Abstract
This study exploited alloreactivity of natural killer (NK) cells for augmenting the recognition of human acute myeloid leukemia (AML). To circumvent the inhibitory effect of killer immunoglobulin receptor (KIR) signaling, we generated NK-cell lines with single KIR specificities for major human leukocyte antigen (HLA) class I allotypes. We demonstrated efficient cytolysis of KIR-HLA class I–mismatched primary AML blasts even at low effector-to-target ratios. To define the impact of tumor-associated activating NKG2D-ligands (NKG2D-L), 66 AML patients at diagnosis were analyzed. NKG2D-L were selectively expressed on monoblastic cells in AML M4 and M5 yet absent or weakly expressed on myeloblastic cells in all AML subtypes. Paucity of cell-surface NKG2D-L was not the result of shedding because levels of soluble ULBP1 ligand measured in AML plasma were in the normal range. Notably, purified NKG2D-L+ monoblastic cells were more susceptible to NK-mediated killing than NKG2D-L− myeloblastic cells. Accordingly, induction of cell-surface NKG2D-L by treatment with the histone deacetylase inhibitor, valproic acid, rendered cells more sensitive to NK cytolysis. These data suggest that adoptive transfer of selected populations of alloreactive HLA class I–mismatched NK cells in combination with pharmacologic induction of NKG2D-L merits clinical evaluation as novel approaches to immunotherapy of human AML.
Introduction
Improved outcome of acute myeloid leukemia (AML) after stem-cell transplantation across the human leukocyte antigen (HLA) class I barrier highlighted a potential of natural killer (NK) cells in recognition and elimination of residual malignant cells by the graft-versus-leukemia (GVL) effect in the absence of graft-versus-host disease (GVHD).1,2 NK cells develop rapidly from transplanted progenitor cells but display numerous phenotypic abnormalities and a functional immaturity, which may limit their effectiveness in tumor rejection in vivo.3,4 Adoptive transfer of mature NK cells in the early posttransplantation period is therefore a rational approach aimed at providing the patient with the benefit of competent cytotoxic effectors.5
NK cells are innate immunity CD56+CD3− lymphocytes, the function of which is regulated by the integration of signals delivered from multiple inhibitory and activating receptors.6 The killer immunoglobulin-like receptors (KIRs), which are clonally distributed in the NK-cell repertoire, recognize allelic groups of HLA class I molecules on target cells and are dominant determinants of NK-cell function.7 NK cytotoxicity is inhibited on recognition of “self” HLA class I molecules, whereas absence or loss of HLA class I can render cells susceptible to NK-mediated lysis. In the transplant setting from haploidentical stem-cell donor, alloreactivity of NK cells is provided by the KIR-HLA class I mismatch.8 In addition to sensing the missing self HLA class I molecules, NK cells need to be stimulated through engagement of activating NK receptors by specific cell-surface ligands expressed by target cells.9,10 NKG2D is one of the best characterized receptors required for NK-mediated tumor immunosurveillance.11 In mice, NKG2D function can protect the host from tumor initiation and also eradicate existing tumors expressing the H60 and Rae-1 ligands.12,13 The human NKG2D ligands (NKG2D-L) include 2 families of proteins belonging to major histocompatibility complex class I (MIC)–related molecules, and UL-16 binding proteins (ULBP).14,15 In epithelial tissue, the surface expression of NKG2D-L is up-regulated in response to cellular stress, including heat shock, DNA damage, and stalled DNA replication, which are common in human cancer.16,17 A notion that NKG2D-L play a key role as tumor-associated ligands rendering cells susceptible to NK-mediated lysis, has encouraged pharmacologic approaches to achieve the NKG2D-L induction.18,19
Understanding the molecular interactions regulating the NK function has uncovered numerous escape mechanisms preventing recognition of leukemic blasts by NK cells. Unlike in solid tumors, which frequently down-regulate HLA class I molecules, malignant leukemic cells carry high levels of HLA class I having a protective effect.20 NKG2D-L are absent or expressed at low levels in the majority of patients with acute leukemia,21-23 thereby preventing efficient activating interactions. An additional route of tumor evasion from NK immunosurveillance is through proteolytic shedding of NKG2D-L molecules,21,24 the soluble forms of which systemically down-regulate NKG2D receptor expression and lower the ability of NK cells to recognize tumor cells.24,25 These immune escape mechanisms involving both inhibitory and activating receptor-ligand interactions may diminish the clinical GvL effect of NK cells used as cellular immunotherapy.
The aim of this study is to achieve efficient NK-mediated cytolysis of AML by integrating effects provided by the missing KIR-HLA class I interaction and by the NKG2D receptor-ligand recognition. We show that expression of NKG2D-L is dependent on the type of AML blasts and that pharmacologic up-regulation of NKG2D-L levels by the histone deacetylase (HDAC) inhibitor, valproic acid, leads to a significantly increased NK-mediated lysis of AML cells. The alloreactive effect of NK cells can be optimized by using NK-cell lines with single-KIR specificities mismatched with respect to HLA class I allotype of target tumor cells. Clinical implementation of these strategies may enhance the therapeutic impact of NK alloreactivity against human AML.
Methods
Leukemia patients and healthy donors
Patients presenting with primary AML (n = 55), secondary AML (n = 11), chronic myelomonocytic leukemia (CMML; n = 3), and healthy controls (n = 25) were included in the study. The diagnosis and definition of AML subtypes M1-M5 and CMML were based on morphologic, cytogenetic, and immunophenotypic criteria. The blast content was 59% plus or minus 3.5% (mean ± SEM). Peripheral blood samples from patients and healthy donor controls were collected in compliance with the guidelines of the Ethical Committees of the University Hospitals in Basel and Warsaw. Informed consent was obtained in accordance with the Declaration of Helsinki.
Cell lines
C1R-neo and C1R-ULBP1 transfectants21 (kind gift of A. Steinle, Eberhard-Karls University Tübingen, Tübingen, Germany) were cultured in RPMI-1640 containing 10% fetal calf serum and 1 mg/mL Geneticin (all from Invitrogen, Carlsbad, CA). The lymphoblastic cell line 721.221 and 221-B*5801 (221-Bw4), 221-Cw*0304 (221-C1), and 221-Cw*0401 (221-C2) transfectants26 (kind gift of P. Parham, Stanford University, Stanford, CA) were cultured as above, except that Geneticin was omitted.
