In this issue of Blood, Takeda and colleagues demonstrate that genetic inactivation of murine prolyl hydroxylase domain proteins (PHDs) leads to erythrocytosis, which suggests that these enzymes repress erythropoietin (epo) and red-cell production by inactivation of the transcription factor complex hypoxia-inducible factor (HIF).
Based on microarray experiments performed in endothelial cells, it has been estimated that more than 2% of all human genes are regulated by HIF.1 Importantly, HIF is involved in regulation of the oxygen transport capacity of the blood because it stimulates epo production when oxygen delivery to the kidney and the liver is reduced. Further effects of HIF target-gene activation include promotion of angiogenesis and regulation of vascular tone. Obviously, all these features help tissues adapt to hypoxia.
HIF is a heterodimer composed of a labile α-subunit and a constitutively expressed β-subunit. Three α-subunits have been defined: HIF-1α and HIF-2α share several, though not all, regulatory mechanisms and effects. HIF-3α is not so well characterized. Oxygen sensitivity is conferred to the HIF complex by rapid degradation of HIF-α subunits in the presence of normal levels of oxygen. Two distinct proline residues undergo oxygen-dependent hydroxylation by enzymes termed prolyl hydroxylase domain proteins (PHD1, PHD2, and PHD3). The modified HIF-α subunits bind von-Hippel-Lindau protein, are polyubiquitinated, and are then degraded within minutes by the proteasome. In addition, hydroxylation of an asparagine residue in the C-terminal transactivation domain leads to inhibition of transcription cofactor binding, and thus reduces target-gene expression.
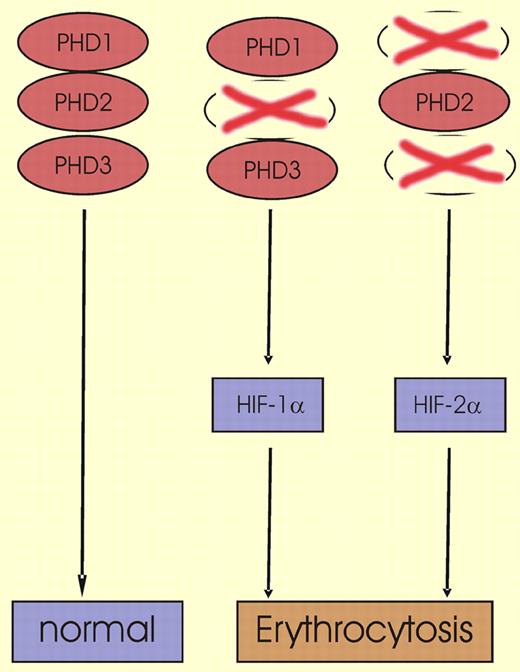
The HIF-α PHDs were identified in 2001,2,3 and although many studies have since been performed in vitro and in cell-culture systems, data on hydroxylase function in vivo have remained scarce. Recently, Takeda and coauthors reported that genetic deletion of either PHD1 or PHD3 leads to a normal phenotype in mice; in contrast, the inactivation of PHD2 is lethal between embryonic day 12.5 and 14.5 because of defective placental and cardiac development.4 The same group of investigators has now addressed the question as to whether inactivation of distinct PHDs has an impact on erythropoiesis. In adult mice, such deficiencies indeed had characteristic effects on HIF-1α, HIF-2α, and erythropoiesis (as shown in the figure). While genetic inactivation of PHD1 or PHD3 did not affect hematologic parameters, double knockout of both PHD1 and PHD3 led to moderate erythrocytosis. Remarkably, in this situation HIF-2α was induced in the liver but not in the kidney, HIF-1α was not detectable, and serum epo levels were decreased. When deletion of PHD2 was induced after birth by means of a tamoxifen-inducible Cre-recombinase system, severe erythrocytosis with hematocrit values of up to 83% occurred, and the life-span of the animals was significantly reduced. In the PHD2-deficient mice, induction of HIF-1α in kidney and liver as well as epo elevation in the serum were observed, while HIF-2α remained undetectable.
In wild-type mice, 3 prolyl hydroxylase domain proteins (PHD1, PHD2, and PHD3) that are critical for HIF-α degradation are present. HIF-α levels are kept at low levels and erythropoiesis is normal. When the animals are deficient for PHD2, HIF-1α is up-regulated and erythropoiesis is stimulated. In contrast to PHD2-deficient mice, PHD1 and PHD3 double-knockout mice show activation of HIF-2α, which also leads to erythrocytosis.
In wild-type mice, 3 prolyl hydroxylase domain proteins (PHD1, PHD2, and PHD3) that are critical for HIF-α degradation are present. HIF-α levels are kept at low levels and erythropoiesis is normal. When the animals are deficient for PHD2, HIF-1α is up-regulated and erythropoiesis is stimulated. In contrast to PHD2-deficient mice, PHD1 and PHD3 double-knockout mice show activation of HIF-2α, which also leads to erythrocytosis.
These data are well in line with recent reports that demonstrate a moderately elevated hematocrit value in a patient who is heterozygous for a PHD2 mutation,5 and that RNA interference directed against PHD2 is sufficient to induce HIF-1α in a cell-culture setting.6 Taken together, these reports suggest that PHD2 keeps HIF-1 activity at a low level in vitro and in vivo, and that the 2 remaining PHDs cannot act as a substitute for this function of PHD2. On the other hand, the report by Takeda and coauthors indicates that PHD1 and PHD3 regulate HIF-2α, which has been previously reported to promote epo production in murine liver.7 The current paper clearly adds to our understanding of the interplay between PHDs, HIFs, and epo, which collectively maintain red blood cell homeostasis.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■