Neutrophils have a very short half-life in the circulation, undergoing rapid death by apoptosis, but a number of agents can either delay or accelerate the rate at which these cells undergo death. TNFα can exert opposing, concentration-dependent effects on neutrophils to either accelerate their apoptosis or enhance their survival. We show that TNFα greatly increases the rate of turnover of Mcl-1, an antiapoptotic protein that plays a key role in neutrophil survival. In contrast to Mcl-1 turnover in control- or granulocyte-macrophage colony-stimulating factor (GM-CSF)–treated neutrophils that occurs via the proteasome, TNFα-accelerated Mcl-1 turnover occurs via activation of caspases. Mcl-1–depleted cells thus have accelerated rates of apoptosis. While TNFα had no effect on MCL-1 transcription, it induced expression of another antiapoptotic molecule, BFL-1. Low concentrations of TNFα (≤ 1 ng/mL) stimulated BFL-1 expression, whereas higher concentrations (≥ 10 ng/mL) triggered caspase-dependent acceleration of Mcl-1 turnover. These opposing effects on 2 separate antiapoptotic systems of neutrophils explain the divergent effects of TNFα on neutrophil apoptosis and have important implications for understanding how TNFα may affect immune function in inflammatory diseases.
Introduction
Apoptosis of neutrophils, followed by their safe removal via phagocytosis, is essential for the resolution of inflammation and prevention of tissue damage. Circulating neutrophils have a very short half-life of only a few hours in the circulation, but signals such as adhesion, transmigration, hypoxia, and cytokines can all delay their apoptosis and extend their lifespan, thereby enhancing their ability to function during inflammatory challenge.1 Once their function is complete, then death signals (eg, death receptor engagement or cytokine depletion) trigger apoptosis and phagocytic removal of apoptotic neutrophils by macrophages or endo/epithelial cells. Many inflammatory conditions are considered to result from defective apoptosis of immune cells, and so understanding the molecular processes that either control neutrophil death or survival could have therapeutic potential.2
The antiapoptotic protein, Mcl-1, plays a major role in controlling the rate at which neutrophils undergo apoptosis.3,4 Cellular levels of Mcl-1 are regulated by its rate of transcription, but also by the rate of turnover of the protein.5 Mcl-1 has a very short half-life in cells and tissues of approximately 3 hours,5,6 being degraded by the proteasome. Agents that delay apoptosis in cells such as cytokines, can stimulate small, but significant increases in MCL-1 mRNA,5 but their major effects are often to enhance cellular protein levels by delaying its rate of turnover.7,8 This enhanced Mcl-1 stability occurs via signaling pathways involving Raf, PI3K, GSK, and ERK.7,–9 Alternatively, some agents can significantly increase the rate of Mcl-1 turnover and thereby enhance neutrophil apoptosis. For example, sodium salicylate treatment decreases the half-life of Mcl-1 to approximately 1.5 hours, and under these conditions, turnover occurs via caspase cleavage rather than proteasomal degradation.10
There is much interest in understanding the role of TNFα in the regulation of inflammation, in view of the fact that remarkable benefits of anti-TNF therapy are often observed in diseases such as rheumatoid arthritis11 and inflammatory bowel disease.12 TNFα is reported to regulate neutrophil apoptosis,13,,–16 but differential effects are seen in that it can accelerate death in a subpopulation of cells but delay apoptosis in the subpopulation that escapes this initial death signal.17 It has been proposed that these differential effects are concentration dependent, with high concentrations acting as a death signal, but lower concentrations stimulating apoptosis delay.18 In this report, we show that these opposing, concentration-dependent effects of TNFα on neutrophil apoptosis are due to differential effects on neutrophil survival mechanisms. The rapid cell death occurs via accelerated, caspase-dependent Mcl-1 degradation, while survival may be mediated by enhanced expression of another antiapoptotic molecule, Bfl-1.
Methods
This study was approved by the NHS COREC [National Health Service Central Office for Research Ethics Committee] and the University of Liverpool Committee on Research Ethics.
Materials
Polymorphprep was from Axis-Shield (Oslo, Norway); RPMI 1640 medium and TRIzol reagent were from GibcoBRL (Paisley, United Kingdom). FCS, luminol, cycloheximide, PMA, and peroxidase-linked goat antimouse antibodies were from Sigma (Poole, United Kingdom). Cytokines used were granulocyte-macrophage colony-stimulating factor (GM-CSF; Glaxo, Greenford, United Kingdom) and TNFα (Calbiochem, Nottingham, United Kingdom). Caspase inhibitor Z-VAD-FMK and proteasome inhibitor MG-132 were from Calbiochem. FITC-conjugated CD15 and CD16 antibodies and Mcl-1 and Bax antibodies were from BD Biosciences (Oxford, United Kingdom); GAPDH antibody was from Abcam (Cambridge, United Kingdom); caspase antibodies were from Cell Signaling Technology (Boston, MA); while peroxidase-linked donkey antirabbit antibodies, enhanced chemiluminescence (ECL) detection kit, and Amplify were from Amersham (Amersham, United Kingdom). The Dual Luciferase Assay System, pGL-3 Basic reporter vector, pRL-SV40 (Rluc), AMW reverse transcriptase, and dNTP mix were from Promega (Southampton, United Kingdom). Rapid Romanowsky stain was from HD Supplies (Ayelsbury, United Kingdom). MitoTracker red was from Invitrogen (Paisley, United Kingdom). RNeasy mini kit and Quantitect SYBR Green polymerase chain reaction (PCR) kit were from Qiagen (Crawley, United Kingdom). Oligonucleotides were from MWG-Biotech AG (Ebersberg, Germany). Atlas nylon arrays were from Clontech (Mountain View, CA).