Flow cytometry (fluorescence-activated cell sorter) and monoclonal antibodies
Fresh heparinized peripheral blood (50-200 μL) was stained with mouse monoclonal antibodies (mAbs) conjugated with fluorescein isothiocyanate (FITC), phycoerythrin (PE), peridin chlorophyll protein, or allophycocyanin (APC) followed by lysis of red blood cells with fluorescence-activated cell sorter (FACS) lysis buffer (BD Biosciences, San Jose, CA). Conjugated mAbs against human CD3, CD45, CD56, CD158b, and control mouse IgG1 (all from BD Biosciences), CD158a (Beckman Coulter, Marseille, France), and CD158e (BD Biosciences and R&D Systems, Abingdon, United Kingdom) were used. Staining with unconjugated mouse mAbs against ULBP1 (M295), ULBP2 (M311), ULBP3 (M551) (all at 10 μg/mL; kind gift of D. Cosman, Amgen, Seattle, WA), MICA/B and HLA class I (both at 10 μg/mL; BD Biosciences) was followed by staining with secondary goat α-mouse IgG-FITC (Jackson ImmunoResearch, West Grove, PA). At least 100 000 events were acquired using FACS (FACSCalibur; BD Biosciences), and analysis was performed using CellQuest Pro software (BD Biosciences). Expression level of NKG2D-L was defined as the mean fluorescence intensity ratio (MFI-R) of values obtained with specific mAbs divided by values given by secondary or control mAbs.
Purification and culture of AML cells
Mononuclear cells were isolated from peripheral blood by Ficoll-Histopaque (Sigma-Aldrich, St Louis, MO) and cryopreserved until use. AML and CMML cell subpopulations were purified by sorting according to the side scatter and CD45 staining using FACSVantage (BD Biosciences). AML cells (5 × 106/mL) were cultured for 2 days in serum-free X-Vivo 10 medium (Cambrex, Verviers, Belgium) supplemented with cytokines, bovine serum albumin, insulin, and transferin, as described,27 without and with valproic acid (VA) at 1 mM (Orfiril; Desitin, Liestal, Switzerland).
Purification and culture of NK cells
NK cells were obtained from peripheral blood mononuclear cells of a healthy donor (HLA A29/A23, B35/B44, Cw04/Cw12; KIR haplotype B) and purified by CD3+ cell depletion followed by CD56+ cell-selection with antibody-conjugated immunomagnetic microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) to a purity of more than 98%. Cells were stained with mAbs against CD158a-PE, CD158b-FITC, and CD158e-APC and single KIR-expressing NK cells were sorted using FACSVantage. Sorted cells were cultured in 24-well plates containing 2 mL of Iscove modified Dulbecco medium and 5% human AB+ serum and supplemented with IL-2 (100 U/mL; kind gift of Novartis, Basel, Switzerland) and phytohemagglutinin (1 μg/mL, Murex Biotech, Dartford, United Kingdom) in the presence of irradiated allogeneic monononuclear cells.28 After 500- to 1000-fold expansion during 14 to 21 days, the purity of NK cells was defined by staining with anti-CD56, -CD3, and respective -CD158 mAbs. NK-cell lines with the purity of more than 97% were cryopreserved.
Cellular cytotoxicity assays
The cytotoxic activity of NK-cell lines was tested by chromium release assay as described.22 Briefly, 2 × 106 target cells were labeled with 250 μCi 51Cr (Amersham, Little Chalfont, United Kingdom) in Iscove modified Dulbecco medium containing 10% fetal calf serum for 2 hours at 37°C, followed by incubation with NK cells for 4 hours at 37°C at the indicated effector-to-target (E/T) ratios. For blocking experiments, effector cells were preincubated with anti-NKG2D M585 mAb (kind gift of D. Cosman) at 10 μg/mL or mouse IgG1κ (BD Biosciences) for 1 hour at 37°C. Maximum 51Cr release was determined with target cells lysed in 0.1% Triton-X. Percentage of cytotoxicity was calculated as follows: 100 × (experimental release − spontaneous release)/(maximum release − spontaneous release). Experiments were performed in triplicate.
Analysis of soluble ULBP1
An ULBP1-specific enzyme-linked immunosorbent assay (ELISA) was developed to detect soluble ULBP1 (sULBP1) in plasma of AML patients and healthy blood donors. Plasma samples were diluted 1:1 with 50 mM Tris-HCL, pH 6.8, containing 1% Triton X-100 and incubated for 30 minutes at 37°C. Recombinant human (rh) ULBP1/Fc (R&D Systems) was used as standard. Supernatants and cell lysates prepared in 50 mM Tris-HCl, pH 8.0, containing 0.15% saponin and 1% Triton X-100,29 from C1R-ULBP1 and C1R-neo cells were used as controls. ELISA plates (Maxisorp; Nunc, Roskilde, Denmark) were coated with the capture anti-ULBP1 M295 mAb at 5 μg/mL in phosphate-buffered saline (PBS) overnight at 4°C, followed by blocking with ELISA buffer (2% bovine serum albumin, 0.2% Tween 20 in PBS) for 90 minutes at room temperature (RT) and washing. Samples were added in duplicates for 90 minutes at RT. After washing, biotinylated detection antibodies BAF1380 (R&D Systems) were added at 0.5 μg/mL in ELISA buffer for 1 hour at RT. ELISA was developed based on streptavidine alkaline phosphatase substrate system (Sigma-Aldrich), and the absorbance was measured at 405 nm with a microplate reader (Spectra MAX 190; Bucher Biotec, Basel, Switzerland). Detection range of ULBP1 was 0.2 to 25 ng/mL. For measurement of sMICA, we performed a sandwich ELISA using the capture anti-MICA M673 mAb (5 μg/mL, Amgen), the detecting biotinylated Ab BAF1300 (0.5 μg/mL, R&D Systems), and rh MICA/Fc (R&D Systems) as standard. Plasma samples were diluted in ELISA buffer. Detection range of sMICA was 0.2 to 50 ng/mL.
Size exclusion chromatography was performed using a Superose 12-column (10 × 300 mm) equilibrated in PBS containing 250 mM NaCl, with 200 μL human plasma from a healthy peripheral blood donor containing 78 ng/mL sULBP1, as determined by ULBP1 ELISA. Fractions of 0.5 mL were collected in PBS containing 250 mM NaCl and tested in duplicate by ELISA for the presence of sULPB1. To determine the molecular weight of sULBP1, the column was calibrated with a set of molecular weight standards (6.5-63 kDa, Sigma-Aldrich; and Amersham, Freiburg, Germany).
