Abstract
Platelets are an abundant source of CD40 ligand (CD154), an immunomodulatory and proinflammatory molecule implicated in the onset and progression of several inflammatory diseases, including systemic lupus erythematosus (SLE), diabetes, and cardiovascular disease. Heretofore considered largely restricted to activated T cells, we initiated studies to investigate the source and regulation of platelet-associated CD154. We found that CD154 is abundantly expressed in platelet precursor cells, megakaryocytes. We show that CD154 is expressed in primary human CD34+ and murine hematopoietic precursor cells only after cytokine-driven megakaryocyte differentiation. Furthermore, using several established megakaryocyte-like cells lines, we performed promoter analysis of the CD154 gene and found that NFAT, a calcium-dependent transcriptional regulator associated with activated T cells, mediated both differentiation-dependent and inducible megakaryocyte-specific CD154 expression. Overall, these data represent the first investigation of the regulation of a novel source of CD154 and suggests that platelet-associated CD154 can be biochemically modulated.
Introduction
CD154, also referred to as CD40 ligand (CD40L), gp39, and TRAP, is a member of the tumor necrosis factor (TNF) ligand superfamily that plays a critical role in the modulation of humoral and cellular immunity.1 CD154 is a type II membrane protein originally thought to be restricted to activated T cells.1,2 CD154 mediates immunomodulatory effects, via binding to its cognate receptor CD40, which is widely expressed on cells of the adaptive immune compartment including B cells, monocytes/macrophages, and dendritic cells.3 CD154/CD40 interaction is essential for the activation, maturation, and development of the high-affinity humoral response, as well as for enhancing antigen presentation leading to efficient T-cell function.1,2,4 The importance of the CD154/CD40 axis on adaptive immune responses is evident by the immunocompromised phenotype observed in patients with genetic CD154 deficiency.5
An emerging role for the CD154/CD40 axis is the modulation of inflammation. Elevated serum levels of a biologically active, soluble form of CD154 (sCD154) is observed in patients with a variety of chronic and acute inflammatory diseases including systemic lupus erythematosus (SLE), type I and II diabetes, glomerular nephritis, and coronary heart disease.2,6-10 Soluble CD154 is shown to trigger proinflammatory and prothrombotic activity in major vessels via ligation of CD40 in smooth muscle and endothelium.11-13 Mach and colleges showed a direct role for CD154 on atheroma development and altering phenotype.14-16 Additional studies implicate CD154 as a key modulator of other inflammatory diseases such as SLE and diabetes. Interruption of CD154 activity via immunosuppressive and antibody-based therapies have been shown to improve symptoms and slow disease progression.10,17-20 The emerging role for CD154 in vascular inflammation makes understanding the source and regulation of CD154 an important link to the design of possible anti-inflammatory therapies. While CD154 was traditionally associated with activated T cells, subsequent studies showed platelets, anucleate cells traditionally associated with hemostasis, were an abundant source of membrane-bound and soluble CD154.21,22
Because CD154 expression was believed to be restricted to T cells, the vast majority of studies have focused on understanding the transient, inducible CD154 expression in the context of T-cell activation. A consensus of CD154 regulation studies in both primary T cells and T-cell lines demonstrates a requirement of activation induced T cell–associated nuclear factor of activated T cells (NFAT) transcription factors.23-25 NFATs are a family of calcineurin-dependent transcription factors essential for the T-cell activation-induced regulation of several other cytokines imperative for T-cell growth and function, including interleukin-2 (IL-2), Fas ligand, CD154, and tumor necrosis factorα (TNFα).24-29 The central role of NFAT in the modulation of T cell–specific cytokines such as IL-2 and CD154 is the basis for the immunosuppressive activity of the calcineurin inhibitors cyclosporine A (CsA) and FK506 and has lead to the use of such compounds for the treatment of autoimmune inflammatory disorders such as SLE.17,23,30-32
Given the abundance of platelets, the discovery that platelet-associated CD154 can mediate prothrombotic and proinflammatory effects on vascular cells strongly implicates a role for platelets in onset and progression of inflammatory diseases. The present study represents the first investigation into the regulation of CD154 in platelet precursor cells, megakaryocytes. We observed that CD154 expression is restricted to the megakaryocyte lineage in nonlymphoid hematopoietic cells. Furthermore, our promoter analysis indicates that differentiation-enhanced transcription of CD154 is at least in part mediated via NFAT, transcription factors associated with activated T cells. The demonstration that CsA suppresses CD154 expression in megakaryocytes has potential clinical implications in that a novel and abundant source of CD154 can be potentially modulated.
Methods
Cell lines and reagents
K562, Meg-01, CMK, HL-60, Jurkat, and Hela cell lines were purchased from ATCC (Manassas, VA) and were routinely maintained in RPMI 1640, supplemented with 10% heat-inactivated fetal calf serum (FCS), 10 mM l-glutamine, and 100 μg/mL Pen-Strep at 37°C and 5% CO2 (complete medium). Phorbol 13-myristate (PMA), ionomycin, and CsA were all purchased from Sigma-Aldrich (St Louis, MO). Megakaryocyte differentiation of K562, CMK, and Meg-01 was induced by culturing in the presence of 10 nM PMA for 4 days was induced by culturing as previously described.33 For the transient activation studies, various cell types were stimulated with 1 μg/mL ionomycin, 1 μg/mL PMA, or in combination in the presence or absence of CsA (1 μg/mL) for 2 hours. Where described, K562 cells were also differentiated along the erythroblast lineage by culturing in the presence of 100 mM Hemin (Sigma-Aldrich) for 96 hours.