Cell isolation and culture
Neutrophils were isolated using Polymorphprep (as described in the manufacturer's instructions).19,–21 Contaminating erythrocytes were removed by hypotonic lysis. Neutrophils were routinely examined for purity and viability using trypan blue exclusion, which were more than 97% and more than 98%, respectively, immediately after isolation. Purity was confirmed using morphologic analysis of cytospins, and CD15 and/or CD16 expression.22,23 Purified neutrophils were resuspended in RPMI 1640 with 25 mM HEPES, supplemented with 10% FCS and 1 mM l-glutamine, at 5 × 106 cells/mL and cultured at 37°C in a humidified incubator. Cytokines and inhibitors were added as indicated. HeLa cells were cultured and maintained in MEM supplemented with 10% FCS and nonessential amino acids and incubated at 37°C in a humidified incubator with 5% CO2.
Measurements of apoptosis
Following culture, a 20-μL aliquot of neutrophil suspension (105 cells) was made up to 200 μL with RPMI 1640, and cells were cytocentrifuged using a Shandon Cytospin3 (Runcorn, Cheshire, United Kingdom). Romanowsky staining of cytospins allowed apoptosis to be scored by morphology. This method correlates well with other markers of apoptosis.24 In addition, apoptosis was determined by measurement of FITC–annexin V binding and flow cytometry as described previously.8
Mitochondrial staining
Neutrophils (106) were incubated in 2 mL supplemented media in 35-mm glass-bottomed Iwaki culture dishes (Barlow Scientific, Stone, United Kingdom), and 25 nM MitoTracker red (CM-H2XRos) was used to monitor the mitochondrial membrane potential. Cells were incubated on a gas- and temperature-controlled microscope stage, and fluorescence was visualized using a LSM510 confocal microscope (Carl Zeiss, New York, NY) at 1024 × 1024 pixel resolution through a 63× Plan Apochromat (NA 1.4) objective with 2 times averaging. Excitation was at 543 nm and emission collected through a 585-nm long-pass filter.
Western analysis
Following culture, 106 cells were rapidly lysed in boiling, reducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer containing aprotinin (20 μg/mL), leupeptin (20 μg/mL), pepstatin (10 μg/mL), and PMSF (400 μg/mL): samples were immediately boiled for 5 minutes with occasional vortexing, and stored at −80°C until use. SDS-PAGE and electrotransfer to PVDF membranes were performed as described.25 Primary antibodies used were anti–Mcl-1; anti–caspase-3, -8, -9, -10; and anti-GAPDH. Horseradish peroxidase–conjugated secondary antisera used were donkey antirabbit IgG and goat antimouse IgG. Bound antibodies were detected using the ECL system. Densitometry on carefully exposed blots (to avoid film saturation) was performed with Image 1.44 VDM software (National Institutes of Health, Bethesda, MD). Ponceau S–stained actin on membranes after electrotransfer and levels of GAPDH were measured to confirm equivalence of loading of neutrophil samples.
MCL-1 and BFL-1: luciferase reporter gene assay
The human MCL-1 promoter cloned into the pGL3-Basic luciferase reporter system was used to measure rates of MCL-1 transcription, as described previously.26 The human BFL-1 promoter (a kind gift from Dr D. Walls, School of Biotechnology, Dublin City University) cloned into pGL2-Basic luciferase reporter system was used to measure BFL-1 transcription.27 In addition, 1 μg pRL-SV40 (luciferase driven by SV40 early enhancer/promoter) was added to each transfection as an internal control. HeLa cells were seeded at 2 × 105 cells/2 mL in 35-mm culture dishes and allowed to adhere overnight. Cells were then transfected using Fugene 6 (Welwyn Garden City, United Kingdom) in a 2:1 ratio (2 μL:1 μg DNA) and incubated overnight. After transfection, cells were incubated for 30 minutes at 37°C in the presence or absence of the NF-κB inhibitor parthenolide (10 μM) before 24-hour incubation with the indicated stimulants (GM-CSF [50 U/mL], phorbol ester [10 nM] or TNFα [10 ng/mL]), at 37°C in a humidified incubator with 5% CO2. Growth media were removed and the cells washed with PBS (lacking Mg2+/Ca2+) before lysis of cells with buffer supplied in the kit. Luciferase activity was assayed (as described in the manufacturer's instructions). Transfected cells were harvested after 24-hour incubation and cells were lysed in 100 μL passive lysis buffer provided with the kit; cell debris was pelleted by centrifugation. Cell lysate (20 μL) was used for each luciferase assay using a Berthold Lumistar luminometer (BMG, Offenburg, Germany). Peak values (which are proportional to luciferase concentration) were recorded. Transfection efficiencies were normalized to Renilla luciferase activity. All values were converted to fold luciferase activities by comparison with the activity measured of the control plasmid (pGL-3 Basic). All experiments were performed in triplicate and data shown represent mean values with error bars showing standard deviations.
