Abstract
The Aiolos transcription factor, member of the Ikaros family of zinc finger proteins, plays an important role in the control of mature B lymphocyte differentiation and proliferation, and its function appears to be modulated through alternative splicing. To assess Aiolos isoform role in humans' pathologies, we studied Aiolos variant distribution and expression in mature B lymphoproliferative disorders (chronic lymphocytic leukemia [CLL] and other B-cell lymphomas). We demonstrated that more than 80% of expressed Aiolos in normal as well as in malignant B cells is of the hAio1 type, and we showed for the first time a homogeneous overexpression of the total amounts of Aiolos transcripts in the B cells of CLL patients, independently of ZAP-70 and IgVH mutational status prognosis factors. This up-regulation of Aiolos, confirmed at protein level, seems independent of Aiolos promoter H3K9 acetylation and H3K4 trimethylation.
Introduction
Mature B-cell neoplasms, including chronic lymphocytic leukemia (CLL), represent frequent malignancies of mature B cells whose cell derivation and pathogenesis are unknown.1,2 Here we focused on Aiolos, a hematopoietic chromatin remodeler and transcription factor of the Ikaros family that plays critical roles in B-cell differentiation,3-6 and we examined whether a deregulation of its expression was an integral part of malignant transformation in CLL and other B-cell lymphomas.
Methods
Subjects and B-cell isolation
Cryopreserved peripheral blood mononuclear cell (PBMC) samples were collected and informed consent obtained in accordance with the Declaration of Helsinki from CLL or other B-cell lymphoma patients followed at the Hematological Department of Pitié-Salpêtrière Hospital, according to a protocol approved by the institutional ethic committee. Diagnosis was assessed according to the recent World Health Organization classification. In all cases, blood sample collection was performed at the time of diagnosis. Fresh blood from healthy donors was collected by the Etablissement Francais du Sang (France), and PBMCs were frozen after density gradient isolation. Total RNA extraction was done after B cells isolation by anti-CD19 magnetic beads (Dynal Biotech, Oslo, Norway).
Reverse-transcribed quantitative polymerase chain reaction analysis
RNA (500 ng) was reversed transcribed and amplified, either by Titanium one-step RT-PCR (Clontech, Mountain View, CA) or by Superscript II (Invitrogen, Carlsbad, CA) and quantitative polymerase chain reaction (PCR) done using predeveloped TaqMan gene expression assays (Applied Biosystems, Foster City, CA).
Chromatin immunoprecipitation
Anti-CD19 Dynal beads were detached by DETACHaBEAD before proceeding to chromatin cross-link. Chromatin immunoprecipitation (ChIP) assays were carried out as previously described7 with primers spanning a 2-kb region upstream and downstream of the Aiolos translation initiation site.
Western blot analysis
Western blot analysis was carried out as previously described.8
Results and discussion
The hAio1 transcript represents more than 80% of the Aiolos isoforms in B cells
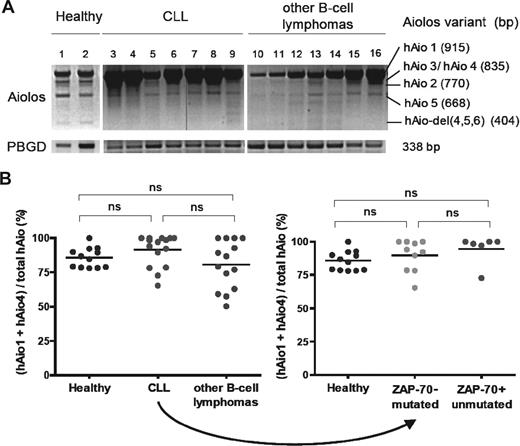
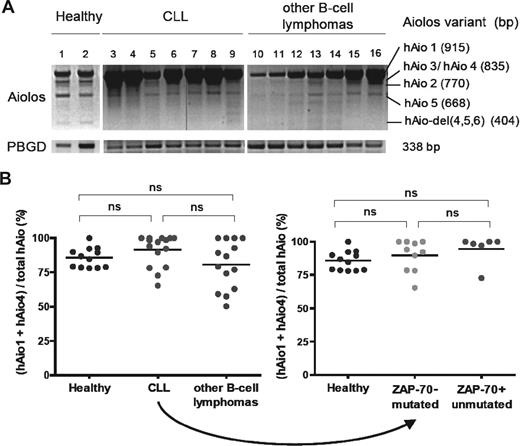
We first investigated the expression pattern of different Aiolos isoforms in PBMCs of patients diagnosed with mature B-cell malignancies (17 CLL, 8 mantle zone, and 2 marginal zone lymphomas; Figure 1A; Table 1). Analysis of healthy donors revealed the presence of 6 Aiolos variants. Five corresponded to hAio1, hAio2, hAio3/4, and hAio5 as previously described.9 A new shorter variant was observed and identified by direct sequencing of the PCR products as hAio-del(4,5,6), lacking the 4 N-terminal zinc fingers. No major differences in the distribution of Aiolos isoforms were detected between healthy donors and CLL/other B-cell lymphoma patients, except for the absence of Aio-del(4,5,6). In all cases, hAio1 was the most abundant mRNA species, although varied according to lymphocytes quantities. The shorter variants were equal or slightly decreased in patients compared with controls.
