Abstract
Homozygous mutations in HAX1 cause an autosomal recessive form of severe congenital neutropenia (CN). By screening 88 patients with CN, we identified 6 additional patients with HAX1 mutations carrying 4 novel mutations. Of these, 2 affect both published transcript variants of HAX1; the other 2 mutations affect only transcript variant 1. Analysis of the patients' genotypes and phenotypes revealed a striking correlation: Mutations affecting transcript variant 1 only were associated with CN (23 of 23 patients), whereas mutations affecting both transcript variants caused CN and neurologic symptoms, including epilepsy and neurodevelopmental delay (6 of 6 patients). In contrast to peripheral blood, transcript variant 2 was markedly expressed in human brain tissue. The clinical phenotype of HAX1 deficiency appears to depend on the localization of the mutation and their influence on the transcript variants. Therefore, our findings suggest that HAX1 isoforms may play a distinctive role in the neuronal system.
Introduction
Severe congenital neutropenia (CN) is a rare primary immunodeficiency syndrome and comprises a heterogeneous group of inherited disorders.1,2
Patients with CN who have an autosomal-dominant mode of inheritance frequently show heterozygous mutations in the gene encoding neutrophil elastase (ELA2).3 In patients with autosomal recessive CN we have recently identified homozygous mutations in the antiapoptotic gene HAX1.4 A total of 3 nonsense mutations (p.Trp44X, p.Arg86X, and p.Gln190X) were found in several patients from the Middle East and Sweden, respectively.4
HAX1 is a molecule with predominantly mitochondrial localization, controlling the integrity of the inner mitochondrial membrane potential.5 In addition, HAX1 has been identified as an interacting partner of multiple viral and cellular proteins with diverse functions. For example, HAX1 interacts with bile export proteins (MDR1, MDR2, and BSEP),6 the alpha subunit of the G protein G13,7 phospholamban,8 and cortactin,9 suggesting that its role may not be limited to control apoptosis at the level of mitochondria.
To search for novel mutations and to establish first phenotype-genotype correlations, we systematically sequenced HAX1 in patients with CN.
Methods
Patients were referred to us by an established network of European and international physicians. Informed consent was obtained in accordance with the Declaration of Helsinki. The study was approved by the institutional review board (IRB) at Hannover Medical School. Genomic sequencing of HAX1 followed a previously published protocol.4
For the detection of HAX1 splice variants, we used reverse transcription and polymerase chain reaction (PCR) of a fragment of HAX1 cDNA spanning nucleotides 162 to 687 of the full-length cDNA (GenBank accession no. NM_006118).10 Human brain total RNA was purchased from Ambion (Warrington, United Kingdom).
HAX1 protein was detected by Western blot analysis using a monoclonal anti-HAX1 antibody (clone 52; BD Transduction Laboratories, Erembodegem, Belgium), followed by a secondary horseradish peroxidase (HRP)–linked antibody and detection by enhanced chemiluminescence (GE Healthcare, Little Chalfont, United Kingdom).
Results and discussion
We sequenced HAX1 in a cohort of 88 patients with CN without ELA2mutations. The patients fulfilled the criteria of early onset of severe bacterial infections due to sustained neutropenia (absolute neutrophil count [ANC] < 0.5 × 109/L [<500/μL]). We detected 29 patients with mutations in HAX1, including the 23 patients of the previously reported cohort.4 We also sequenced the HAX1 gene in a control group of 88 patients with CN who had ELA2 mutations. Confirming our previous data,4 no patient had a mutation in both genes.
A total of 26 (90%) of the 29 patients carrying HAX1 mutations were of Middle-Eastern descent. However, HAX1 mutations are also found in patients of European descent (eg, in patients from the original pedigree described by Kostmann).4,11
Here, we identify 4 novel homozygous nonsense or frameshift mutations in HAX1 (Table 1; Figure 1A), all leading to a premature stop codon. All new mutations were detected in patients of Middle-Eastern descent (patients 1-3 from Turkey; patient 4 from Iran).