Quantitative real-time PCR assay
Neutrophils were left untreated or treated with 10 ng/mL TNFα for the indicated period after which they were lysed and total RNA was extracted using TRIzol before cleaning with RNeasy mini kit. cDNA was synthesized using AMW reverse transcriptase. Quantitative real-time PCR analysis was carried out on the Corbett Rotorgene RG3000 Detection System using the Quantitect SYBR Green PCR kit as recommended by the manufacturer (Corbett Research, Sydney, Australia). Each sample was tested in triplicate and cDNA species were amplified using real-time quantitative reverse-transcription (qRT)–PCR, and this was carried out to measure the relative levels of MCL-1, BAX, and BFL-1 gene expression using the comparative cycle threshold (Ct). The values for β-actin were used to normalize the gene expression data. The relative value of target gene expression was calculated from the standardized target gene and β-actin amplified curves.
The cycling conditions were 95°C/15 minutes with a melt of 95°C/15 seconds on first cycle and 40 cycles of 95°C/6 seconds, 60°C/25 seconds, and 72°C/1 minute. The cycle threshold was determined to provide the optimal standard curve values (0.98 to 1.0). Specific primers were Mcl-1 (forward, 5′-GGGCAGGATTGTGACTCTCATT-3′; reverse 5′-GATGCAGCTTTCTTGGTTTATGG-3′); Bax (forward, 5′-CCCACCAGCTCTGAGCAGATC-3′; reverse, 5′-CAGTTGAAGTTGCCGTCAGAA-3′); Bfl-1 (forward, 5′-CGGCATCATTAACTGGGGAAG-3′; reverse, 5′-TGGTCAACAGTATTGCTTCAGGA-3′), and β-actin (forward, 5′-AAGGATTCCTATGTGGGCGAC-3′; reverse, 5′-CGCGGTTGGCCTTGGGGTTCA-3′).
Microarray analysis
Atlas nylon arrays for apoptosis genes were used to analyze RNA samples of neutrophils incubated for 3 hours in the presence and absence of TNFα (10 ng/mL). Gene expression analysis was performed pairwise using control neutrophil samples versus TNFα-stimulated neutrophils. RNA isolation and reverse transcription were performed as previously described; labeling (33P-ATP) and hybridization were performed per the manufacturer's instructions. Nylon arrays were exposed to GP/LE phosphor screens (Molecular Dynamics, Sunnyvale, CA) for 24 hours, and scanned using a Storm PhosphorImager (Molecular Dynamics). IMAGEQUANT (Molecular Dynamics) or ATLASIMAGE 2.01 (Clontech) was used for quantitation of signal intensities.
Statistics
Datasets were analyzed using the Student t test.
Results
Effects of TNFα on neutrophil apoptosis and Mcl-1 levels
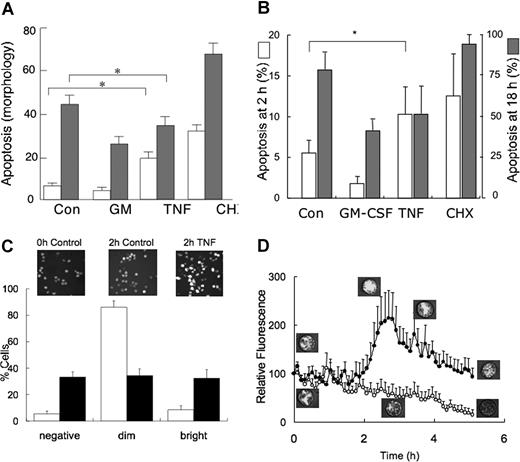
Figure 1A shows that when used at a concentration of 10 ng/mL, TNFα enhanced neutrophil apoptosis, compared with control suspensions, as assessed by morphology at 6 hours. However, after 20-hour incubation with TNFα, apoptosis was significantly less than that observed in untreated control suspensions, demonstrating the dual effects of this cytokine on neutrophil apoptosis: rapid induction of apoptosis in a subset of cells, but delayed apoptosis in those cells that survive this initial death signal. Figure 1A also shows rates of neutrophil apoptosis when incubated in the presence of GM-CSF (which significantly delays apoptosis) or the protein synthesis inhibitor cycloheximide (which significantly accelerates apoptosis). Similar results were obtained when apoptosis was determined by measuring the binding of FITC–annexin V (Figure 1B). Confocal microscopy of mitochondrial membrane potential (MitoTracker red staining) revealed that these kinetics of apoptosis observed in the population of neutrophils to TNFα reflected considerable cell:cell heterogeneity in response to this cytokine (Figure 1C). As control neutrophils aged in culture, their mitochondrial membrane potential gradually decreased (designated as “dim” in Figure 1C), presumably as they lost function and became committed to apoptosis. When 10 ng/mL TNFα was added to neutrophils, cells responded in 1 of 2 contrasting ways. First, a subpopulation of cells showed accelerated loss of membrane population such that the “dim” population was significantly greater than that of controls (TNFα: 33% ± 5%; control: 5% ± 2%, P < .01). Second, a subpopulation of neutrophils exhibited significantly enhanced mitochondrial membrane potential in response to TNFα (“bright” in Figure 1C), which presumably delays their apoptosis. This heterogeneous response to TNFα and kinetics of changes in mitochondrial membrane potential is illustrated in Figure 1D, which shows the time course of changes in average fluorescence intensities of 6 cells that lose their mitochondrial membrane potential (and become committed to apoptosis) and 6 cells that undergo a transient enhancement of mitochondrial membrane potential, which then slowly declines.