hAio1 is the most abundant Aiolos isoform in normal and malignant B cells. (A) Representative results of Aiolos isoform expression in 2 healthy subjects (1 and 2), 7 CLL (3-9), and 7 other B-cell lymphomas (10-16) patients of the set 1 after one-step RT-PCR amplification of PBMC RNA with the following primers: Aiolos forward 5′-GGCAGCGACATGGAAG-3′ (exon 1), Aiolos reverse 5′-TAGCTGATGGCGTTATTGATGG-3′ (exon 8). Porphobilinogen deaminase (PBGD) is used as internal control with the following primers: PBGD forward 5′-CTGGTAACGGCAATGCGGCT-3′, PBGD reverse 5′-GCAGATGGCTCCGATGGTGA-3′. The vertical line in band Aiolos is the result of a malfunction of the UV machine. (B) Ratio of absolute Aiolos isoform quantities obtained using reverse-transcribed quantitative polymerase chain reaction between primer pair/probe couple 1 (Applied Biosystems, Hs00232635_m1, exons 2-3 junction, hAio1-5 + hAio-del)(4,5,6) and primer pair/probe couple 2 (Applied Biosystems, Hs00918017_m1, exons 5-6 junction, hAio1 + hAio4) of each subgroup of patients and healthy subjects of the set 2 (B cells). Distinct PCR efficiencies between Aiolos TaqMan gene expression assays were corrected by absolute quantifications of known quantities of pcDNA3.1-hAio1 construct. Differences between groups were tested using one-way ANOVA and Tukey multiple comparison tests (Prism4.0c software; ns indicates nonsignificant, ie, P > .05).
hAio1 is the most abundant Aiolos isoform in normal and malignant B cells. (A) Representative results of Aiolos isoform expression in 2 healthy subjects (1 and 2), 7 CLL (3-9), and 7 other B-cell lymphomas (10-16) patients of the set 1 after one-step RT-PCR amplification of PBMC RNA with the following primers: Aiolos forward 5′-GGCAGCGACATGGAAG-3′ (exon 1), Aiolos reverse 5′-TAGCTGATGGCGTTATTGATGG-3′ (exon 8). Porphobilinogen deaminase (PBGD) is used as internal control with the following primers: PBGD forward 5′-CTGGTAACGGCAATGCGGCT-3′, PBGD reverse 5′-GCAGATGGCTCCGATGGTGA-3′. The vertical line in band Aiolos is the result of a malfunction of the UV machine. (B) Ratio of absolute Aiolos isoform quantities obtained using reverse-transcribed quantitative polymerase chain reaction between primer pair/probe couple 1 (Applied Biosystems, Hs00232635_m1, exons 2-3 junction, hAio1-5 + hAio-del)(4,5,6) and primer pair/probe couple 2 (Applied Biosystems, Hs00918017_m1, exons 5-6 junction, hAio1 + hAio4) of each subgroup of patients and healthy subjects of the set 2 (B cells). Distinct PCR efficiencies between Aiolos TaqMan gene expression assays were corrected by absolute quantifications of known quantities of pcDNA3.1-hAio1 construct. Differences between groups were tested using one-way ANOVA and Tukey multiple comparison tests (Prism4.0c software; ns indicates nonsignificant, ie, P > .05).
