprotein B, human herpesvirus 5, is compared with the consensus sequence used for BLAST searching. Solid lines between the 2 represent identity. Dotted lines represent conserved substitutions. “X” represents an amino acid position that could not be identified from the phage display data. (B) A similar comparison of the consensus motif from patient 20, motif 1 to the UL-48 gene product sequence.
Sequence comparisons. (A) A short segment from the native sequence of glycoprotein B, human herpesvirus 5, is compared with the consensus sequence used for BLAST searching. Solid lines between the 2 represent identity. Dotted lines represent conserved substitutions. “X” represents an amino acid position that could not be identified from the phage display data. (B) A similar comparison of the consensus motif from patient 20, motif 1 to the UL-48 gene product sequence.
We then tested the assumption that the peptide consensus sequences in Figure 3A and B are associated with the patients' paraproteins. Polyclonal antibodies that are generated in the normal course of an immune response would not have resulted in a consensus sequence, since each antibody binds to a different peptide sequence, precluding the identification of a consensus. Nonetheless, for a first proof-of-principle, we set out to formally demonstrate the link between the paraproteins and binding to the peptide sequence expressed on the surface of specific phage clones. To address this question, we performed a phage immunoblot experiment (Figure 4). A phage immunoblot will identify the immunoglobulins in the patients' sera that caused these particular phage clones to be selected. We first separated serum proteins on a high-resolution agarose gel and then transferred the proteins (in replicate lanes) to a nitrocellulose membrane. We then probed the nitrocellulose membranes with purified phage clones corresponding to each motif (Figure 2), as immunoblot probes. Electrophoretic bands were detected with anti-M13 cpVIII antibody–horseradish peroxidase conjugate.
Serum protein electrophoretic analysis and phage immunoblot, patients 12 and 20. The paraproteins are evident on serum protein electrophoresis (SPEP) pattern (lane 1). The anode (positive pole) is toward the top, with albumin (ALB) being the most anodal serum protein visible in the gel. The serum paraprotein is denoted with →. For patient 12 (lefthand gel), lane 2 is probed with phage clone 20-41, which expresses a peptide associated with the AD-2S1 epitope of HCMV glycoprotein B. Lane 3 is an irrelevant phage clone. For patient 20, lanes 2 to 4 are replicates of lane 1, blotted onto a nitrocellulose membrane, and immunostained with various probes. Lane 2 was immunostained with a human IgG-specific antibody conjugate. Lanes 3 and 4 were immunostained with the indicated phage clones, which are also listed in Figure 2. Clone 20-61 is derived from motif 1, containing the UL-48 gene product paraprotein epitope. Sera are undiluted in lane 1, and diluted 1:100 for lanes 2 to 4. Appl pt denotes the gel application point, which sometimes yields a faint artifactual band.
Serum protein electrophoretic analysis and phage immunoblot, patients 12 and 20. The paraproteins are evident on serum protein electrophoresis (SPEP) pattern (lane 1). The anode (positive pole) is toward the top, with albumin (ALB) being the most anodal serum protein visible in the gel. The serum paraprotein is denoted with →. For patient 12 (lefthand gel), lane 2 is probed with phage clone 20-41, which expresses a peptide associated with the AD-2S1 epitope of HCMV glycoprotein B. Lane 3 is an irrelevant phage clone. For patient 20, lanes 2 to 4 are replicates of lane 1, blotted onto a nitrocellulose membrane, and immunostained with various probes. Lane 2 was immunostained with a human IgG-specific antibody conjugate. Lanes 3 and 4 were immunostained with the indicated phage clones, which are also listed in Figure 2. Clone 20-61 is derived from motif 1, containing the UL-48 gene product paraprotein epitope. Sera are undiluted in lane 1, and diluted 1:100 for lanes 2 to 4. Appl pt denotes the gel application point, which sometimes yields a faint artifactual band.