In vitro differentiation of human CD34+ and mouse LSK hematopoietic progenitor cells
CD34+ cells were obtained by magnetic bead separation from granulocyte colony-stimulating factor (G-CSF) immobilized healthy consenting donors and released for use in research under general tissue procurement institutional review board approval from the University of Iowa Blood bank. Informed consent was obtained in accordance with the Declaration of Helsinki. To induce differentiation along the megakaryocyte lineage, the CD34+ cells were cultured for 14 days in StemSpan serum-free medium supplemented with recombinant human interleukin-3 (IL-3), stem cell factor (SCF), and thrombopoietin (TPO), all purchased from Stem Cell Technologies (Vancouver, Canada) The percentage of CD41/61+ cells was determined by flow cytometry. Separation of the CD41+ and CD41− populations was performed using CD41+ magnetic bead separation (Miltenyi Biotech, Auburn, CA), according to the manufacturer's instructions. Mouse Lin−, Sca-1+, c-Kit+ (LSK) hematopoietic stem cells were isolated from C57BL/6 bone marrow using magnetic bead positive and negative selection (Miltenyi). Purified LSK cells were cultured for 12 days in Stem span medium containing recombinant TPO, interleukin-11, IL-3, and SCF (PeproTech, Rocky Hill, NJ). Primary murine megakaryocytes were isolated from mouse bone marrow and culture-derived megakaryocytes were isolated from nonmegakaryocyte cells using anti-CD41 microbeads (Miltenyi).
Transient transfections and luciferase assays
For transfection of differentiated K562, Meg-01, and CMK cells, cells were added to a 12-well plate at a density of 0.5 × 106 cells/well and differentiated with the addition of PMA. After 4 days, DNA was transfected by Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) using the manufacturer's protocol. Each transfection used 3.2 μg of reporter plasmid along with 0.5 μg of pSv-βGal as an internal standard. DIMRE C (Invitrogen) was used for the transfection of untreated cells, following the manufacturer's protocol. Twenty-four hours after transfection, the cells were lysed in 200 μL/well of Reporter Lysis buffer (Promega, Madison, WI). Luciferase activity was assessed by mixing 40 μL of extract with 100 μL of luciferase substrate (Promega) and immediately reading the sample in a Monolight 2010 luminometer (BD Biosciences, Franklin Lakes, NJ). β-galactosidase activity was determined by mixing 20 μL of extract with galactolyte β-galactosidase substrate (Tropix, Bedford, MA), incubating for 1 hour at 25°C, adding 300 μL of galactolyte accelerator (Tropix), and reading the sample in the luminometer. Luciferase activity was normalized by dividing the mean luciferase relative light units (RLU) by the mean β-galactosidase RLU.
Quantitative real-time RT-PCR
Total cellular RNA was isolated from undifferentiated and differentiated cells using the RNAeasy kit (Qiagen, Valencia, CA). First strand synthesis was performed using Superscript III reverse transcriptase (Invitrogen) in a reaction using 2 μg of total RNA primed with random hexamers following manufacturer's instructions. Two microliters of each reverse transcription reaction was subjected to real-time quantitative polymerase chain reaction (PCR) using proprietary TaqMan primer and probe sets for human and mouse CD154, human CD41, and 18s rRNA (ABI, Foster City, CA). For each sample, 3 PCRs were performed. The resulting relative increase in reporter fluorescent dye emission was monitored by the TaqMan system (GeneAmp 5700 sequence detection system and software; PerkinElmer, Norwalk, CT). The level of CD154 (or CD41) mRNA, relative to 18s rRNA, was calculated using the formula relative mRNA expression = 2 − (Ct of CD154 − Ct of 18s rRNA), where Ct is the threshold cycle value.
CD154 promoter-luciferase constructs
The −1800 bp to +65 bp (relative to the transcription start site) 5′ regulatory region of the human CD154 gene was amplified from K562 genomic DNA by PCR using Pfu high-fidelity DNA polymerase (Stratagene, La Jolla, CA) and the forward and reverse primers 3′-GTAAATTATGGTGATCGGCAGGTCAGG-5′ and 3′-GTAGAGGCTGAGGTGTCAAGGACGG-5′. The amplified fragment was A-tailed by a 15-minute, 72°C incubation reaction containing Taq polymerase and dATP. The A-tailed fragment was then T/A cloned into TOPO 2.1 PCR vector (Invitrogen). Orientation and fidelity of the fragment was confirmed by sequencing using the M13Rev and T7 primer sites flanking the multicloning site (MCS) of the TOPO 2.1 PCR vector. The fragment was then excised with HindIII and XhoI and subcloned upstream of the luciferase cDNA segment into the pGL-3 basic luciferase reporter vector (Promega). Luciferase reporter constructs containing 5′ truncations of the CD154 promoter were developed using the common downstream primer with an engineered HindIII site and XhoI containing upstream primers corresponding to the described truncations. The full-length −1800-bp construct served as the template. The XhoI/HindIII-restricted fragments were cloned into the similarly restricted pGL-3 basic vector. Reporter constructs containing site-specific mutations were generated using the PCR-based methods. The fidelity of all final vectors was confirmed by sequencing.