Immunoprecipitation/Western analysis of ULBP1
Fractions 21 to 25 from the size exclusion chromatography, selected according to the ULBP1 ELISA, were pooled and subjected to immunoprecipitation with anti-ULBP1 M295 mAb, or mouse IgG1κ (BD Biosciences) or anti-NKG2D M585 mAb (10 μg each) as controls. Lysates from C1R-neo and C1R-ULBP1 transfected cells (1 mg protein) were subjected to immunoprecipitation with anti-ULBP1 M295 mAb (3.5 μg). Immunoprecipitates of the chromatography fractions and C1R cells were incubated with protein A-Sepharose 4B beads (Sigma, 50 μL/sample) for 4 hours at 4°C. For Western analysis, proteins were separated on sodium dodecyl sulfate-polyacrylamide gels (12%), blotted onto nitrocellulose, and ULBP1 was detected with biotinylated anti-ULBP1 BAF1380 (R&D Systems) Ab at 0.1 μg/mL. As a positive control rh ULBP1/Fc chimeric protein (3 ng) was loaded. Western blots were revealed with streptavidin-horseradish peroxidase (R&D Systems) and chemoluminescent substrate (SuperSignal; Pierce, Rockford, IL).
Statistical analysis
GraphPad software (San Diego, CA) and Student t tests were used to analyze NKG2D-L levels in AML and cytolysis of AML cells by NK cells.
Results
NKG2D ligands are preferentially expressed by monoblastic leukemic cells in AML M4 and M5
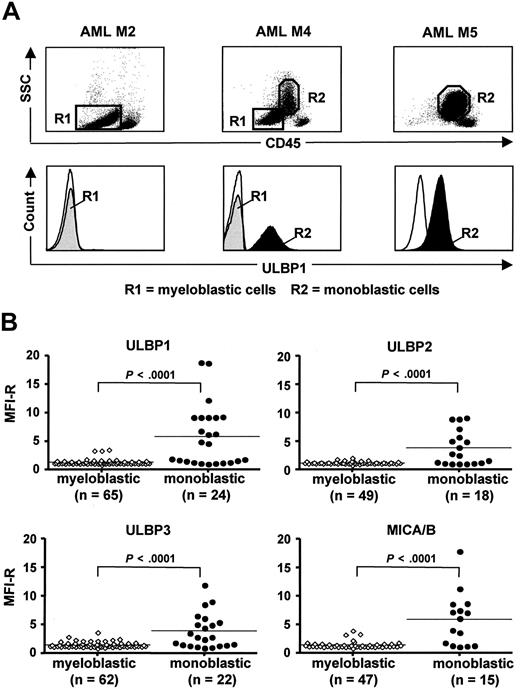
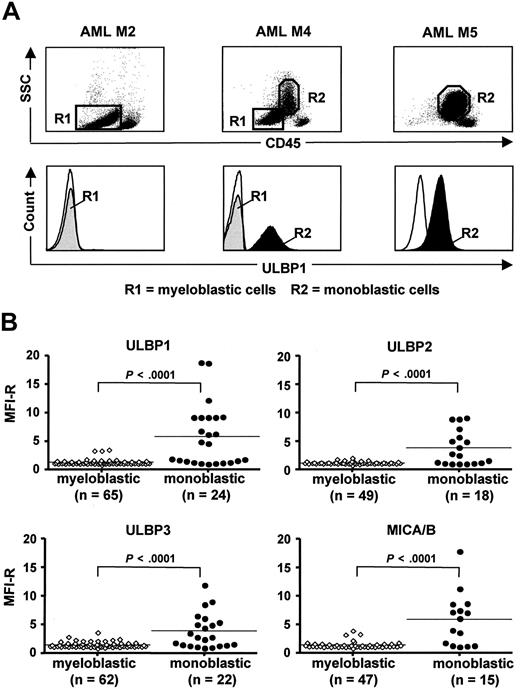
According to previous studies in AML, including our own, leukemic blasts from most patients have a NKG2D-L–negative/low phenotype, and sporadically, NKG2D-L–positive cases can be found.21-23 To elucidate the pattern of NKG2D-L expression in AML, we performed a detailed FACS analysis of cell-surface ULBP and MIC ligands in 66 patients at diagnosis (Figure 1). The myeloid blast populations, analyzed according to side scatter and CD45 expression level, were further subdivided as myeloblastic CD45-dim (R1) and monoblastic CD45-intermediate (R2), which were clearly distinct from a population of CD45-bright residual normal lymphocytes (Figure 1A top panels). In AML M1-M3, in which the myeloblastic CD45-dim cells constitute the only blast population, NKG2D-L were absent or low (MFI-R of 1.0-2.0) in a majority of cases (ULBP1-negative M2 is shown in Figure 1A). In AML M4 and M5, containing CD45-dim and CD45-intermediate blast populations in different proportions, there was a marked difference in NKG2D-L expression dependent on the type of leukemic cells: myeloblastic cells were mostly ligand-negative, similar to M1-M3, whereas monoblastic cells were frequently ligand-positive (Figure 1A bottom panels). The results on ULBP1, ULBP2, ULBP3, and MICA/B expression are summarized in Figure 1B (details on NKG2D-L expression in 66 individual AML patients are given in Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article); an additional 3 patients with CMML were included in the study because of the presence of both myeloblastic and monoblastic cells. The myeloblastic cells displayed one or more NKG2D-L in only 11 of 69 (16%) patients and only at low levels (MFI-R up to 3.9). In contrast, the monoblastic cells expressed the ligands in 17 of 24 (71%) patients and at significantly higher levels (MFI-R up to 18.8; P < .001). This pattern indicates that NKG2D-L expression is related to the subtype of myeloid leukemia and that NKG2D-L surface density is higher on monoblastic cells that have undergone oncogenic transformation at later stages of myeloid differentiation.
Expression of cell surface NKG2D-L by myeloblastic and monoblastic cells in AML and CMML. (A) FACS analysis of ULBP1 expression by AML peripheral blood cells of 3 patients with AML M2, M4, and M5. Gating of myeloblastic CD45-dim (R1) and monoblastic CD45-intermediate (R2) is shown in top panels, according to side scatter (SSC) and CD45 expression level. Histograms of ULBP1 expression by myeloblastic cells (R1, gray area) and monoblastic cells (R2, black area) are shown in bottom panels. (Thin line) Staining with secondary mAb. (B) Summary of ULBP1, ULBP2, ULBP3, and MICA/B expression levels, defined as MFI-R, by monoblastic and myeloblastic cells in all analyzed AML (n = 66) and CMML (n = 3) patients (for details on individual patients, see Table S1). Horizontal bars show the median values. (P < .001, significant difference in NKG2D-L expression levels by 2 types of leukemic blasts.)