To confirm the predominance of hAio1 variant, we performed quantitative analysis of Aiolos isoforms on CD19-positive peripheral blood cells in a new group of 16 CLL, 1 prolymphocytic leukemia, 4 mantle zone, 6 marginal zone, 1 follicular, and 2 atypical lymphomas, and 12 healthy donors (Table 2). We calculated the ratio of the copy numbers for (hAio1 + hAio4)/total Aiolos. Although homogeneous in controls, with a mean value of 85.8%, the ratios were more fluctuating in patient samples rising from 50.4% to 100% (Figure 1B). However, mean values (91.6% and 80.7%, for CLL and other B-cell lymphomas, respectively) were not significantly different from healthy subjects (P = .083). The 2 CLL subgroups, with indolent or aggressive disease as determined by ZAP-70 expression and IgVH mutational status, showed similar Aiolos isoform distribution (89.8% and 94.7% respectively, P = .211).
In contrast to Ikaros,10-12 these results did not reveal any significant overexpression of Aiolos dominant negative isoforms in mature B-cell lymphoproliferative disorders compared with healthy donors and demonstrated the predominance of the hAio1 variant in normal and malignant B cells. A slight imbalance between dominant negative and anti-oncogenic Aiolos isoforms, as observed in some of our patients, might be involved in the pathogenesis of B-cell malignancies, by disturbing Aiolos subcellular localization and its association with histone deacetylase (HDAC)–containing complexes.6
Aiolos is up-regulated in B cells of CLL patients
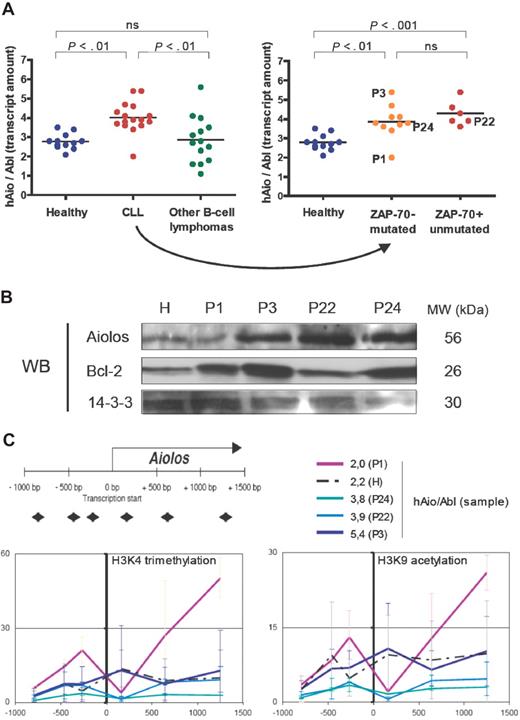
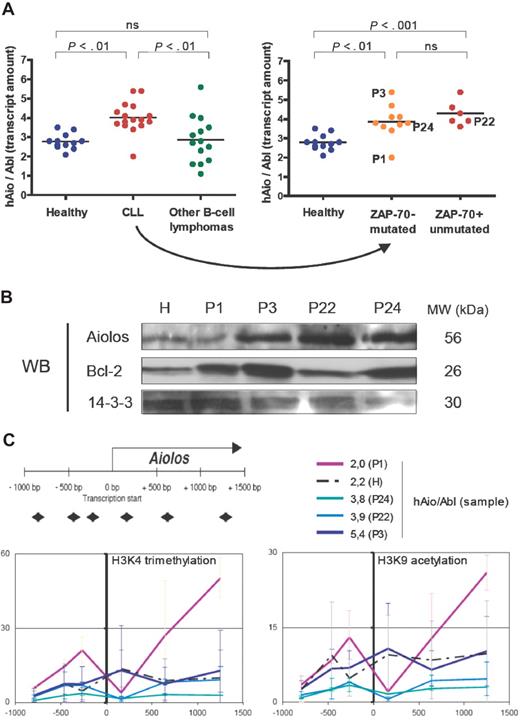
Considering the total amounts of Aiolos transcripts expressed by B cells of healthy donors and patients, we observed a homogeneous 1.5-fold increase of Aiolos transcripts in CLL samples (except for patient P1) compared with healthy donors (Figure 2A), whereas Aiolos was highly fluctuating among other B-cell lymphoma samples (ANOVA on distinct pathologies P = .001, ANOVA on distinct CLL subgroups P = .001). We analyzed Aiolos protein expression in a healthy donor (H) and 4 CLL patients showing different Aiolos expression levels: P1 (hAio/Abl = 2.0), P22 (hAio/Abl = 3.9), P24 (hAio/Abl = 3.8), and P3 (hAio/Abl = 5.4) (Figure 2C). We confirmed Aiolos up-regulation in P3, P22, and P24 (Figure 2B).