Electrophoretic mobility shift assay and supershift analysis
Sense and antisense oligonucleotides corresponding to distal and proximal regions of the CD154 promoter were synthesized by Integrated DNA Technologies (Coralville, IA). The double-stranded oligonucleotides were 5′-end labeled by mixing 2 pmol of double-stranded oligonucleotide with 3.7 × 105 Bq (10 μCi) of [32P]ATP (GE Healthcare, Little Chalfont, United Kingdom), 8 units of T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) in 1× polynucleotide kinase (PNK) buffer and incubating for 1 hour at 37°C. After incubation, the end-labeled double-stranded oligonucleotides were purified from free adenosine triphosphate (ATP) by passing over a Sephadex G50 column (GE Healthcare).
A total of 3 μL nuclear extract corresponding to 5 to 10 μg protein was mixed with 1 μL radiolabeled oligonucleotide (50 000 cpm) in a reaction mix containing 1× binding buffer (10 mM Tris [tris(hydroxymethyl)aminomethane], 1 mM EDTA [ethylenediaminetetraacetic acid], 1 mM dithiothreitol, 100 mM KCl, 10% [vol/vol] glycerol), and 1 μg poly(dI-dC) (GE Healthcare) as a nonspecific inhibitor in a final volume of 25 μL for 30 minutes at 25°C. The samples were resolved on a 6% polyacrylamide gel in Tris borate EDTA that was prerun for 30 minutes. For the supershift analyses, anti-NFATc, anti-NFATp, or nonimmune Ig control antibodies, purchased from Santa Cruz Biotechnology (Santa Cruz, CA), were added to the reaction mixtures during the 20 minutes' incubation prior to electrophoresis. The gel was visualized on a STORM PhosphorImager (GE Healthcare).
Microscopy, immunoprecipitation, and Western blotting
Cells and washed human platelets were lysed in RIPA buffer with freshly added protease inhibitors (Sigma), followed by a high-speed spin in remove cellular debris. The lysates were precleared for 2 hours at 4°C by the addition of protein A/G plus-agarose beads (Santa Cruz Biotechnology). Immunoprecipitation was carried out overnight using mouse anti–human CD154 monoclonal IgG (TRAP1) (Santa Cruz) and protein A/G beads at 4°C. All samples were run under reducing conditions on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) using precast 10% NUPAGE polyacrylamide gels (Invitrogen) and electrophorectically transferred to nitrocellulose membranes (BioRad, Hercules, CA). The blots were treated 2 hours with blocking buffer (5% nonfat dry milk and 0.05% Tween-20 in Tris-buffered saline [TBS]). CD154 was detected using overnight exposure to rabbit antihuman CD154 polyclonal IgG (H-215, Santa Cruz Biotechnology), followed by 1 hour of incubation with biotinylated goat antirabbit antibody (Zymed/Invitrogen), then finally with horseradish peroxidase (HRP)–conjugated strepavidin (Zymed/Invitrogen). The blots were developed using enhanced chemiluminescence (ECL) chemoluminescent detection system (GE Healthcare) and exposed to Kodak Hypermax film (Eastman Kodak, Rochester, NY) for a few seconds. Quantity of CD154 protein in the lysates of the various human cells was determined by enzyme-linked immunosorbent assay (ELISA; Bender Medsystems, Burlingame, CA). To visualize altered cellular morphology before and after megakaryocyte-specific differentiation, CMK cells were grown and treated in a 6-well tissue culture plate for the time point indicated. After treatment, the medium was aspirated and the cells were air dried and fixed before performing Giemsa stain (LeukoStat stains) according to manufacturer's instructions (Fischer Scientific, Pittsburgh, PA). All staining was performed at room temperature. The cells were visualized at 250× magnification using a VistaVision inverted microscope (VWR, West Chester, PA), and the images were acquired as JPEGs with a Canon A630 Powershot digital camera. The images were processed using Adobe Photoshop Elements 3.0 software.