Effects of TNFα on neutrophil apoptosis. (A) Neutrophils were incubated for 6 hours (□) or 20 hours (▩) in the absence and presence of GM-CSF (50 U/mL), TNFα (10 ng/mL), or CHX (10 μg/mL), prior to measurement of apoptosis by assessment of morphology. There was a significant difference (*P < .05) between apoptosis of controls and TNFα-treated neutrophils showing initial increase in neutrophil apoptosis, whereas at 20 hours it acted as a survival factor. Values shown are means (± SD, n = 5). (B) Neutrophils were incubated for either 2 hours or 18 hours as indicated, and apoptosis was determined by measuring the binding of FITC–annexin V and flow cytometry. Values shown are means (± SD, n = 4) and * shows a P value of less than .015 between 2-hour control and 2-hour plus TNFα. (C) Neutrophils were stained with MitoTracker red after culture for 2 hours in the absence (control, □) or presence (10 ng/mL, ■) of TNFα. Graph shows percentage of cells that had either lost (negative) or had decreased (dim) or enhanced (bright) fluorescence, compared with that measured in the cell population at time zero. Insert shows representative confocal images. Values shown are means (± SD, n = 5). (D) Mean fluorescence of 6 individual cells whose fluorescence either decreased or was enhanced following TNFα (10 ng/mL) treatment, together with representative confocal images of MitoTracker red fluorescence.
Effects of TNFα on neutrophil apoptosis. (A) Neutrophils were incubated for 6 hours (□) or 20 hours (▩) in the absence and presence of GM-CSF (50 U/mL), TNFα (10 ng/mL), or CHX (10 μg/mL), prior to measurement of apoptosis by assessment of morphology. There was a significant difference (*P < .05) between apoptosis of controls and TNFα-treated neutrophils showing initial increase in neutrophil apoptosis, whereas at 20 hours it acted as a survival factor. Values shown are means (± SD, n = 5). (B) Neutrophils were incubated for either 2 hours or 18 hours as indicated, and apoptosis was determined by measuring the binding of FITC–annexin V and flow cytometry. Values shown are means (± SD, n = 4) and * shows a P value of less than .015 between 2-hour control and 2-hour plus TNFα. (C) Neutrophils were stained with MitoTracker red after culture for 2 hours in the absence (control, □) or presence (10 ng/mL, ■) of TNFα. Graph shows percentage of cells that had either lost (negative) or had decreased (dim) or enhanced (bright) fluorescence, compared with that measured in the cell population at time zero. Insert shows representative confocal images. Values shown are means (± SD, n = 5). (D) Mean fluorescence of 6 individual cells whose fluorescence either decreased or was enhanced following TNFα (10 ng/mL) treatment, together with representative confocal images of MitoTracker red fluorescence.
These changes in rates of apoptosis were associated with changes in the levels of Mcl-1, a key regulator of apoptosis in neutrophils.2,,,–6 Figure 2A shows that levels of this antiapoptotic protein declined as neutrophils underwent apoptosis. In line with previous reports, GM-CSF partly prevented this decline, whereas cycloheximide accelerated the loss of this protective protein.8 These observations are explained by the facts that (1) in the presence of cycloheximide to block protein synthesis, the protein undergoes rapid, constitutive turnover and (2) GM-CSF increases cellular levels by delaying this turnover.8 TNFα treatment of neutrophils resulted in decreased levels of Mcl-1, compared with control levels, but remarkably, Mcl-1 was barely detectable in cells treated with both TNFα and cycloheximide. These results would indicate that TNFα treatment accelerates the rate of turnover of Mcl-1 above the rate of constitutive turnover, a result that was confirmed in Figure 2B. In these experiments, the half-life of Mcl-1 in control cells (time required for a 50% decrease in the levels of protein remaining in cycloheximide-treated cells) was just more than 3 hours, while in the presence of TNFα this half-life was decreased to approximately 1.5 hours.
Changes in Mcl-1 protein stability induced by TNFα. (A) Neutrophils were incubated in the presence of the protein synthesis inhibitor cycloheximide (CHX) at 10 μg/mL, in the presence or absence of TNFα (10 ng/mL) or GM-CSF (50 U/mL). Protein lysates were prepared after 2-hour (▩) and 5-hour (■) incubation, and Mcl-1 levels were detected by Western blotting. Top panel shows a representative blot of Mcl-1 protein and GAPDH protein as a loading control (after 5-hour incubation), while the bottom panel shows mean data (± SD) of relative Mcl-1 levels (taking the signal at time zero as 100%) of 3 separate experiments. (B) Neutrophils were incubated in the presence of cycloheximide alone or with TNFα, and at 1-hour intervals, protein lysates were prepared for analysis of Mcl-1 levels by Western blotting. Inset shows representative Western blots (plus GAPDH levels as loading control), while graph shows mean values of repeat experiments (± SD, n = 5).