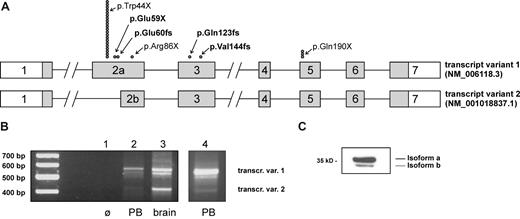
HAX1 transcript variants and summary of known mutations in patients with CN. (A) The intron-exon structure with the coding region (▒) for both isoforms of the HAX1 gene is shown. ● indicates the position of the mutations; each symbol represents a single patient (mutations were homozygous in all patients). Novel mutations are printed in bold letters. (B) Expression of transcript variants in peripheral blood cells (PB) and brain tissue. cDNAs from peripheral blood mononuclear cells (lanes 2 and 4) and from human brain (lane 3) have been amplified as described in “Methods.” A distinct band for transcript variant 2 could be detected only after secondary PCR (lane 4). The identity of transcript variants 1 and 2 has been confirmed by sequencing. The band migrating between the 2 variants consisted of heteroduplices of the 2 variants as confirmed by sequencing. (C) Expression of HAX1 isoforms in the HL-60 cell line. Western blot analysis was performed with total-cell lysate.
HAX1 transcript variants and summary of known mutations in patients with CN. (A) The intron-exon structure with the coding region (▒) for both isoforms of the HAX1 gene is shown. ● indicates the position of the mutations; each symbol represents a single patient (mutations were homozygous in all patients). Novel mutations are printed in bold letters. (B) Expression of transcript variants in peripheral blood cells (PB) and brain tissue. cDNAs from peripheral blood mononuclear cells (lanes 2 and 4) and from human brain (lane 3) have been amplified as described in “Methods.” A distinct band for transcript variant 2 could be detected only after secondary PCR (lane 4). The identity of transcript variants 1 and 2 has been confirmed by sequencing. The band migrating between the 2 variants consisted of heteroduplices of the 2 variants as confirmed by sequencing. (C) Expression of HAX1 isoforms in the HL-60 cell line. Western blot analysis was performed with total-cell lysate.
Patient 1 (p.Glu60fs) presented with severe neutropenia shortly after birth and has been successfully treated with recombinant granulocyte colony-stimulating factor (G-CSF) from age 7 years on. At the age of 14 years, a somatic CSF3R mutation was detected. However, no additional cytogenetic aberrations could be detected, and the patient is in stable condition.
Patient 2 (p.Gln123fs) developed a myelodysplastic syndrome (MDS) at the age of 7 years and was subsequently treated by bone marrow transplantation. In addition to the hematologic phenotype, the patient showed mental and psychomotor retardation and had several episodes of seizures. Neurologic diagnostics revealed pathologic electroencephalography but normal magnetic resonance imaging of the brain.
The clinical presentation of patient 3 (p.Val144fs) has previously been reported.12 This patient is remarkable for the development of acute lymphoblastic leukemia at age 7 months. Noteworthy, the patient had several episodes of seizures starting at the age of 3 months, indicating a neurologic phenotype in this patient. No abnormalities were detected by cranial computer tomography (CT) and ultrasonography.
Patient 4 (p.Glu59X) was diagnosed at the age of 3.5 years after a long history of severe recurrent infections that led to necrosis of the nasal cartilage. No neurologic abnormality was reported in this patient.
Heterozygosity of the parents has been confirmed in all patients except for patient 1, for whom no samples of the parents were available.
Patients with CN show a predisposition to develop clonal myeloid disorders (MDS/acute leukemia), a finding that is not limited to patients with ELA2 mutations. Patient 3 developed acute lymphoblastic leukemia (ALL) in infancy, and patient 2 developed MDS early in life. Moreover, mutations in CSF3R that are specific for patients with CN and have been implicated with leukemic development13 have been detected in 2 of the 6 new patients. While these new patients confirm that HAX1 deficiency constitutes a preleukemic condition, it remains to be shown whether the risk to develop a clonal hematopoietic disorder is comparable between patients with HAX1 and ELA2 mutations.