Results
CD154 expression is associated with the megakaryocyte phenotype
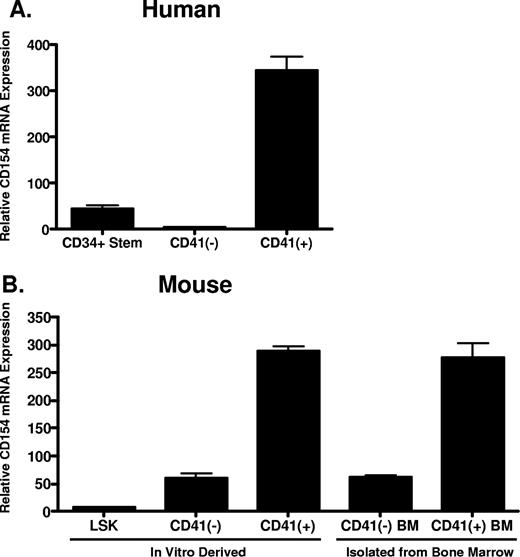
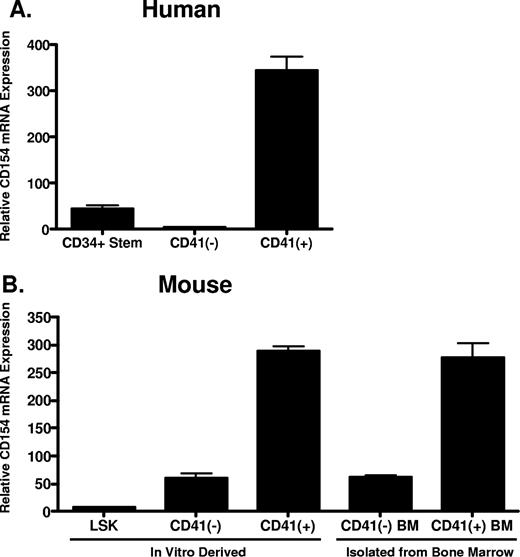
Given that platelets are anucleate and possess little capacity for de novo protein synthesis, we hypothesized that platelets acquire CD154 from megakaryocytes during thrombogenesis. Reinforcing this hypothesis is the previous observation that platelets express full-length, membrane-bound CD154, making uptake and incorporation into platelet secretory granules from serum unlikely.21,22 To address this hypothesis, we first assessed CD154 expression in primary murine megakaryocytes and both human and murine hematopoietic stem cells before and after cytokine-driven in vitro megakaryocyte differentiation. Human CD34+ hematopoietic progenitor cells were differentiated in vitro to mature megakaryocytes using culture conditions described in “In vitro differentiation of human CD34+ and mouse LSK hematopoietic progenitor cells.” After 14 days, approximately 70% of the differentiated cells expressed CD41, a cell surface integrin whose expression is restricted to megakaryocytes (data not shown). Following culture, the CD41+ cells were isolated from the non–CD41-expressing cells and levels of CD154 mRNA in the original CD34+ stem cells, and the differentiated CD41+ and CD41− cells were compared by quantitative real-time RT-PCR. As shown in Figure 1A, CD154 mRNA expression is enhanced approximately 300-fold in human hematopoietic stem cells following in vitro megakaryocytic differentiation. We also assessed CD154 expression in primary murine megakaryocytes (CD41 + BM) and in vitro megakaryocyte differentiated hematopoietic progenitor LSK cells (Lin−, Sca-1+, and c-Kit+), isolated from mouse bone marrow. Figure 1B shows that, similar to human megakaryocytes; high-level CD154 mRNA expression correlates with the CD41+ cell population. Moreover, only low-level CD154 expression is observed in the CD41-depleted differentiated cells. These data indicate CD154 is constitutively expressed in primary mouse megakaryocytes and appears to be associated with culture-derived differentiated cells bearing the megakaryocyte phenotype.
CD154 mRNA expression in primary hematopoietic progenitor cells and megakaryocytes. (A) Relative CD154 mRNA levels in primary human CD34+ hematopoietic progenitor cells, the CD41-negative and CD41-positive cell populations after in vitro differentiation as described in “In vitro differentiation of human CD34+ and mouse LSK hematopoietic progenitor cells.” (B) Relative CD154 mRNA expression in mouse hematopoietic stem cells (LSK), the CD41-positive and -negative cell populations after in vitro differentiation, and primary CD41-positive and -negative cells isolated from mouse bone marrow. The bars represent means plus or minus the standard error from 3 independent experiments.
CD154 mRNA expression in primary hematopoietic progenitor cells and megakaryocytes. (A) Relative CD154 mRNA levels in primary human CD34+ hematopoietic progenitor cells, the CD41-negative and CD41-positive cell populations after in vitro differentiation as described in “In vitro differentiation of human CD34+ and mouse LSK hematopoietic progenitor cells.” (B) Relative CD154 mRNA expression in mouse hematopoietic stem cells (LSK), the CD41-positive and -negative cell populations after in vitro differentiation, and primary CD41-positive and -negative cells isolated from mouse bone marrow. The bars represent means plus or minus the standard error from 3 independent experiments.