Expression of cell surface NKG2D-L by myeloblastic and monoblastic cells in AML and CMML. (A) FACS analysis of ULBP1 expression by AML peripheral blood cells of 3 patients with AML M2, M4, and M5. Gating of myeloblastic CD45-dim (R1) and monoblastic CD45-intermediate (R2) is shown in top panels, according to side scatter (SSC) and CD45 expression level. Histograms of ULBP1 expression by myeloblastic cells (R1, gray area) and monoblastic cells (R2, black area) are shown in bottom panels. (Thin line) Staining with secondary mAb. (B) Summary of ULBP1, ULBP2, ULBP3, and MICA/B expression levels, defined as MFI-R, by monoblastic and myeloblastic cells in all analyzed AML (n = 66) and CMML (n = 3) patients (for details on individual patients, see Table S1). Horizontal bars show the median values. (P < .001, significant difference in NKG2D-L expression levels by 2 types of leukemic blasts.)
Soluble ULBP1 is less abundant than soluble MICA in AML plasma
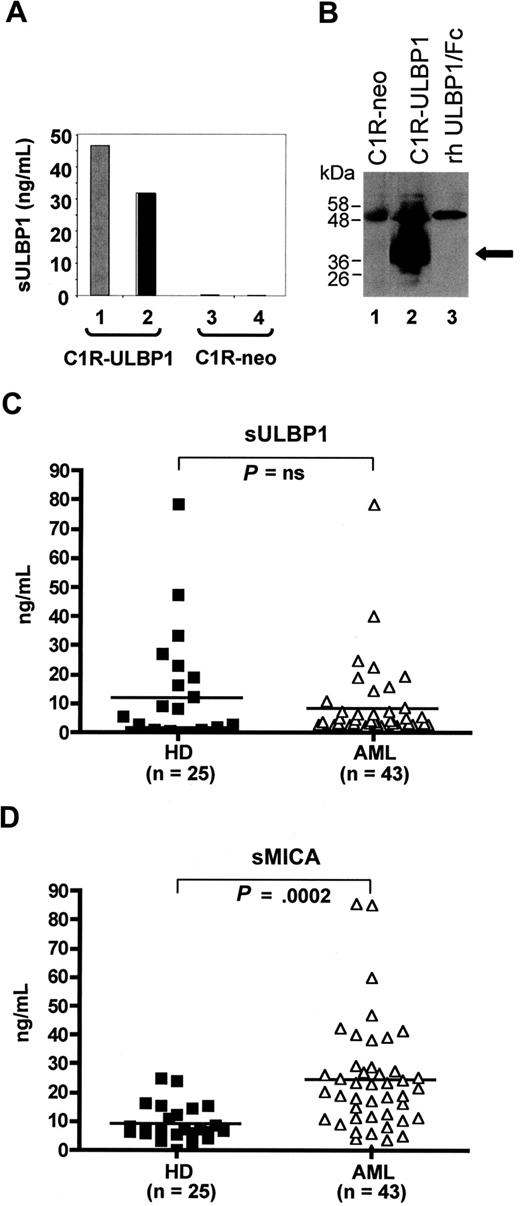
We considered the possibility that absence or low levels of membrane-bound NKG2D-L is the result of shedding from the cell surface. Proteolytic release of NKG2D-L has been described with MIC molecules, and recently with ULBP2, in various human tumors including hematopoietic malignancies.21,30,31 Here, we established the ULBP1 ELISA to detect sULBP1 in human plasma (Figure 2). The specificity of the assay was demonstrated in control experiments in which high levels of ULBP1 were found in cell lysates and culture supernatants of C1R-ULBP1 transfectant cells, exceeding 200- to 300-fold the levels of endogenous ULBP1 in mock-transfected C1R-neo cells (Figure 2A). Using ULBP1-specific mAbs, the immunoprecipitation and Western analysis of cell lysates (Figure 2B) from C1R-ULBP1 transfectants revealed a band of approximately 40 kDa, probably reflecting a full-length glycosylated ULBP1 molecule (lane 2), which was absent in C1R-neo control cells (lane 1). In human plasma, ULBP1 was found to exist as a 63 kDa complex, formed by 20 kDa portions corresponding to the ULBP1 extracellular domain (Figure S1).
Identification of soluble ULBP1 (sULBP1) in AML plasma. (A) ULBP1 ELISA was established (see “Analysis of soluble ULBP1”) and used to detect sULBP1 in C1R-ULBP1 cells (lane 1, culture supernatant gray bar; lane 2, cell lysate, black bar) but not in control C1R-neo cells (lanes 3 and 4). (B) Immunoprecipitation and Western analysis of sULBP1 in cell lysates of C1R-neo control cells (lane 1) C1R-ULBP1 cells (lane 2) and rh ULBP1/Fc (55 kDa; 3 ng, lane 3). Black arrow indicates the full-length ULBP1 molecule of approximately 40 kDa. (C,D) Levels of sULBP and sMICA in plasma from AML patients (n = 43; open symbols) and healthy donors (HD; n = 25; closed symbols), as determined by ELISA. Horizontal bars show the median values. Not significant (P = not significant) or highly significant (P < .001) difference in sULBP1 and sMICA levels in AML versus HD, respectively.
Identification of soluble ULBP1 (sULBP1) in AML plasma. (A) ULBP1 ELISA was established (see “Analysis of soluble ULBP1”) and used to detect sULBP1 in C1R-ULBP1 cells (lane 1, culture supernatant gray bar; lane 2, cell lysate, black bar) but not in control C1R-neo cells (lanes 3 and 4). (B) Immunoprecipitation and Western analysis of sULBP1 in cell lysates of C1R-neo control cells (lane 1) C1R-ULBP1 cells (lane 2) and rh ULBP1/Fc (55 kDa; 3 ng, lane 3). Black arrow indicates the full-length ULBP1 molecule of approximately 40 kDa. (C,D) Levels of sULBP and sMICA in plasma from AML patients (n = 43; open symbols) and healthy donors (HD; n = 25; closed symbols), as determined by ELISA. Horizontal bars show the median values. Not significant (P = not significant) or highly significant (P < .001) difference in sULBP1 and sMICA levels in AML versus HD, respectively.