Changes in Mcl-1 protein stability induced by TNFα. (A) Neutrophils were incubated in the presence of the protein synthesis inhibitor cycloheximide (CHX) at 10 μg/mL, in the presence or absence of TNFα (10 ng/mL) or GM-CSF (50 U/mL). Protein lysates were prepared after 2-hour (▩) and 5-hour (■) incubation, and Mcl-1 levels were detected by Western blotting. Top panel shows a representative blot of Mcl-1 protein and GAPDH protein as a loading control (after 5-hour incubation), while the bottom panel shows mean data (± SD) of relative Mcl-1 levels (taking the signal at time zero as 100%) of 3 separate experiments. (B) Neutrophils were incubated in the presence of cycloheximide alone or with TNFα, and at 1-hour intervals, protein lysates were prepared for analysis of Mcl-1 levels by Western blotting. Inset shows representative Western blots (plus GAPDH levels as loading control), while graph shows mean values of repeat experiments (± SD, n = 5).
Regulation of TNFα–induced Mcl-1 turnover
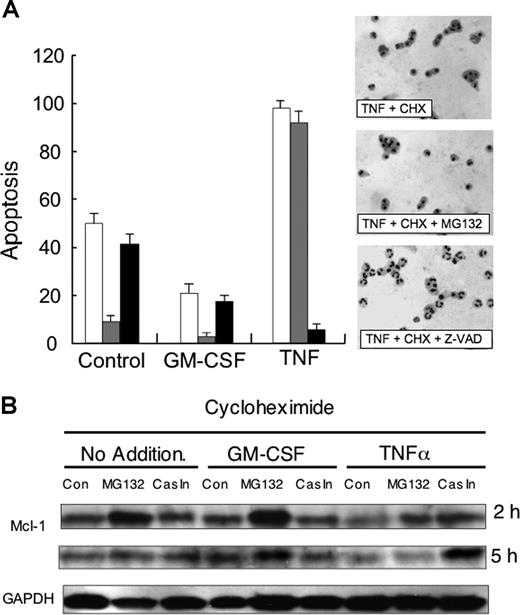
These changes in Mcl-1 levels in the presence of cycloheximide were paralleled by changes in the rate of neutrophil apoptosis. After 3-hour incubation with cycloheximide, neutrophil apoptosis was approximately 50% of the cell population, but this apoptosis was significantly blocked by the addition of the proteasome inhibitor, MG-132 (Figure 3A). The addition of GM-CSF to cycloheximide-treated cells slowed the rate of apoptosis, and again, the proteasomal inhibitor blocked this apoptosis, in line with preventing Mcl-1 turnover. In contrast, the pan-caspase inhibitor, Z-VAD, had no significant effect on control- or GM-CSF–regulated apoptosis under these conditions. In line with its ability to greatly enhance the rate of Mcl-1 turnover, TNFα treatment resulted in 100% apoptosis of cycloheximide-treated cells by 3-hour incubation (Figure 3A). However, in complete contrast to constitutive and GM-CSF–regulated apoptosis, the proteasomal inhibitor had no effect on TNFα-enhanced apoptosis, but instead the pan-caspase inhibitor, Z-VAD, blocked apoptosis. These results would suggest that TNFα triggers a new pathway of caspase-dependent apoptosis.
Role of caspases in TNFα-induced apoptosis and Mcl-1 turnover. (A) Neutrophils were incubated in the presence of cycloheximide (10 μg/mL) alone, or with TNFα (10 ng/mL) or GM-CSF (50 U/mL) in the absence (□) or presence of either MG132 (50 μM, ▩) or Z-VAD-FMK (Cas In 50 μM, ■). After 3-hour incubation, samples were removed and apoptosis was determined by morphology. Side panel shows representative cytospins. Typical result of 3 separate experiments. (B) Neutrophils were incubated as described for panel A but after 2 hours and 5 hours, protein lysates were analyzed by Western blotting for levels of Mcl-1 or GAPDH. Typical result of 3 separate experiments.
Role of caspases in TNFα-induced apoptosis and Mcl-1 turnover. (A) Neutrophils were incubated in the presence of cycloheximide (10 μg/mL) alone, or with TNFα (10 ng/mL) or GM-CSF (50 U/mL) in the absence (□) or presence of either MG132 (50 μM, ▩) or Z-VAD-FMK (Cas In 50 μM, ■). After 3-hour incubation, samples were removed and apoptosis was determined by morphology. Side panel shows representative cytospins. Typical result of 3 separate experiments. (B) Neutrophils were incubated as described for panel A but after 2 hours and 5 hours, protein lysates were analyzed by Western blotting for levels of Mcl-1 or GAPDH. Typical result of 3 separate experiments.