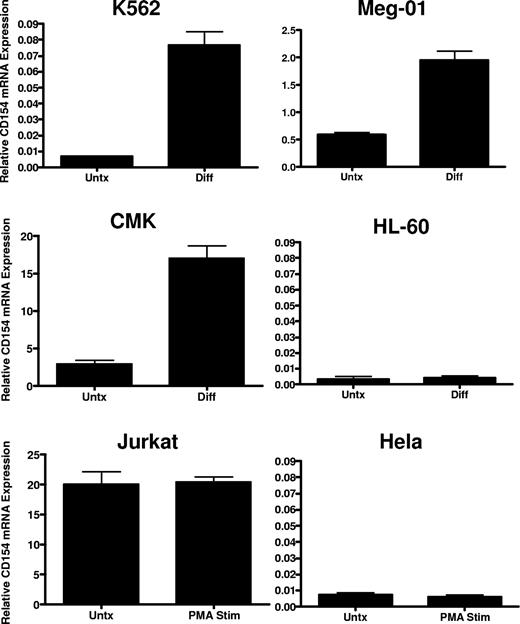
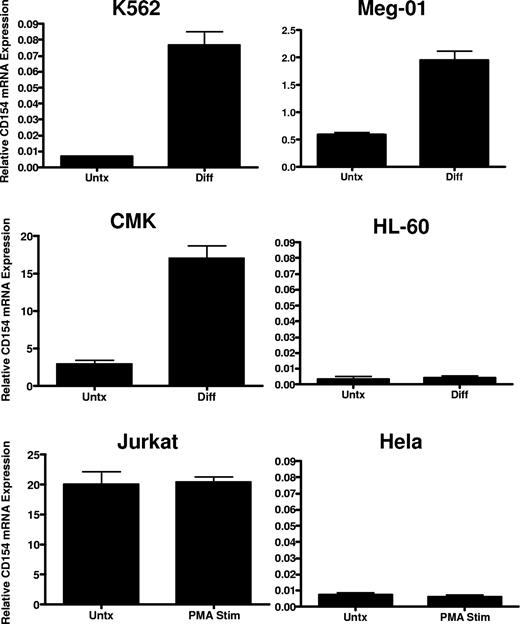
To verify lineage-specific expression and to better characterize regulation in megakaryocytes, we assessed CD154 mRNA expression in a panel of human leukemia cell lines possessing the capacity to undergo megakaryocyte-specific differentiation via chronic protein kinase C (PKC) activation. Cell lines displaying bipotential, myeloblastic phenotype (K562), premegakaryoblastic phenotype (Meg-01), and early megakaryocyte phenotype (CMK) were assessed for CD154 mRNA and protein expression before and after induction of megakaryocyte differentiation using 4-day exposure in the presence of 10−8 M PMA. As a control for nonspecific, differentiation-mediated expression, CD154 expression was also assessed in HL-60 before and after macrophage differentiation with PMA, and in Jurkat thymoma cells before and after 6 hours of stimulation with PMA. As a negative control, nonhematopoietic lineage expression of CD154 was also assessed in the epithelial Hela cell line before and after PMA exposure. Figure 2 shows variation in lineage-specific basal expression among the cell lines with relatively high-level expression observed in the Jurkat, Meg-01, and CMK lines cells and undetectable CD154 mRNA levels in the K562, Hela, and HL-60 cell lines before differentiation. CD154 mRNA is enhanced in K562, Meg-01, and CMK lines, but not in the HL-60, Jurkat, or Hela lines after PMA exposure. Furthermore, we observe that CD154 mRNA expression is enhanced approximately 10- to 20-fold after maturation to a mature megakaryocyte phenotype (Meg-01 and CMK). In contrast, CD154 mRNA, nearly undetectable in undifferentiated K562, is increased 2- to 3-fold after transition from early myeloid to an early megakaryocyte phenotype. Finally, no increase in CD154 mRNA was observed in the macrophage-differentiated HL-60 cells or the PMA-activated Hela cells. To verify that increased mRNA levels correlate with increased CD154 protein expression and to assess relative CD154 expression in all of the cell lines, ELISA was used to quantify the expression of CD154 protein before and after differentiation. To eliminate potential cell line–associated artifacts, CD154 protein was also assessed in culture-derived primary human megakaryocytes (CD41+). The data in Table 1 show that in agreement with the observed mRNA levels, primary CD41+ cells abundantly express CD154 protein at levels near the activated Jurkat T cells. Both undifferentiated Meg-01 and CMK cells express relatively high basal levels of CD154 protein, which further increase after differentiation. In contrast, CD154 protein was detected in K562 only after megakaryocyte differentiation. As expected, CD154 protein expression in differentiated HL-60 and Hela remains below the limits of detection.
CD154 mRNA expression in differentiated hematopoietic cell lines. Relative CD154 mRNA levels in the indicated cell lines before and after 4-day PMA exposure in the case of K562, Meg-01, CMK, and HL-60, and before (Untx) and after (Diff) 6 hours of PMA exposure in Jurkat and Hela cells. The bars represent mean plus of minus the standard error from 3 independent experiments.
CD154 mRNA expression in differentiated hematopoietic cell lines. Relative CD154 mRNA levels in the indicated cell lines before and after 4-day PMA exposure in the case of K562, Meg-01, CMK, and HL-60, and before (Untx) and after (Diff) 6 hours of PMA exposure in Jurkat and Hela cells. The bars represent mean plus of minus the standard error from 3 independent experiments.