Patient 12 has a single, well-defined paraprotein by serum protein electrophoresis (Figure 4 lane 1, lefthand gel, denoted with an arrow). As expected, phage clone 20-41 (that expresses the glycoprotein B peptide motif) binds to the paraprotein on an immunoblot. As a negative control, an irrelevant phage clone 20-61 is nonimmunoreactive. Therefore, we conclude that the paraprotein binds directly to the peptide insert, since the irrelevant clone is otherwise identical.
Patient 20 also has a single clinically apparent paraprotein on SPEP, shown on the right-hand gel of Figure 4 lane 1 (denoted with an arrow). Consequently, we could not explain the fact that we obtained 2 completely different motifs, identifying 2 different proteins (Figure 2). To answer this question, we probed replicate lanes of the nitrocellulose membrane with either phage clone 20-61 (lane 4, representing motif 1, expressing the UL-48 gene product peptide) or phage clone 20-41 (lane 3, representing motif 2, expressing the AD-2S1 epitope of glycoprotein B). Figure 4 illustrates that the 2 phage clones bind to different monoclonal immunoglobulins of patient 20, migrating to distinct gel positions. This finding resembles a biclonal gammopathy, a common clinical occurrence. Clone 20-61 (motif 1, having the UL-48 sequence) binds to the paraprotein. Phage clone 20-41 (motif 2, having the glycoprotein B sequence) binds to a doublet band that represents a separate serum monoclonal immunoglobulin. The doublet probably represents a monomer and (noncovalently associated) dimer forms of the same paraprotein, a frequent occurrence in serum protein electrophoresis. Therefore, patient 20's 2 consensus sequences are associated with 2 distinct monoclonal immunoglobulins, only one of which is the clinically apparent paraprotein seen in lane 1. This finding demonstrates the presence of minor clonal bands to the same virus in MM that were not previously appreciated using conventional serum protein electrophoresis. These minor bands represent the secreted monoclonal antibody from a less prevalent lymphocyte clone, raising the intriguing possibility that it may be a coexisting premalignant clone. In this way, antigen-specific immunoblots may offer new insights in analyzing paraproteins in gammopathies.
In order to confirm the prediction derived from the phage clone sequencing data, we performed solid-phase immunoassays and agarose gel immunoblots with HCMV whole virus, viral lysates, and synthetic peptides. HCMV is a common virus to which the majority of the population is exposed. Consequently, most people have at least low titers of serum antibodies to CMV. There are two methods by which we distinguished background (polyclonal) immunoreactivity from that of the paraprotein. By diluting the sera out beyond the usual concentrations for standard serologic assays, we can diminish the immunoreactivity associated with normal background and preferentially detect the paraproteins, which are present in high concentrations. HCMV immunoblots provide an even greater level of specificity because we can compare the electrophoretic mobility of the HCMV-binding immunoglobulins to the mobility of the paraproteins.
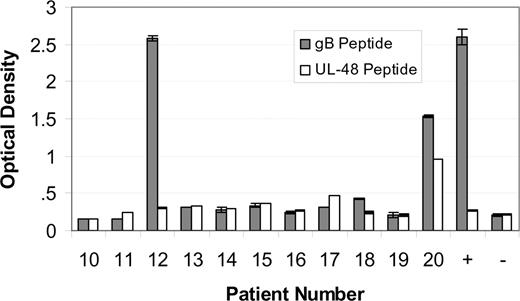
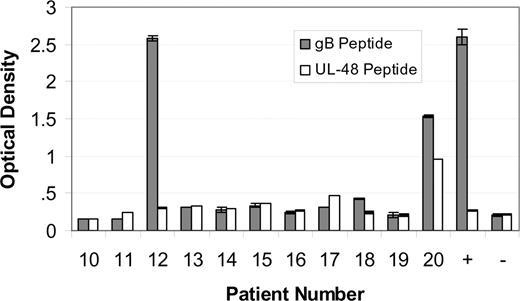
Sera from patients with MM designated 10 through 20 were tested for immunoreactivity to the AD-2S1 epitope of glycoprotein B using a synthetic peptide as the antigen. Sera were diluted out to 1:1250, so as to detect only those serum antibodies in high concentration. Only patients 12 and 20 were immunoreactive (Figure 5 solid bars). The same patients with MM were also tested for immunoreactivity to the N-terminus (amino acids 1-20) of the UL-48 gene product, using a synthetic peptide as the analyte. Only patient 20's serum sample was immunoreactive (Figure 5 open bars), confirming the E-MAP prediction. Sera from other patients with myeloma, randomly chosen because they were accrued at approximately the same time (and therefore have adjacent number assignments), were included as specificity controls.