TNFα–mediated caspase activation
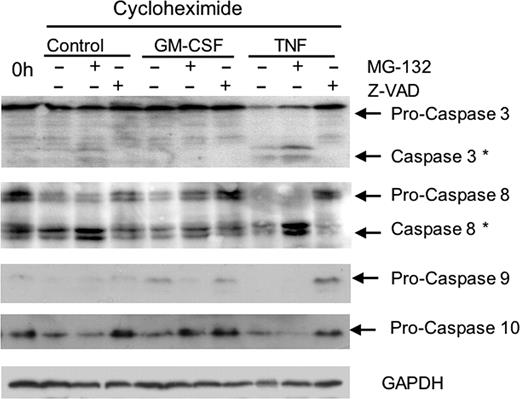
It was then necessary to determine whether TNFα accelerated Mcl-1 turnover via caspase-dependent processes. Neutrophils were incubated for 2 and 5 hours with cycloheximide in the presence and absence of GM-CSF or TNFα, together with the proteasomal inhibitor MG-132 or the pan-caspase inhibitor, Z-VAD. Figure 3B shows that both constitutive and GM-CSF–delayed turnover of Mcl-1 were partly blocked by the proteasomal inhibitor, but the pan-caspase inhibitor did not protect against degradation under these conditions. In contrast, TNFα-accelerated Mcl-1 turnover was largely proteasome independent but caspase dependent. We then confirmed that TNFα triggers caspase activation in neutrophils by measuring levels of either procaspases or activated (cleaved) caspases in Western blots following treatment. As can be seen in Figure 4, TNFα resulted in greatly increased levels of activation of a number of caspases, shown as decreased levels of procaspases-3, -8, -9, and -10 and increased levels of cleaved (active) caspases-3 and -8. Note that we could not detect any cleaved (active) caspase-9, presumably because the activated enzyme is unstable and our anti–caspase-10 antibody detected only pro–caspase-10. The pan-caspase inhibitor, Z-VAD (but not MG-132), could prevent activation of these caspases following TNFα treatment.
Activation of caspases by TNFα. Neutrophils were incubated in the presence of cycloheximide (10 μg/mL), alone (control), or with TNFα (10 ng/mL) or GM-CSF (50 U/mL) in the presence or absence MG132 (50 μM) and Z-VAD-FMK (50 μM). After 5-hour incubation, samples were removed and levels of caspases-3, -8, -9, and -10 (or GAPDH as a loading control) were detected by Western blotting. Typical result of 3 separate experiments.
Activation of caspases by TNFα. Neutrophils were incubated in the presence of cycloheximide (10 μg/mL), alone (control), or with TNFα (10 ng/mL) or GM-CSF (50 U/mL) in the presence or absence MG132 (50 μM) and Z-VAD-FMK (50 μM). After 5-hour incubation, samples were removed and levels of caspases-3, -8, -9, and -10 (or GAPDH as a loading control) were detected by Western blotting. Typical result of 3 separate experiments.
These experiments conclusively show that the accelerated rate of neutrophil apoptosis induced by TNFα results from activation of caspase-dependent turnover of Mcl-1. However, these data do not explain the protective effect of TNFα on the subpopulation of cells that survive this initial, rapid induction of cell death. Clearly, other survival mechanisms must operate.
Effects of TNFα on neutrophil gene expression
We therefore measured global changes in expression of cell death and survival genes induced by TNFα using a macroarray. TNFα induced only small changes in expression of only a small number of genes of interest, examples of which are shown in Figure 5A. For example, TNFα treatment did not alter the levels of mRNA for caspase-3 or Mcl-1, but significantly increased levels of mRNA for BFL-1, an antiapoptotic protein that has been implicated previously in neutrophil survival. These results were confirmed in a luciferase reporter gene assay (Figure 5B). Whereas MCL-1 transcription was enhanced by GM-CSF and the phorbol ester, PMA, TNFα had no effect on MCL-1 transcription. Furthermore, neither constitutive nor activated transcription of MCL-1 appeared to involve NF-κB, as the inhibitor parthenolide had no effect on reporter gene activity. In contrast, TNFα (and PMA) stimulated the transcription of BFL-1 via an NF-κB–dependent pathway, whereas GM-CSF had no effect on transcription of this gene. These results were confirmed by real-time RT-PCR, showing that BFL-1 mRNA, but not BAX or MCL-1 mRNA, was enhanced by treatment of neutrophils with TNFα (Figure 5C).
Effects of TNFα on mRNA levels. (A) mRNA samples of freshly isolated neutrophils and neutrophils incubated in the presence of TNFα (10 ng/mL) for 3 hours were determined using Atlas nylon arrays. Selected spots shown are those of caspase-3, MCL-1, and BFL-1. Typical result of 1 of 3 separate experiments using neutrophils from different donors. (B) HeLa cells were transfected with luciferase vectors driven by either the BFL-1 or MCL-1 promoters. Twenty-four hours after transfection, the cells were incubated in the presence or absence of GM-CSF (50 U/mL), TNFα (10 ng/mL), or PMA (10 μg/mL) with (▩) or without (■) the NF-κB inhibitor parthenolide (10 μM). Values shown are means (± SD, n = 3). (C) Neutrophils were incubated in the presence of TNFα (10 ng/mL), and RNA samples were isolated at the indicated time points. Levels of transcripts for BAX (■), BFL-1 (●), and MCL-1 (○) mRNA were measured using quantitative real-time PCR (normalized to GAPDH levels). Values shown are means (± SD, n = 3).