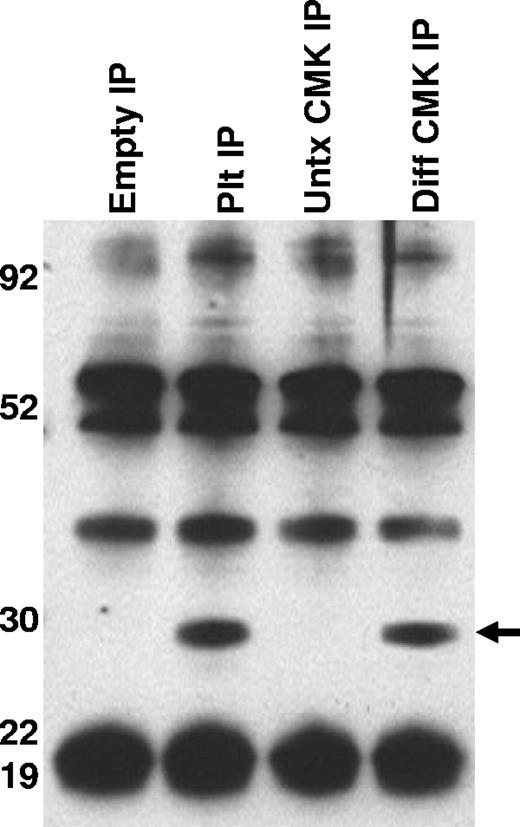
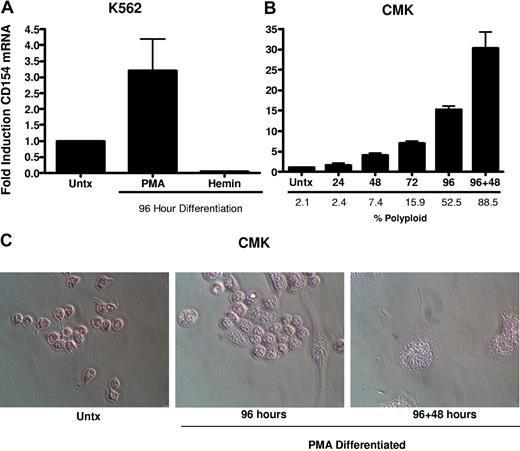
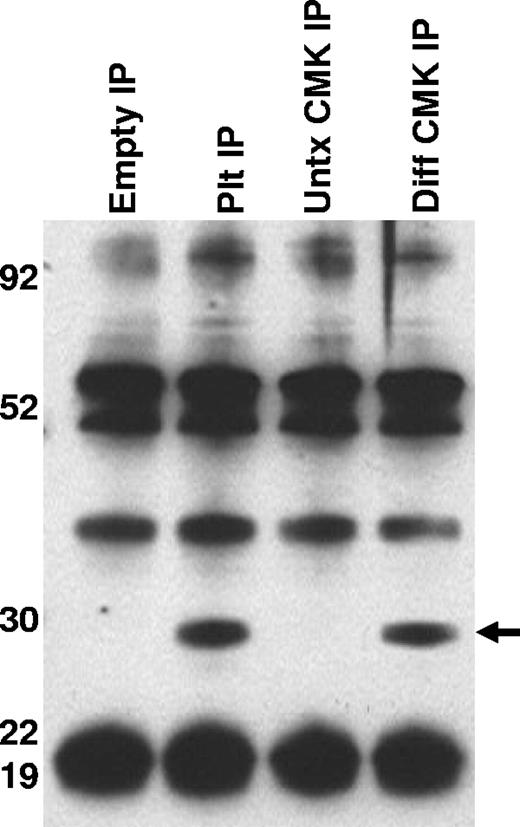
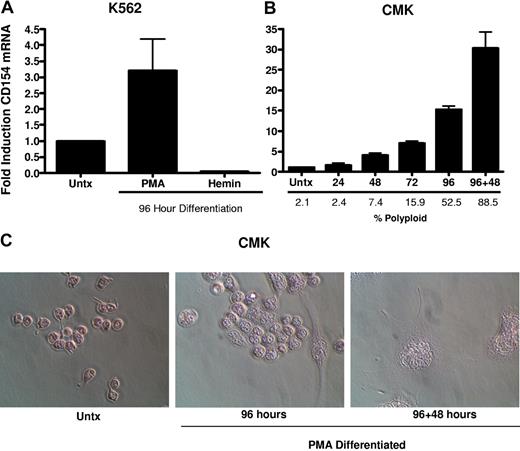
To better characterize the CD154 protein expressed in platelets and differentiated megakaryocytes, CD154 protein was immunoprecipitated from washed human platelets, undifferentiated and PMA-differentiated CMK cells, then analyzed by Western blotting. Figure 3 shows that both platelets and the differentiated CMK cells express a similar 27-kDa protein. Taken together, these data indicate megakaryocyte-restricted expression in the nonlymphoid hematopoietic cells and suggests CD154 expression correlates with differentiation stage of the megakaryocytes. To confirm that CD154 expression is megakaryocyte specific, we exploited the ability of K562 to differentiate to an erythroblast lineage via exposure to hemin (100 mM) or to megakaryocytic differentiation via exposure to PMA. The data in Figure 4A indicate CD154 expression is suppressed after erythroblastic-specific differentiation. While the PKC agonist PMA is well established as an inducer of megakaryocyte differentiation in some myeloid and premegakaryocyte cell lines, it is evident that the observed CD154 gene induction could be an artifact of a nonspecific inducer. For this reason, we wanted to determine expression kinetics and to distinguish between differentiation-driven versus PMA-driven expression of CD154. For this, CMK cells were assessed for CD154 mRNA expression at various time points after PMA exposure and then at 48 hours after removal of PMA stimulation. As a control for mature megakaryocyte differentiation, cells were monitored for polyploidy by observation by light microscopy and counting of multinucleated cells per given field. Figure 4B shows that CD154 mRNA levels gradually increase over 96 hours, corresponding with more cells displaying a mature megakaryocyte phenotype. Moreover, both percent of polyploid cells and CD154 mRNA levels continued to increase 48 hours after removal of PMA. Figure 4C shows the alterations in CMK cellular phenotype following 96 hours of PMA treatment and an additional 48-hour incubation in the absence of PMA after 96 hours of PMA stimulation. The delayed expression kinetics of CD154 mRNA coupled with the enhanced gene expression after removal of the stimulus suggests that CD154 expression correlates with the megakaryocyte phenotype. Overall, these data, when considered with our previous observations in primary cells, appear to establish CD154 as part of the megakaryocyte gene program.