Immunoreactivity of paraproteins. Immunoreactivity of paraproteins from patients 10 to 20 with a synthetic peptide corresponding to the AD-2S1 epitope of glycoprotein B human herpesvirus 5 ( ) or with a synthetic peptide corresponding to the amino terminus (amino acids 1-20) of the UL-48 gene product (
) or with a synthetic peptide corresponding to the amino terminus (amino acids 1-20) of the UL-48 gene product ( ). MM patient sera were diluted 1:1250 so as to fall within the assay linear range. An HCMV-immunoreactive serum was used as a positive (+) control. A nonreactive serum was used as a negative control (-). Since patients 12 and 20 are our index patients, we randomly included MM patients 10 to 20 in the immunoassay, for comparison. The patient selection is random, since the patient number designation is assigned in order of specimen acquisition. Samples were measured in duplicate. The mean plus or minus the standard deviation are shown.
). MM patient sera were diluted 1:1250 so as to fall within the assay linear range. An HCMV-immunoreactive serum was used as a positive (+) control. A nonreactive serum was used as a negative control (-). Since patients 12 and 20 are our index patients, we randomly included MM patients 10 to 20 in the immunoassay, for comparison. The patient selection is random, since the patient number designation is assigned in order of specimen acquisition. Samples were measured in duplicate. The mean plus or minus the standard deviation are shown.
Immunoreactivity of paraproteins. Immunoreactivity of paraproteins from patients 10 to 20 with a synthetic peptide corresponding to the AD-2S1 epitope of glycoprotein B human herpesvirus 5 ( ) or with a synthetic peptide corresponding to the amino terminus (amino acids 1-20) of the UL-48 gene product (
) or with a synthetic peptide corresponding to the amino terminus (amino acids 1-20) of the UL-48 gene product ( ). MM patient sera were diluted 1:1250 so as to fall within the assay linear range. An HCMV-immunoreactive serum was used as a positive (+) control. A nonreactive serum was used as a negative control (-). Since patients 12 and 20 are our index patients, we randomly included MM patients 10 to 20 in the immunoassay, for comparison. The patient selection is random, since the patient number designation is assigned in order of specimen acquisition. Samples were measured in duplicate. The mean plus or minus the standard deviation are shown.
). MM patient sera were diluted 1:1250 so as to fall within the assay linear range. An HCMV-immunoreactive serum was used as a positive (+) control. A nonreactive serum was used as a negative control (-). Since patients 12 and 20 are our index patients, we randomly included MM patients 10 to 20 in the immunoassay, for comparison. The patient selection is random, since the patient number designation is assigned in order of specimen acquisition. Samples were measured in duplicate. The mean plus or minus the standard deviation are shown.