Effects of TNFα on mRNA levels. (A) mRNA samples of freshly isolated neutrophils and neutrophils incubated in the presence of TNFα (10 ng/mL) for 3 hours were determined using Atlas nylon arrays. Selected spots shown are those of caspase-3, MCL-1, and BFL-1. Typical result of 1 of 3 separate experiments using neutrophils from different donors. (B) HeLa cells were transfected with luciferase vectors driven by either the BFL-1 or MCL-1 promoters. Twenty-four hours after transfection, the cells were incubated in the presence or absence of GM-CSF (50 U/mL), TNFα (10 ng/mL), or PMA (10 μg/mL) with (▩) or without (■) the NF-κB inhibitor parthenolide (10 μM). Values shown are means (± SD, n = 3). (C) Neutrophils were incubated in the presence of TNFα (10 ng/mL), and RNA samples were isolated at the indicated time points. Levels of transcripts for BAX (■), BFL-1 (●), and MCL-1 (○) mRNA were measured using quantitative real-time PCR (normalized to GAPDH levels). Values shown are means (± SD, n = 3).
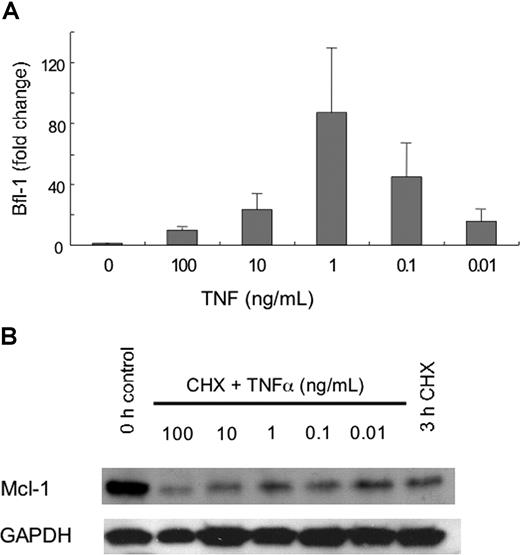
Previous work has reported that the differential effects of TNFα on neutrophil apoptosis are concentration dependent,18 low concentrations having antiapoptotic effects but higher concentrations stimulating cell death. We then determined if the differential effects of TNFα that we observed on neutrophil Mcl-1 turnover and BFL-1 expression were concentration dependent. Figure 6A shows that the accelerated turnover of Mcl-1 occurred only at concentrations of TNFα of 10 ng/mL and higher, while maximal increases in BFL-1 transcription occurred at TNFα concentrations of 0.1 and 1 ng/mL (Figure 6B).
Effects of TNFα on relative levels of Bfl-1 and Mcl-1. Neutrophils were incubated in the presence or absence of the indicated concentrations of TNFα, or cycloheximide (CHX, 10 μg/mL) for 3 hours. (A) Protein lysates were prepared and Mcl-1 levels were detected by Western blotting. A representative blot of Mcl-1 protein levels from 3 separate experiments is shown. (B) RNA samples were processed and quantitative real-time PCR was performed. Values shown are means (± SD, n = 3)
Effects of TNFα on relative levels of Bfl-1 and Mcl-1. Neutrophils were incubated in the presence or absence of the indicated concentrations of TNFα, or cycloheximide (CHX, 10 μg/mL) for 3 hours. (A) Protein lysates were prepared and Mcl-1 levels were detected by Western blotting. A representative blot of Mcl-1 protein levels from 3 separate experiments is shown. (B) RNA samples were processed and quantitative real-time PCR was performed. Values shown are means (± SD, n = 3)
Discussion
TNFα exerts numerous effects on a variety of cell types, varying dramatically from inducing cell death in some cells but conversely promoting cell survival in others. These pleiotropic effects may involve its interaction with different receptors (TNFR55 and TNFR75) and how these are coupled to intracellular signaling pathways that regulate cell function.28 Neutrophils appear somewhat unusual in that TNFα can exert these opposing effects on death or cell survival on subpopulations of cells in what appears to be a concentration-dependent process.17,18 In this report, we show, for the first time, that these opposing effects on neutrophil apoptosis are due to differential effects on elements of the antiapoptotic machinery of these cells. We propose that cell death (at concentrations of 10 ng/mL and higher) occurs by rapid, caspase-dependent turnover of the antiapoptotic protein Mcl-1, whereas cell survival occurs via TNFα-mediated activation of Bfl-1 expression, another antiapoptotic survival mechanism. Notably, when de novo synthesis is blocked (by cycloheximide), 100% of the cells become apoptotic within 2 hours of TNFα treatment. This clearly demonstrates that active gene expression is required to maintain neutrophil survival following TNFα treatment. In addition, the data presented here show, for the first time, that the population of blood neutrophils contains subpopulations of cells that differ markedly in their responsiveness to a single concentration of TNFα: at a concentration of 10 ng/mL, a subpopulation of neutrophils undergoes rapid loss of mitochondrial membrane potential (and becomes committed to apoptosis), whereas another subpopulation displays enhanced mitochondrial membrane potential that is beneficial to apoptosis delay. The reason for this altered responsiveness is unknown, but it could represent cells in the blood of different ages following their release from the bone marrow, or functional heterogeneity based upon, for example, altered expression of death receptors or altered levels of proapoptotic or antiapoptotic proteins in individual cells.