Characterization of CD154 expressed in platelets and megakaryocytes. CD154 protein was immunoprecipitated from lysates prepared from washed human platelets, untreated and PMA-differentiated CMK cells. Also included is the immunoprecipitate from the lysis buffer alone (Empty IP). The relative molecular weight is noted on the left side of the image. One representative experiment of 2 performed is shown.
Characterization of CD154 expressed in platelets and megakaryocytes. CD154 protein was immunoprecipitated from lysates prepared from washed human platelets, untreated and PMA-differentiated CMK cells. Also included is the immunoprecipitate from the lysis buffer alone (Empty IP). The relative molecular weight is noted on the left side of the image. One representative experiment of 2 performed is shown.
CD154 expression is differentiation dependent and restricted to the megakaryocyte phenotype. (A) Relative CD154 mRNA expression in K562 cells driven to megakaryocyte or erythrocyte differentiation with PMA or hemin, respectively. (B) Kinetics of CD154 mRNA expression in PMA-differentiated CMK cells. After 96 hours of PMA exposure, the CMK cells were harvested, washed, and replated in fresh medium in the absence of PMA and allowed to incubate an additional 48 hours (96 + 48). The bars represent means plus or minus the standard error from 3 independent experiments. (C) Micrographs of CMK cells after the described treatment. Original magnification 250× for all panels.
CD154 expression is differentiation dependent and restricted to the megakaryocyte phenotype. (A) Relative CD154 mRNA expression in K562 cells driven to megakaryocyte or erythrocyte differentiation with PMA or hemin, respectively. (B) Kinetics of CD154 mRNA expression in PMA-differentiated CMK cells. After 96 hours of PMA exposure, the CMK cells were harvested, washed, and replated in fresh medium in the absence of PMA and allowed to incubate an additional 48 hours (96 + 48). The bars represent means plus or minus the standard error from 3 independent experiments. (C) Micrographs of CMK cells after the described treatment. Original magnification 250× for all panels.
Mechanisms of megakaryocyte-specific CD154 regulation
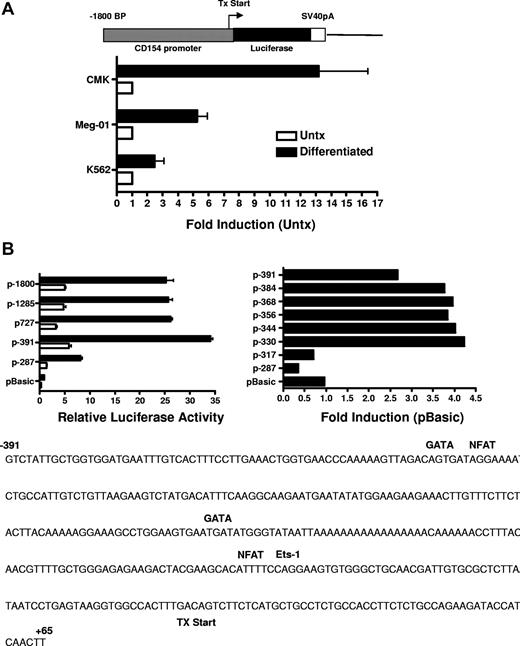
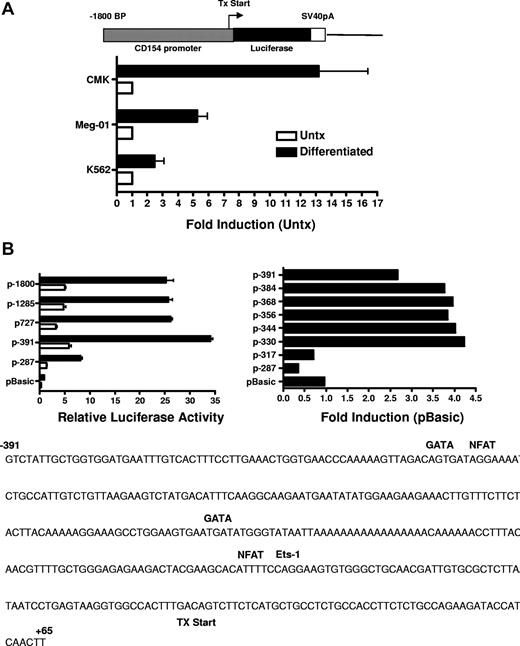
Previous studies investigating CD154 expression in T cells demonstrated CD154 is rapidly up-regulated (within 4 hours) following T-cell receptor (TCR) engagement.34 Transient transfection studies using CD154 promoter constructs indicate TCR signaling promotes a rapid increase in promoter activity and that NFAT and NFκB are important mediators of promoter activity following T-cell activation. Based on our previous studies showing enhanced steady-state levels of CD154 mRNA after several days of stimulation, we next ascertained if CD154 promoter activity is enhanced in megakaryocytes. For these experiments, the 5′ regulatory region of CD154, −1800 to +65 bp relative to the transcription start site was isolated and cloned into the pGL3 basic luciferase reporter vector. The resulting vector (p1800CD154-luc) was transfected into untreated and 96 hours of PMA-differentiated CMK, Meg-01, and K562 cells. Figure 5A shows enhanced transcriptional activity of the CD154 promoter in the MK-differentiated cells. These data show enhanced transcriptional activity of the CD154 promoter in the megakaryocytes.