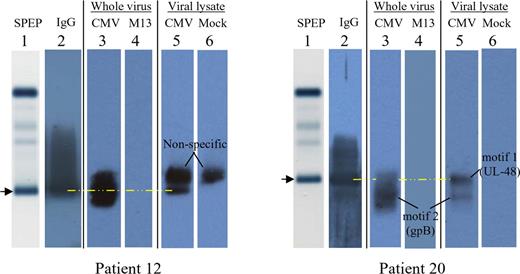
As a further confirmation, we tested whether the paraprotein bands visible on agarose gel electrophoresis bind to an HCMV lysate or intact HCMV. As in Figure 4, the sera of both patients 12 and 20 show a single paraprotein (Figure 6 lane 1, arrows). The normal background of polyclonal immunoglobulins is absent, a common finding in MM. A more sensitive immunoblot reveals the presence of trace IgG immunoglobulins (Figure 6 lane 2). In order to assess HCMV immunoreactivity, an agarose gel immunoblot method was used (lanes 3-6).13,14 For patient 12, the paraprotein band (lane 1, arrow) binds to both intact HCMV virions (lane 3) and an HCMV lysate (lane 5). With intact virions, viral membrane proteins such as glycoprotein B are accessible for antibody binding.
Agarose gel immunoblots, demonstrating the specific paraproteins responsible for immunoreactivity of patients 12 and 20 against HCMV. Patient 12, left; patient 20, right. Lane 1 depicts the serum protein electrophoresis (SPEP) pattern of major serum protein bands, as stained with amido black, without any protein transfer onto nitrocellulose. The image is that of the gel itself. The arrow and yellow dashed line denote the gel position of the serum paraprotein. The images for lanes 2 to 6 were scanned from a photographic film, after exposure to a nitrocellulose membrane. For lane 2, the membrane was probed for the presence of human IgG. For lanes 3 and 4, nitrocellulose membranes were precoated (prior to protein transfer) with inactivated, density-gradient purified HCMV whole virion (lane 3) or the antigenically unrelated M13 virus (lane 4). For lanes 5 and 6, nitrocellulose membranes were precoated with an HCMV lysate (lane 5) or a mock lysate derived from uninfected cells (lane 6). Each patient's image therefore represents a composite. For patient 12, the nonspecific band that is present in both the HCMV lysate lane (lane 5) and mock lysate lane (lane 6) does not comigrate with the paraprotein (lane 1,  ).
).
Agarose gel immunoblots, demonstrating the specific paraproteins responsible for immunoreactivity of patients 12 and 20 against HCMV. Patient 12, left; patient 20, right. Lane 1 depicts the serum protein electrophoresis (SPEP) pattern of major serum protein bands, as stained with amido black, without any protein transfer onto nitrocellulose. The image is that of the gel itself. The arrow and yellow dashed line denote the gel position of the serum paraprotein. The images for lanes 2 to 6 were scanned from a photographic film, after exposure to a nitrocellulose membrane. For lane 2, the membrane was probed for the presence of human IgG. For lanes 3 and 4, nitrocellulose membranes were precoated (prior to protein transfer) with inactivated, density-gradient purified HCMV whole virion (lane 3) or the antigenically unrelated M13 virus (lane 4). For lanes 5 and 6, nitrocellulose membranes were precoated with an HCMV lysate (lane 5) or a mock lysate derived from uninfected cells (lane 6). Each patient's image therefore represents a composite. For patient 12, the nonspecific band that is present in both the HCMV lysate lane (lane 5) and mock lysate lane (lane 6) does not comigrate with the paraprotein (lane 1,  ).
).
The analysis for patient 20 (Figure 6 righthand side) is more complex because there is a paraprotein, immunoreactive with the UL-48 gene product, as well as a minor monoclonal band, immunoreactive with glycoprotein B. The HCMV immunoblots reveal that the paraprotein aligns with the restricted band in the HCMV lysate lane (lane 5) but is not immunoreactive with intact HCMV virions (lane 3). This is expected, since the UL-48 gene product is a nuclear capsid protein and not present on the viral membrane. With intact HCMV virions, the paraprotein cannot penetrate the viral membrane and bind to an internal protein, such as the UL-48 gene product. By contrast, the minor monoclonal band denoted “motif 2” binds to both the HCMV lysate (lane 5) as well as intact virions (lane 3). Since motif 2 relates to glycoprotein-B specificity, binding to intact virions is expected. These findings collectively indicate that the 2 patients' paraproteins are HCMV immunoreactive. Since the peptide sequences identified by E-MAP are specific to HCMV and do not exist in other herpesviruses, the observed immunoreactivity is HCMV specific.