To date, most HAX1-deficient patients identified show the p.Trp44X mutation (21 [72%] of 29), and their phenotype appears to be limited to neutropenia. However, HAX1 deficiency may also cause a phenotype in the nervous system ranging from mild cognitive defects to severe developmental delay and/or epilepsy. Of the newly described patients of this report, 2 (patient 2, with mutation p.Val144fs; patient 3, with mutation p.Gln123fs) show neurologic symptoms; the patient from our previous report harboring mutation p.Arg86X mutation4 suffered from a seizure disorder and developmental delay.14 These patients did not show any abnormalities using brain imaging techniques. The mutation present in the members of the original Kostmann pedigree (p.Gln190X)4 is also associated with measurable cognitive defects.15,16 Recently, another patients with CN who had mutation p.Arg86X has been described who was characterized by severe develop-mental delay and epilepsy without abnormalities in magnetic resonance imaging.17
A transcript variant of HAX1 has been identified in cDNA libraries18 that uses an alternate in-frame splice site producing a markedly shortened exon 2 (exon 2b; Figure 1A). The truncation of the deduced protein from splice variant 2 (isoform b) does not affect the HAX1 PEST polypeptide sequences and the putative transmembrane domain of the protein. However, putative Bcl-2 and Nip3 homology domains BH1 and BH2 are partially affected by this deletion.5 Up to now, it was not clear whether the splice variants code for 2 proteins with different functions and whether they can (partially) compensate for each other.
We tested whether defined HAX1 mutations correlated with the presence of neurologic symptoms. Interestingly, HAX1 mutations affecting exclusively the full-length transcript (p.Trp44X, p.Glu59X, p.Glu60fs) lead to CN without neurologic symptoms (23 of 23 tested). In contrast, mutations affecting both isoforms (p.Arg86X, p.Gln123fs, p.Val144fs, and p.Gln190X) are associated with an additional neurologic phenotype (6 of 6 tested). Thus, a striking genotype-phenotype correlation could be established (P < .001 using the Fisher exact test).
We could confirm the expression of both transcripts in cells from peripheral blood and in brain tissue (Figure 1B). The identity of both transcript variants was confirmed by sequencing. The translation of transcript variant 2 into a smaller protein isoform could be confirmed by Western blot analysis in cell lines (Figure 1C). Interestingly, expression of the smaller transcript variant 2 was much stronger in the brain compared with in peripheral blood cells (Figure 1B; lanes 2 and 3), where a distinct band of the smaller transcript was detected only after secondary PCR (lane 4).
These data suggest that the presence of isoform b is critical for neuronal function. Interestingly, most recent findings from a Hax1 knock-out model in mice revealed an antiapoptotic role of Hax1 in lymphocytes and neurons.19 Future studies are needed to elucidate functional properties of HAX1 isoforms in hematopoietic and neuronal cells of mice and men, respectively. Taken together, our data suggest that HAX1 plays a previously unrecognized role in the central nervous system. Patients with HAX1 mutations affecting both isoforms have a more severe phenotype and should be assessed for additional neurologic problems.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are indebted to the patients and their families. We thank K. Clodi, R. Dopfer, A. Fahimzad, B. Neophytou, M. Schmid, A. Schulz, and all physicians who referred and registered patients to the Severe Chronic Neutropenia International Registry (SCNIR) and provided clinical information. We also would like to acknowledge the excellent technical assistance of Jana Diestelhorst, Marly Dalton, and Yvonne Peter.
This work was supported by grants from the Federal Ministry of Education and Research (German Network on Congenital Bone Marrow Failure Syndromes), the German José-Carreras Foundation, and the DFG (KliFo 110).
Authorship
Contribution: M. Germeshausen designed and performed research and wrote the manuscript; M. Germeshausen, M. Grudzien, and B.G. did the work on HAX1 transcript variants; C.Z. collected data in the SCN registry and classified the patients; H.A. analyzed one of the patients in this study; S.Y. and N.R. provided samples and clinical data for this study; M.B. analyzed data and wrote the manuscript; K.W. initiated and supervised the project; and C.K. designed research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karl Welte, Pediatric Hematology and Oncology, Medizinische Hochschule Hannover, Carl-Neuberg-Str. 1, 30625 Hannover, Germany; e-mail: welte.karl.h@mh-hannover.de.