There is now much evidence demonstrating the central role of Mcl-1 in regulating neutrophil survival.2,,–5 This is an unusual antiapoptotic member of the Bcl-2 family, with some unique properties that distinguish it from other family members. Mcl-1 has a large N-terminal domain, not present in other family members, that confers many of its specialized properties.29 Motifs within this N-terminal domain are targets for posttranslational modifications and give the molecule the unique ability to be acutely up- or down-regulated within minutes of the cell receiving a particular stimulus. This region of the protein also contains a number of PEST domains, and the protein is ubiquitinated by its own E3 ligase (MULE) that targets the protein for proteasomal degradation.30 Indeed, proteasomal inhibition can prevent Mcl-1 turnover, and hence such treatment delays both constitutive and GM-CSF–delayed neutrophil apoptosis. The domain also contains 2 caspase-cleavage sites,31 and caspase-dependent cleavage of Mcl-1 is seen after triggering of apoptosis by agents such as bisindolmaleimide and sodium salicylate.10,32 Also present in this region are a number of potential phosphorylation sites. Some of these remain undefined, but phosphorylation of Thr163 stabilizes the protein, while phosphorylation of Ser159 destabilizes the protein and accelerates its rate of turnover.9,33 Kinases such as Raf, Ras, PI3K, Akt, ERK, and GSK are responsible for these modifications that alter the rate of Mcl-1 turnover and hence its ability to protect against apoptosis.
We show here that TNFα-induced degradation of Mcl-1 occurs via caspase-dependent processes as shown by the fact that the pan-caspase inhibitor Z-VAD blocks TNFα-induced turnover and protects against apoptosis. Furthermore, TNFα induced activation of caspases-3, -8, -9, and -10; this activation itself was prevented by Z-VAD. Previous reports have suggested caspase-independent mechanisms of TNFα induced neutrophil death,34,–36 based on the fact that Z-VAD may actually enhance TNFα-induced apoptosis. However, these effects of Z-VAD are strictly concentration dependent.37 The concentrations used in our studies clearly block TNFα-stimulated, caspase-mediated neutrophil apoptosis, and much higher concentrations are required to observe these unusual effects of Z-VAD.
Once we had shown that TNFα-induced neutrophil apoptosis results from the acceleration of Mcl-1 turnover, we then set out to understand the mechanism of TNFα-induced survival. Neutrophils are unusual in that they are devoid of the more widely expressed antiapoptotic molecules, Bcl-2 and Bcl-XL.5 These latter proteins are relatively stable within cells and hence not as suited to switching activity (by changes in turnover rate) as Mcl-1. This may explain why neutrophils express Mcl-1 rather than Bcl-2 and Bcl-XL: rapid changes in Mcl-1 function via posttranslational modification permit neutrophils to switch cell fate very rapidly from survival to death in response to external signals. We then sought to identify neutrophil genes whose expression was activated by TNFα, using a macroarray. To our surprise, very few genes involved in cell death and survival were regulated by TNFα in neutrophils. TNFα did not alter mRNA levels for Mcl-1 or a range of death proteins, but did stimulate a significant increase in mRNA for another antiapoptotic molecule, Bfl-1. It is well established that Bfl-1 expression is TNFα regulated via an NF-κB–dependent process and can protect against apoptosis.38,–40 Enhanced Bfl-1 expression has been reported previously in cytokine-treated neutrophils,41 but we clearly show here that Bfl-1 and Mcl-1 expression is independently regulated, strongly suggesting that these 2 antiapoptotic systems may function cooperatively to protect against neutrophil apoptosis, in response to different signaling pathways. However, unlike studies using murine cells,42 defining the role of Bfl-1 in human neutrophils continues to be frustrated by the lack of a specific antibody that can unambiguously identify the protein in Western blots.
In summary, we show that the proapoptotic effects of TNFα on human neutrophils result from stimulation of a caspase-dependent process of accelerated Mcl-1 turnover. However, TNFα can also trigger expression of the antiapoptotic Bfl-1. These effects of TNFα are very much concentration dependent: higher concentrations stimulating Mcl-1 degradation, and lower concentrations triggering Bfl-1 expression. These concentration-dependent effects will have profound implications on how TNFα may regulate neutrophil function in vivo and hence how anti-TNFα therapy may affect immune function.43 At concentrations of TNFα of 1 ng/mL or less, the major effect is antiapoptotic and so blocking its function in disease is predicted to accelerate neutrophil apoptosis and thereby help resolve inflammation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the North West Cancer Research Fund, Aintree Arthritis Research Trust, and Wyeth for financial support.
Authorship
Contribution: S.W.E. designed the study, analyzed data, and wrote the paper; A.C. performed experiments and analyzed data; and R.J.M. analyzed data and wrote paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steven W. Edwards, Biosciences Building, Crown Street, University of Liverpool, Liverpool L69 7ZB, United Kingdom; e-mail: s.w.edwards@liv.ac.uk.