Discussion
This is the first technique for identifying the cognate antigen for an otherwise uncharacterized antibody. Until now, the literature on paraproteins includes descriptions of paraprotein targets that were identified by chance clinical associations. They include individual case reports of paraproteins binding to the p24 gag protein of HIV,15 cytomegalovirus,16 or streptolysin-O,17,18 all of which were identified after serologic assays on the patients came back with unexpectedly strong positive results. Unlike these previous reports, our new method allows us to identify the target antigen without any clinical clues or serendipitous laboratory findings from a serologic assay. The E-MAP technique incorporates 2 innovations that facilitate accurate protein prediction. First, we developed modifications to the phage-panning protocol that result in longer and more accurate consensus sequences for the paraprotein's epitope. In addition, we incorporated a new bioinformatic analysis technique, called pairwise analysis.5 Pairwise analysis involves looking for any 2 patients' paraproteins that bind to the exact same protein. This assumes that the antigenic stimulus for paraproteins is common, and is targeted by many patients' paraproteins. By making this assumption, pairwise analysis allows us to test this hypothesis by identifying proteins that contain both epitopes, from each paraprotein. Two sequences, converging on the same protein, help achieve sufficient information content from the sequence prediction analysis so as to accurately identify protein targets from the nonredundant protein database. From the analysis of the 9 patients' paraprotein consensus binding sequences, the cytomegalovirus hit by 2 patients was the first and most striking. Namely, 2 of the 9 patients' data converged on cytomegalovirus glycoprotein B, and were subsequently experimentally confirmed (Figures 4,Figure 5–6).
Cytomegalovirus represents an important pathogen in humans, as the majority of people have been exposed to it. Following an initial infection, HCMV normally remains in a persistent, latent state within the host, controlled by the host's immune system. The virus is capable of reactivation and shedding, even in seropositive immune-competent individuals. Thus, it likely represents a chronic immune stimulus, fostering the ongoing stimulation and growth of HCMV-specific B and T lymphocytes.
HCMV is known to be a powerful immune stimulus, often resulting in such a profound clonal expansion as to produce paraproteins in otherwise healthy individuals19 as well as in immunosuppressed patients.20 In normal, healthy HCMV-seropositive individuals, HCMV-specific CD8+ T lymphocytes comprise approximately 0.1% of the peripheral blood population, as measured by limiting dilution analysis.21 The proportion of HCMV-reactive lymphocytes increases with age, exacting an increasingly heavy burden in elderly individuals. MHC tetramer analysis of elderly HCMV-seropositive individuals indicates that, on average, approximately 5%22,23 of the CD8+ T lymphocytes may be specific for the HCMV pp65 immunodominant peptide. This figure may underestimate the percentage of T lymphocytes reactive with HCMV proteins since, contrary to previous belief, the T-cell repertoire is not as focused solely on pp65 as was originally thought.23,24 Such a long-lasting, strong immune response to a single agent, years after initial exposure, may be due to chronic repetitive viral reactivation.25,26 The fact that the paraproteins of our 2 patients with myeloma target HCMV is presumably due to transformation of one of these chronically stimulated reactive B lymphocytes.