Having established the specificity of ULBP1 ELISA, we assayed the levels of sULBP1 in plasma from AML patients (n = 43) compared with healthy donors (n = 25; Figure 2C). The average sULBP1 concentration in AML was 8.1 plus or minus 2.1 ng/mL, which was not significantly different from 11.6 plus or minus 3.7 ng/mL in control samples. In 9 of 43 AML patients, plasma levels of sULBP1 were above average values and reached 78.5 ng/mL, but similarly increased sULBP1 was found in 7 of 25 healthy donors. It should be noted that 9 samples with elevated sULBP1 included AML of different subtypes with ULBP1− and ULBP1+ cell surface phenotypes, and there was no correlation to blast count. Next, we compared levels of sULBP1 with soluble MICA (sMICA; Figure 2D). Concentrations of sMICA in AML plasma were significantly higher than in plasma of healthy donors (24.5 ± 2.9 ng/mL vs 9.3 ± 1.2 ng/mL; P < .001), in agreement with previous studies.21 In only one of 43 analyzed samples, both sULBP1 and sMICA levels were simultaneously exceeding the average values (details on sULBP1 and sMICA levels in individual AML plasma samples are given in Table S1). The lack of correlation between sULBP1 and sMICA levels suggests that different mechanisms are responsible for proteolytic cleavage of GPI-linked ULBP and transmembrane MIC ligands. Altogether, the measurement of plasma levels of sULBP1 demonstrated that ULBP1-negative/low phenotype of myeloblastic cells found in the majority of AML patients is not the result of ULBP1 shedding but represents an intrinsic property of AML blasts.
Generation of cytotoxic NK cells with single KIR-HLA class I specificity
Our next goal was to determine the relevance of cell surface NKG2D-L for recognition of AML blasts by NK cells. We took into account that AML blasts express HLA class I (see Table S1), and are therefore subject to dominant signals elicited by KIR-HLA class I interactions. To circumvent the inhibitory signaling which may mask the contribution of activating ligands, we generated NK-cell lines expressing single types of KIRs, each specific for one of the 3 major HLA class I allotypes. Healthy donor-derived purified CD56+CD3− NK cells were stained with mAbs specific for CD158a (KIR2DL1) recognizing HLA-C group 2 alleles, CD158b (KIR2DL2 and KIR2DL3) recognizing HLA-C group 1 alleles, and CD158e (KIR3DL1) specific for HLA-Bw4 alleles.32 The single KIR-expressing CD158a+, CD158b+, or CD158e+ cells, termed subsequently “single-KIR” NK cells, represented small subsets of 14%, 5%, and 2% of donor NK cells, respectively (Figure 3A). The “single-KIR” subsets were purified by FACS sorting and cultured under conditions allowing up to 1000-fold expansion within 2 to 3 weeks28 while maintaining the KIR specificity, thus generating NK-cell lines with 97% to 98% purity with respect to each type of KIR (Figure 3B). The functional specificity of these NK cells was tested against the HLA class I-negative lymphoblastoid 721.221 cell line stably transfected with genes encoding the HLA-Bw4 and HLA-C group 1 and group 2 alleles. CD158a+, CD158b+, and CD158e+ NK-cell lines were highly cytotoxic against parental 721.221 cells, but not HLA class I transfectants matching the KIR specificity (Figure 3C). Subsequently, the activity of “single-KIR” NK-cell lines was tested against HLA class I-deficient leukemic K562 cell targets and against primary AML cells (Figure 3D). Using CD158a+ and CD158e+ cells as an example, we demonstrated that K562 cell lysis was independent of KIR specificity, whereas AML blasts missing the C2 group allele were selectively killed by mismatched CD158a+ cells. These results confirmed the specificity of target cell recognition by “single-KIR” NK-cell lines.
“Single-KIR” NK-cell lines specifically detect HLA class I allotypes. (A) FACS analysis of KIR (CD158a, 158b, and CD158e) expression by healthy donor-derived NK cells stained with a single anti-CD158 mAb (x-axis) versus a mixture of 2 anti-CD158 mAb (y-axis), as indicated. Single KIR-expressing CD158a+, CD158b+, and CD158e+ NK-cell subpopulations of 14%, 5%, and 2%, respectively, are indicated. (B) FACS analysis of CD158a+, CD158b+, and CD158e+ cells after FACS sorting and expansion in culture for up to 21 days revealed a purity of single KIR-expressing NK-cell lines of more than 97%. Numbers on plots are percentages of total CD56+CD3− cells. (C) Cytotoxicity of CD158a+, CD158b+, and CD158e+ NK-cell lines against 721.221 control cells (open symbols) and 221 transfectants with HLA class I allotypes belonging to group C1, C2, and Bw4 (closed symbols) at the indicated effector-to-target (E/T) ratios. (D) Cytotoxicity of CD158a+ and CD158e+ NK-cell lines against K562 cells (filled and open circles, respectively) and against leukemic blasts from a patient with AML M2 (filled and open squares, respectively). The HLA class I allotype of the patient is indicated.
“Single-KIR” NK-cell lines specifically detect HLA class I allotypes. (A) FACS analysis of KIR (CD158a, 158b, and CD158e) expression by healthy donor-derived NK cells stained with a single anti-CD158 mAb (x-axis) versus a mixture of 2 anti-CD158 mAb (y-axis), as indicated. Single KIR-expressing CD158a+, CD158b+, and CD158e+ NK-cell subpopulations of 14%, 5%, and 2%, respectively, are indicated. (B) FACS analysis of CD158a+, CD158b+, and CD158e+ cells after FACS sorting and expansion in culture for up to 21 days revealed a purity of single KIR-expressing NK-cell lines of more than 97%. Numbers on plots are percentages of total CD56+CD3− cells. (C) Cytotoxicity of CD158a+, CD158b+, and CD158e+ NK-cell lines against 721.221 control cells (open symbols) and 221 transfectants with HLA class I allotypes belonging to group C1, C2, and Bw4 (closed symbols) at the indicated effector-to-target (E/T) ratios. (D) Cytotoxicity of CD158a+ and CD158e+ NK-cell lines against K562 cells (filled and open circles, respectively) and against leukemic blasts from a patient with AML M2 (filled and open squares, respectively). The HLA class I allotype of the patient is indicated.