An Aiolos overexpression in CLL patients who seem independent of Aiolos promoter epigenetic modifications. (A) Total amounts of Aiolos transcripts in each subgroup of patients and healthy subjects of the set 2 (Table 2) obtained by reverse-transcribed quantitative polymerase chain reaction of B-cell RNA by the primer pair/probe couple 1 and normalized to the Abelson gene (Abl, Applied Biosystems, Hs00245445_m1). Absolute quantification of Aiolos and Abelson was done using known quantities of pcDNA3.1-hAio1 construct and Daudi total RNA, respectively. Differences between groups were tested using one-way ANOVA and Tukey multiple comparison tests (Prism4.0c software; ns indicates nonsignificant, ie, P > .05). (B) Western blot analysis of Aiolos and Bcl-2 expression in a healthy donor (H) and in the CLL patients P1, P3, P22, and P24. 14-3-3 protein is used as internal control. Molecular weights of the proteins are shown. (C) Schematic representation of the position of the primer pairs used to analyze chromatin remodeling and levels of trimethylated H3K4 (Abcam, Cambridge, MA; AB8580, 1/250) and acetylated H3K9 (Upstate Biotechnology, Charlottesville, VA; 07 352, 1/100) obtained in the CLL patients P1, P3, P22, and P24, and in one healthy donor H. The y-axis corresponds to the percentage of immunoprecipitated chromatin. The x-axis corresponds to the position in the promoter of the chromatin modification. The corresponding Aiolos transcript amounts are shown. Primers sequences were as follows: A1 forward 5′-TGGTCACTTCCCCTTTCCTCT-3′, A1 reverse 5′-TCCCAACACCCTCCCTACTG-3′; A2 forward 5′-CAGTTCCCAGAGGGAAAACAAA-3′, A2 reverse, 5′-TTGGTCCAAGTTTTCAGACAGTTG-3′; A3 forward 5′-GACAAAAAGTTCCAGATCTTCCTCA-3′, A3 reverse 5′-GTCAGCCTTGCTTTCTTGGc-3′; A4 forward 5′-GAGAGGCCGAGTAGCCACAG-3′, A4 reverse 5′-TTGCACAGGTTAAGTTTCTCAAAGA-3′; A5 forward 5′-TTGGCATTTGCAGTTCCCTT-3′, A5 reverse 5′-AACCTGATTTTTTGCGCTGG-3′; A6 forward 5′-TGGGCTGTTGTATACTATGGGAAA-3′, A6 reverse 5′-CGGCTAGGAAAAATAGTGTTGGA-3′.
An Aiolos overexpression in CLL patients who seem independent of Aiolos promoter epigenetic modifications. (A) Total amounts of Aiolos transcripts in each subgroup of patients and healthy subjects of the set 2 (Table 2) obtained by reverse-transcribed quantitative polymerase chain reaction of B-cell RNA by the primer pair/probe couple 1 and normalized to the Abelson gene (Abl, Applied Biosystems, Hs00245445_m1). Absolute quantification of Aiolos and Abelson was done using known quantities of pcDNA3.1-hAio1 construct and Daudi total RNA, respectively. Differences between groups were tested using one-way ANOVA and Tukey multiple comparison tests (Prism4.0c software; ns indicates nonsignificant, ie, P > .05). (B) Western blot analysis of Aiolos and Bcl-2 expression in a healthy donor (H) and in the CLL patients P1, P3, P22, and P24. 14-3-3 protein is used as internal control. Molecular weights of the proteins are shown. (C) Schematic representation of the position of the primer pairs used to analyze chromatin remodeling and levels of trimethylated H3K4 (Abcam, Cambridge, MA; AB8580, 1/250) and acetylated H3K9 (Upstate Biotechnology, Charlottesville, VA; 07 352, 1/100) obtained in the CLL patients P1, P3, P22, and P24, and in one healthy donor H. The y-axis corresponds to the percentage of immunoprecipitated chromatin. The x-axis corresponds to the position in the promoter of the chromatin modification. The corresponding Aiolos transcript amounts are shown. Primers sequences were as follows: A1 forward 5′-TGGTCACTTCCCCTTTCCTCT-3′, A1 reverse 5′-TCCCAACACCCTCCCTACTG-3′; A2 forward 5′-CAGTTCCCAGAGGGAAAACAAA-3′, A2 reverse, 5′-TTGGTCCAAGTTTTCAGACAGTTG-3′; A3 forward 5′-GACAAAAAGTTCCAGATCTTCCTCA-3′, A3 reverse 5′-GTCAGCCTTGCTTTCTTGGc-3′; A4 forward 5′-GAGAGGCCGAGTAGCCACAG-3′, A4 reverse 5′-TTGCACAGGTTAAGTTTCTCAAAGA-3′; A5 forward 5′-TTGGCATTTGCAGTTCCCTT-3′, A5 reverse 5′-AACCTGATTTTTTGCGCTGG-3′; A6 forward 5′-TGGGCTGTTGTATACTATGGGAAA-3′, A6 reverse 5′-CGGCTAGGAAAAATAGTGTTGGA-3′.