An important aspect of this work is the experimental confirmation that the peptide sequence and predicted matching protein (from the protein database) match to the paraprotein. HCMV is ubiquitous in the general population, with the majority of people having been exposed to it. Following exposure, there is a normal polyclonal antibody response. Figures 4 and 6 were included to experimentally confirm that the peptide sequences of Figure 1 correspond to the paraprotein seen on serum protein electrophoresis, and not normal (reactive) polyclonal immunoglobulins. For example, Figure 4 demonstrates that the binding to the relevant peptide sequence associated with glycoprotein B in patient 12 exactly colocalizes with the paraprotein. In patient 20, the dominant paraprotein seen on serum protein electrophoresis exactly colocalizes with the UL-48 peptide sequence. Our analysis also revealed a second paraprotein in patient 20 that was glycoprotein-B immunoreactive, albeit at a much lower concentration. Such sharp, narrow bands are characteristic of monoclonal immunoglobulins (paraproteins), not (normal) polyclonal immunoglobulins. There are other experimental methods that can be used to potentially connect the paraprotein with the immunoreactivity to a define target, including purification of the paraprotein on either an HPLC column or with an immunoaffinity antigen adsorbent.
This report is the first application of E-MAP to a human disease. Our previous testing and validation used murine monoclonal antibodies, in a test system.5 In this report, we determined that the consensus sequences of 2 patients' paraproteins converged in identifying cytomegalovirus proteins. From the 7 other patients, at least one other protein target prediction was derived, but we have not yet experimentally validated that prediction. These findings collectively indicate that E-MAP may be a practical tool for identifying protein targets in B-lymphoproliferative disorders such as multiple myeloma. In addition, we believe that E-MAP may be broadly applicable to the investigation of neoplastic, inflammatory, autoimmune, and allergic diseases. This is a first proof-of-concept demonstration for the utility of E-MAP in a clinical disease. The ability of E-MAP to reliably predict protein targets of unknown antibodies will await further application of the method, in additional future clinical applications.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful for the financial support provided by the National Institutes of Health through grants CA94557 and CA106847 from the National Cancer Institute.
National Institutes of Health
Authorship
Contribution: S.R.S. designed and performed research and analyzed and interpreted data; G.B. performed research and analyzed and interpreted data; K.V. performed research; S.A.B. designed research, analyzed and interpreted data, and drafted the manuscript.
Conflict-of-interest disclosure: S.R.S. and S.A.B. both have a financial interest in Medical Discovery Partners. All other authors declare no competing financial interests.
Correspondence: Steven Bogen, Department of Pathology & Laboratory Medicine, Boston University School of Medicine, 715 Albany St, Boston, MA 02118; e-mail: sbogen@bu.edu.







 ) or with a synthetic peptide corresponding to the amino terminus (amino acids 1-20) of the UL-48 gene product (
) or with a synthetic peptide corresponding to the amino terminus (amino acids 1-20) of the UL-48 gene product ( ). MM patient sera were diluted 1:1250 so as to fall within the assay linear range. An HCMV-immunoreactive serum was used as a positive (+) control. A nonreactive serum was used as a negative control (-). Since patients 12 and 20 are our index patients, we randomly included MM patients 10 to 20 in the immunoassay, for comparison. The patient selection is random, since the patient number designation is assigned in order of specimen acquisition. Samples were measured in duplicate. The mean plus or minus the standard deviation are shown.
). MM patient sera were diluted 1:1250 so as to fall within the assay linear range. An HCMV-immunoreactive serum was used as a positive (+) control. A nonreactive serum was used as a negative control (-). Since patients 12 and 20 are our index patients, we randomly included MM patients 10 to 20 in the immunoassay, for comparison. The patient selection is random, since the patient number designation is assigned in order of specimen acquisition. Samples were measured in duplicate. The mean plus or minus the standard deviation are shown.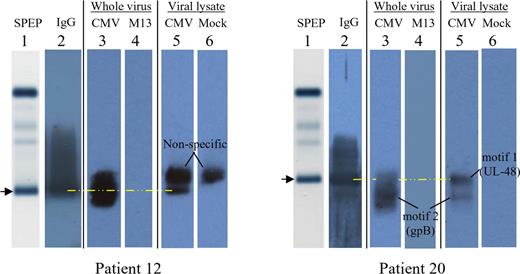
 ).
).