“Single-KIR” NK-cell lines are highly cytotoxic against AML blasts
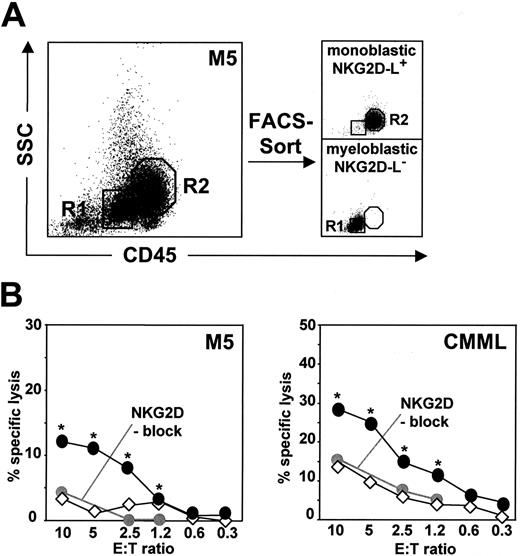
We examined the cytolytic effect of “single-KIR” NK-cell lines against leukemic blasts from 10 patients with various leukemia subtypes characterized by NKG2D-L− and NKG2D-L+ phenotypes (Figure 4). Experiments were performed under defined KIR-HLA class I interactions by selecting both matched and mismatched NK-cell effectors according to HLA class I characteristics of each analyzed patient (for HLA class I haplotypes and predicted alloreactivity of NK cells, see Table S2). At the E/T ratio of 10:1, NK cells carrying mismatched KIRs were highly cytotoxic against leukemic blasts and their effect was significantly more pronounced than cytolysis observed with matched NK cells in case of every patient (Figure 4A). Remarkably, blasts were lysed by KIR-ligand mismatched NK cells even at the low E/T ratio of 1:1 (Figure 4B), underlining a strong cytolytic potential of “single-KIR” cell lines when selected according to HLA class I allotype of each target. There were large interindividual differences in susceptibility to alloreactive NK cells, obscuring the putative differences in lysis of NKG2D-L− and NKG2D-L+ blasts (Figure 4A,B). Therefore, we purified NKG2D-L− myeloblastic cells and NKG2D-L+ monoblastic cells (Figure 5A) and exposed the 2 types of blasts from patients with AML M5 and CMML to KIR-HLA class I-mismatched NK cells (Figure 5B). The lysis of NKG2D-L+ monoblastic cells was significantly more efficient than NKG2D-L− myeloblastic cells over a wide range of E/T ratios (P < .05). This stronger killing of NKG2D-L+ monoblastic cells was reduced to the level of killing of NKG2D-L− myeloblastic cells when NKG2D blocking mAbs were included in the assay (Figure 5B), indicating that expression of cell-surface NKG2D-L is relevant for blast recognition by alloreactive NK cells.
Cytotoxicity of “single-KIR” NK-cell lines against human primary leukemic cells. Cytolysis by CD158a+, CD158b+, and CD158e+ NK-cell lines was tested against cells from 10 AML and CMML patients, at the effector-to-target (E/T) ratios of 10:1 (A) and 1:1 (B). For each patient, NK-cell effectors were selected according to KIR-HLA class I mismatch (■) and KIR-HLA class I match (□), as specified in Table S2. Error bars represent SD. (*P < .01, **P < .001, significant difference between killing by KIR-HLA class I–mismatched and – matched NK cells.)
Cytotoxicity of “single-KIR” NK-cell lines against human primary leukemic cells. Cytolysis by CD158a+, CD158b+, and CD158e+ NK-cell lines was tested against cells from 10 AML and CMML patients, at the effector-to-target (E/T) ratios of 10:1 (A) and 1:1 (B). For each patient, NK-cell effectors were selected according to KIR-HLA class I mismatch (■) and KIR-HLA class I match (□), as specified in Table S2. Error bars represent SD. (*P < .01, **P < .001, significant difference between killing by KIR-HLA class I–mismatched and – matched NK cells.)
Susceptibility of NKG2D-L− and NKG2D-L+ leukemic blasts to “single-KIR” NK-cell lines. (A) Myeloblastic CD45-dim (R1) and monoblastic CD45-intermediate (R2) cells from a patient with AML M5 were purified by FACS-sorting to obtain the NKG2D-L− and NKG2D-L+ blasts (purity > 98%). The same purification procedure was applied to blasts from a patient with CMML (not shown). (B) Specific lysis of purified AML M5 and CMML patient-derived myeloblastic cells (◇) and monoblastic cells (●) by mismatched “single-KIR” NK-cell lines. Blocking α-NKG2D mAbs were preincubated with NK effectors before the cytotoxicity assay at the indicated E/T ratios (●). (* P < .05, significant difference between cytolysis of myeloblastic and monoblastic cells.)
Susceptibility of NKG2D-L− and NKG2D-L+ leukemic blasts to “single-KIR” NK-cell lines. (A) Myeloblastic CD45-dim (R1) and monoblastic CD45-intermediate (R2) cells from a patient with AML M5 were purified by FACS-sorting to obtain the NKG2D-L− and NKG2D-L+ blasts (purity > 98%). The same purification procedure was applied to blasts from a patient with CMML (not shown). (B) Specific lysis of purified AML M5 and CMML patient-derived myeloblastic cells (◇) and monoblastic cells (●) by mismatched “single-KIR” NK-cell lines. Blocking α-NKG2D mAbs were preincubated with NK effectors before the cytotoxicity assay at the indicated E/T ratios (●). (* P < .05, significant difference between cytolysis of myeloblastic and monoblastic cells.)
Valproic acid up-regulates NKG2D-L expression on leukemic blasts and enhances sensitivity to NK-mediated killing
The HDAC inhibitor VA is an antineoplastic drug promoting the myeloid differentiation of leukemic cells.33 A previously reported induction of NKG2D-L in hepatoma cells exposed to VA34 prompted us to investigate the consequences of VA treatment for the NKG2D-dependent recognition and killing of AML cells. The leukemic blasts from AML patients were treated for 2 days with VA, and the cell-surface expression of NKG2D-L and cytolytic effect of “single-KIR” NK-cell lines were examined (Figure 6). VA up-regulated the level of ULBP1 and MICA/B significantly stronger than observed by culturing cells in growth factor-containing medium only (Figure 6A). With 13 analyzed samples (which included 12 patients with NKG2D-L− AML M0-M2, and one patient with MICA/B+ AML M4), the up-regulation of NKG2D-L in response to VA treatment was observed in case of 11 patients and reached 2.5 plus or minus 0.3-fold for ULBP1 (P < .001) and 2.9 plus or minus 0.5-fold for MICA/B (P < .05). The impact of VA-mediated increase of NKG2D-L levels was tested in cytotoxicity assays performed under defined KIR-HLA class I interactions, specific for each patient (Figure 6B). The cytolysis of AML blasts by KIR-HLA class I–mismatched cells was enhanced after treatment with VA, and in one AML case (Figure 6B first middle panel), there was even a substantial induction of killing by matched NK cells. Addition of blocking anti-NKG2D mAbs partly abolished the cytolysis, and the blocking effect was stronger with VA-treated than VA-untreated cells (Figure 6C), indicating the contribution of NKG2D–NKG2D-L interactions to VA-modulated lysis of AML cells. These data demonstrated a role of HDAC inhibitors in enhancing the recognition and cytolysis of AML blasts by alloreactive NK cells.