Consequences of this up-regulation have to be determined. Aiolos has been implicated in the direct control of the bcl-2 gene promoter activity in T cells,13 but its implication in B cells has not been demonstrated so far.14 In our patients, Bcl-2 expression seems independent of Aiolos levels (Figure 2B).15-17 Aiolos also appears to inhibit the threshold of BCR activation in part by modulating tyrosine kinase phosphorylation and calcium release to the cytoplasm.4,14 The 2 Ig-mutated and Ig-unmutated subgroups of CLL showed distinct patterns of response after BCR cross-linking,18,19 whereas we observed a homogeneous CLL profile considering Aiolos expression.
In mouse model, the opposite occurred and B-cell lymphomas were associated with Aiolos inhibition.4 However, consequences of Aiolos overexpression have not been studied so far in this model. It was recently demonstrated that hAio-1 interacts with HDAC-containing complexes and specifically associates with the promoter of SIRT1, a class III histone deacetylase thought to participate in the formation of facultative heterochromatin.6 Aiolos overexpression could therefore have a global impact on acetylation levels and contribute to epigenetic alterations in human as well as in mouse.20 Aiolos up-regulation could also be the consequence of the malignant transformation rather than its cause.
An Aiolos overexpression in CLL patients who seem independent of Aiolos promoter epigenetic modifications
To this day, Pax5,21 SLP-65,22 and Ikaros8 are the only factors described as, directly or indirectly, controlling Aiolos expression. An increase of their quantities in CLL patients may be at the origin of Aiolos up-regulation. Pax5 is differentially expressed in CLL patients with higher levels in the Ig-mutated subgroup.23 We did not observe any variation in Ikaros expression between CLL and healthy samples (data not shown). However, modifications in its subcellular localization and its ability to associate with HDAC-containing complexes may be involved.24 We have recently shown that Aiolos promoter activity is regulated by epigenetic mechanisms.25 To assess the involvement of this type of regulation in the CLL Aiolos up-regulation, we performed ChIP assays on one healthy donor (H) and the 4 CLL patients P1, P3, P22, and P24 (Figure 2C). In our experimental conditions, we observed no significant modifications of the trimethylated H3K4 and acetylated H3K9 euchromatin markers throughout the Aiolos promoter (except at position +1253 bp for P1) between CLL patients and healthy donor. These results highlight a homogeneous transcriptional activity of the Aiolos promoter in normal and malignant cells26 and suggest that other mechanisms, such as mRNA half-life stabilization, could be involved in Aiolos up-regulation. However, we cannot exclude the implication of other types of histone modifications.
In conclusion, we report for the first time an Aiolos expression deregulation in a B-cell malignancy, which does not implicate isoform imbalance but an increase in the Aiolos transcript and protein amounts. The exact causes and consequences of this up-regulation on the survival of CLL B cells and on mechanisms of their malignant transformation have to be determined.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Inserm AVENIR and ARC Funds. M.D. is supported by a predoctoral fellowship from La Ligue Contre Le Cancer.
Authorship
Contribution: M.D. and I.A. performed the experiments; H.M.-B. analyzed the data; A.R. designed the experiments and analyzed the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Angelita Rebollo, Immunologie Cellulaire et Tissulaire Inserm U543, Hôpital Pitié-Salpêtrière, Bâtiment CERVI, 83, Bd de l'Hôpital, 75013 Paris, France; e-mail: rebollo@chups.jussieu.fr.