Increased susceptibility of leukemic blasts to “single-KIR” NK-cell lines after treatment with valproic acid (VA). (A) FACS analysis of ULBP1 and MICA/B expression levels by leukemic blasts from 6 AML patients untreated (black area), and after 2 days treatment with medium alone (black line) or VA (gray area); broken line, isotype-specific mAb staining. (B) Specific lysis of cells treated with medium alone (open symbols) and VA (filled symbols) by KIR-HLA class I–mismatched (squares) and matched NK-cell lines (circles). (*P < .05, **P < .01, significant difference between cytolysis of VA-treated and untreated AML blasts by NK cells.) (C) Reduction of specific lysis of AML cells by α-NKG2D blocking mAbs. KIR-HLA class I–mismatched NK cells were preincubated with blocking mAbs and used as effectors against AML cells cultured in medium without VA (□) and with VA (▒). The effect of α-NKG2D mAbs is the average of results obtained at 3 NK cell/target cell ratios of 10:1 (or 5:1), 2.5:1, and 1.2:1. Lysis after use of α-NKG2D mAbs is shown as  . No blocking was observed with control IgG1 Abs (not shown). Error bars represent SD. (*P < .05, **P < .01, significant difference between lysis of VA-untreated and VA-treated AML blasts in the presence of α-NKG2D mAbs.)
. No blocking was observed with control IgG1 Abs (not shown). Error bars represent SD. (*P < .05, **P < .01, significant difference between lysis of VA-untreated and VA-treated AML blasts in the presence of α-NKG2D mAbs.)
Increased susceptibility of leukemic blasts to “single-KIR” NK-cell lines after treatment with valproic acid (VA). (A) FACS analysis of ULBP1 and MICA/B expression levels by leukemic blasts from 6 AML patients untreated (black area), and after 2 days treatment with medium alone (black line) or VA (gray area); broken line, isotype-specific mAb staining. (B) Specific lysis of cells treated with medium alone (open symbols) and VA (filled symbols) by KIR-HLA class I–mismatched (squares) and matched NK-cell lines (circles). (*P < .05, **P < .01, significant difference between cytolysis of VA-treated and untreated AML blasts by NK cells.) (C) Reduction of specific lysis of AML cells by α-NKG2D blocking mAbs. KIR-HLA class I–mismatched NK cells were preincubated with blocking mAbs and used as effectors against AML cells cultured in medium without VA (□) and with VA (▒). The effect of α-NKG2D mAbs is the average of results obtained at 3 NK cell/target cell ratios of 10:1 (or 5:1), 2.5:1, and 1.2:1. Lysis after use of α-NKG2D mAbs is shown as  . No blocking was observed with control IgG1 Abs (not shown). Error bars represent SD. (*P < .05, **P < .01, significant difference between lysis of VA-untreated and VA-treated AML blasts in the presence of α-NKG2D mAbs.)
. No blocking was observed with control IgG1 Abs (not shown). Error bars represent SD. (*P < .05, **P < .01, significant difference between lysis of VA-untreated and VA-treated AML blasts in the presence of α-NKG2D mAbs.)
Discussion
Relapse is a frequent severe complication in treatment of AML.35 The clinical finding that HLA-nonidentical NK cells can exert the GvL effect and alter the outcome of bone marrow transplantation in AML opened a perspective of cellular immunotherapy with NK cells displaying alloreactivity toward the recipient's HLA type.36,37 In this work, we demonstrate that the clinical impact of NK cells against human AML may be enhanced by selecting for KIR-HLA class I-mismatched subpopulations of NK cells and by combining the cellular therapy with a pharmacologic induction of NKG2D-L to augment the recognition of leukemic blasts by NK cells.
Alloreactivity of NK cells against tumors is based on recognition of tumor-associated activating ligands in the absence of inhibitory KIR engagement with HLA class I. Typically, however, only 2% to 20% of NK cells in peripheral blood carry a single type of a potentially alloreactive KIR.1,23,38 Indeed, the size of a subset mismatched with respect to HLA class I allotype of the AML patient was shown to parallel the degree of NK-cell cytotoxicity against leukemic targets.23 The low content of alloreactive NK cells may explain the requirement of high E/T ratios of up to 100:1 to achieve a pronounced killing of primary AML in vitro.21-23,39 We demonstrated the possibility of generating NK-cell lines expressing a single type of KIR, with CD158a+, CD158b+, or CD158e+ phenotypes, which can be selectively used according to a missing HLA class I allotype on target cells. In such a mismatched setting, every tested AML sample was lysed with a significantly higher efficiency than by NK cells carrying KIRs, which matched the patient's HLA class I. Consequently, prominent cytolysis was achieved using 10:1 and even 1:1 proportion of alloreactive NK cells toward AML targets. Selection for and appropriate choice of “single-KIR” cells eliminates not only nonalloreactive NK-cell subsets, but also hyporesponsive KIR-negative cells,40 from the population of NK effectors. Use of selected competent “single-KIR” NK lines represents an alternative to the postulated use of blocking antibodies against KIRs41,42 to attenuate inhibitory KIR signals. Interestingly, in a mouse model of pulmonary tumors, the tumor load was significantly reduced by a single injection of inhibitory Ly49 ligand-mismatched, but not matched, NK cells, providing evidence for the in vivo effectiveness of alloreactive NK cells with a single inhibitory receptor–ligand incompatibility.43 Although 20% to 60% of “single-KIR” NK-cell lines expressed the inhibitory receptor NKG2A specific for HLA-E,44 elimination of NKG2A-carrying cells to further enhance the lytic potential of alloreactive cells is of no benefit because the NKG2A− phenotype is not stable and selected NKG2A− cells reacquire the receptor on culture (data not shown).
Importantly, after FACS-purification of CD158a+, CD158b+, and CD158e+“single-KIR” NK-cell subsets, we showed an efficient, up to 1000-fold, cell expansion without loss of KIR specificity or expression level in the resulting NK-cell lines. Cytokine-dependent expansion of NK cells is associated with a significant up-regulation of the cell surface expression of the activating receptor NKG2D, which plays an important role in triggering the NK cell–mediated tumor cell lysis.45-47 Accordingly, knocking-down of NKG2D in human NK cells virtually abolishes the killing of leukemic targets.48 The “single-KIR” NK-cell lines, which were cultured in the presence of IL-2, harbored high levels of NKG2D (MFI-R of approximately 100), possibly strengthening the NK cell–target cell recognition. The PHA and cytokine stimulation accompanying the ex vivo expansion is essential for effective alloreactivity because resting “single-KIR” NK cells are poor antileukemic effectors even in the presence of KIR-HLA class I mismatch (data not shown).
Ligands belonging to the ULBP and MIC families are serving as tumor-specific antigens for recognition and destruction in a NKG2D-receptor dependent process. As a newly recognized mechanism of innate immunity, induction of this ligand system represents a response to genomic DNA damage as means to eliminate the precancerous and cancer cells.49 In AML, however, the NKG2D-L+ cases were found only sporadically.21-23 By performing a detailed phenotypic analysis of specific subpopulations of AML blasts, we now show that myeloblastic CD45-dim cells are NKG2D-L−/low in all patients, whereas monoblastic CD45-intermediate cells frequently express high ligand levels. This pattern of ligand distribution indicates that expression of NKG2D-L is preferentially clustered in leukemias of the M4 and M5 subtypes, and provides a strong support to our earlier conclusion that NKG2D-L are acquired at late maturation stages in the myeloid lineage differentiation process.22 This model is reinforced by our current finding that paucity of cell surface ULBP1 was not the result of shedding. Unlike elevated sMICA in AML, sULBP1 levels in AML were high in only a minority of patients, without a clear prevalence according to AML subtype. This pattern is reminiscent of sporadically elevated soluble ULBP2 described recently in sera of leukemic patients and healthy donors.31 The susceptibility of AML to cytotoxic elicited by alloreactive NK cells was highly heterogeneous, and the extent of lysis of individual AML patient-derived blasts did not reflect the cell-surface NKG2D-L levels. This is likely the result of contribution of ligands for other important activating NK-cell receptors, such as molecules interacting with the natural cytotoxicity receptors22 and ligands for DNAM-1.23 These ligand families may also contribute to a low but consistently observed susceptibility of AML to cytolysis by KIR-HLA class I–matched NK-cell lines.
By FACS-based purification of NKG2D-L− myeloblastic cells and NKG2D-L+ monoblastic cells, we showed that NKG2D-L+ cells were significantly more susceptible to NK-mediated killing than NKG2D-L− cells, thus demonstrating the importance of cell-surface NKG2D-L for recognition and killing of leukemic targets. Consequently, we took a pharmacologic approach to achieve the NKG2D-L up-regulation on NKG2D-L−/low AML blasts. Recently, we demonstrated that differentiating drugs, including the HDAC inhibitor trichostatin A, reversing the epigenetic silencing mechanisms can up-regulate the NKG2D-L in the myeloid leukemia cell line HL-60.19 Here we used VA, reported recently as potent inducer of NKG2D ligands in hepatocytes,34 and undergoing clinical trials in myeloid malignancies.33,50 Remarkably, treatment of AML cells with VA produced a significant increase of NKG2D-L levels, resulting in enhanced susceptibility to NKG2D-mediated killing. Of particular importance is that a majority of analyzed AML samples with VA-induced changes belonged to NKG2D-L− AML M0-M2. It is also noteworthy that treatment with VA in some cases even enhanced the killing of blasts by HLA class I–matched NK cells, indicating that activating signals may override the negative KIR signaling, as described with murine NKG2D-L–overexpressing tumor targets.51 According to our preliminary data, VA treatment also has a stimulatory effect on expression level of DNAM-1 ligands on AML cells. These data indicate that VA mediates specific sensitization of malignant AML cells for immune effector mechanisms and may represent a valuable treatment combination in the setting of KIR-HLA class I incompatibility. Based on our in vitro analyses, the pharmacologic application of VA is not likely to inhibit the cytotoxic properties of NK cells but may be associated with inhibitory effect on expansion of adoptively transferred NK cells (data not shown).
Potential clinical benefits of NK cells from HLA-nonidentical donors include not only GvL reactivity but also prevention of GvHD through elimination of dendritic cells.52 Indeed, in a mouse allogeneic transplant model, adoptive transfer of alloreactive NK cells reduced the tumor load along with reducing the GvHD.43 In clinical transplantation, the KIR-HLA class I mismatches may not protect from GvHD when NK cells are derived from unrelated stem-cell donors,53 whereas they may reduce the risk of GvHD in T-cell–depleted haploidentical stem cell transplants.54 Infusions of donor-derived haploidentical NK cells have recently been initiated and proven as safe and of potential clinical benefit in some patients.55,56 Our data indicate that adoptive transfer of haploidentical donor-derived NK cells should be preceded by selection of cells alloreactive toward the recipient's HLA. Subsequent IL-2-driven ex vivo expansion of selected NK cells will yield activated effectors in high numbers to reach effective proportions to the tumor targets in vivo. Use of selected cell subsets will allow reduction of the culture volume during cell expansion and, furthermore, will avoid infusion of large quantities of nonalloreactive bystander NK cells. We propose that appropriate use of alloreactive NK cells in combination with pharmacologic induction of NKG2D-L merits clinical evaluation as novel approaches to prevent relapses of human AML with NK-cell immunotherapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank S. Sendelov for technical help, H. Köller and V. Jäggin for cell sorting, D. Klein for performing the size exclusion chromatography, F. Duong for expertise help with Western analysis, and M. Stern for helpful comments on the manuscript.
This work was supported by grants from the Oncosuisse OCS-01664-02-2005, Swiss National Science Foundation 3100-110511, Krebsliga beider Basel 9/05, Stiftung zur Krebsbekämpfung 216, and Stiftung für Hämatologische Forschung.
National Institutes of Health
Authorship
Contribution: S.D. designed and performed research, analyzed data, and edited the paper; H.H., B.D., A.M.-S., U.S., and U.L. designed and performed research; J.H. designed research and analyzed data; A.G., A.T., M.P., and W.W.-J. provided clinical data; and C.P.K. and A.W.-F. designed research, analyzed data, and wrote and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Aleksandra Wodnar-Filipowicz, Department of Research, University Hospital Basel, Hebelstrasse 20, CH-4031 Basel, Switzerland; e-mail: Aleksandra.Wodnar-Filipowicz@unibas.ch.










 . No blocking was observed with control IgG1 Abs (not shown). Error bars represent SD. (*P < .05, **P < .01, significant difference between lysis of VA-untreated and VA-treated AML blasts in the presence of α-NKG2D mAbs.)
. No blocking was observed with control IgG1 Abs (not shown). Error bars represent SD. (*P < .05, **P < .01, significant difference between lysis of VA-untreated and VA-treated AML blasts in the presence of α-NKG2D mAbs.)