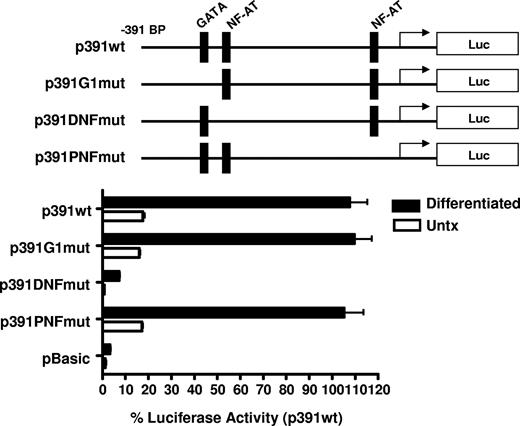
CD154 promoter activity in untreated and PMA-differentiated cell lines. (A) Untreated and 4-day PMA-differentiated CMK, Meg-01, and K562 cells were transfected with the promoter construct shown. Twenty-four hours later, luciferase activity was assessed. Fold induction of luciferase activity in the differentiated cells relative to the untreated cells (Untx) is shown. The bars represent means plus or minus the standard error from 4 independent experiments. (B) Relative luciferase activity of 4-day PMA-differentiated CMK cells transfected with the CD154 promoter-reporter constructs shown. The −391 to +65 nt region of the CD154 with putative transcription factor binding sites is shown. The bars represent means plus or minus the standard error from 4 independent experiments. The sequence of the −391 nt 5′ flanking region with location of the consensus transcription factor binding sites as predicted by Matinspector. The consensus core sequences for GATA-1, NFAT, and Ets-1 are as follows: GATA-1 (GATA), NFAT (TTTTCC) and Ets-1 (GGAA)
CD154 promoter activity in untreated and PMA-differentiated cell lines. (A) Untreated and 4-day PMA-differentiated CMK, Meg-01, and K562 cells were transfected with the promoter construct shown. Twenty-four hours later, luciferase activity was assessed. Fold induction of luciferase activity in the differentiated cells relative to the untreated cells (Untx) is shown. The bars represent means plus or minus the standard error from 4 independent experiments. (B) Relative luciferase activity of 4-day PMA-differentiated CMK cells transfected with the CD154 promoter-reporter constructs shown. The −391 to +65 nt region of the CD154 with putative transcription factor binding sites is shown. The bars represent means plus or minus the standard error from 4 independent experiments. The sequence of the −391 nt 5′ flanking region with location of the consensus transcription factor binding sites as predicted by Matinspector. The consensus core sequences for GATA-1, NFAT, and Ets-1 are as follows: GATA-1 (GATA), NFAT (TTTTCC) and Ets-1 (GGAA)
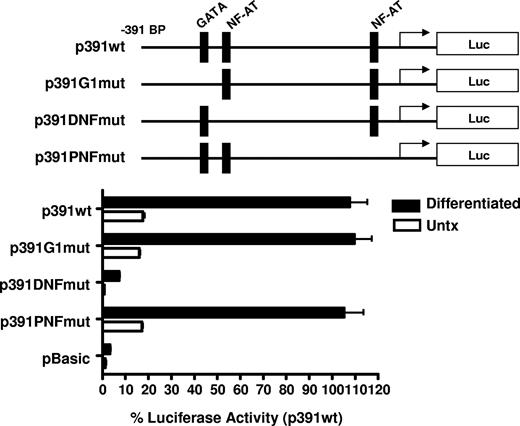
To elucidate the mechanism of increased transcriptional activity, the amount of the 5′ flanking region required for megakaryocyte-associated transcription was determined using 5′ flank deletion constructs. Figure 5B shows transfection of the differentiated CMK cells with a truncation construct consisting of the −391-bp proximal flanking region yielded similar promoter activity of the full-length −1800-bp construct. Further truncation of the 5′ flank to −287 bp effectively abrogated promoter activity, indicating the −390 to −287-bp region is sufficient for megakaryocyte-associated promoter activity. To better resolve the regulatory region required for megakaryocyte-specific regulation, an additional set of 5′ flanking truncation constructs were prepared consisting of approximately 20-bp deletions between the −391- to −290-bp region. We demonstrate transcriptional activity in MK-differentiated cells is diminished by deletion of 20 bp between −331 and −317 bp. This deletion corresponds with the loss of consensus GATA-1 and NFAT elements as revealed by a motif search of the TRANSFAC database (Matinspector). To functionally assess the GATA and the distal and proximal NFAT elements on megakaryocyte-specific CD154 transcription, mutational analysis was performed by generating −391-bp reporter constructs with point mutations to the GATA site or the NFAT sites. The data in Figure 6 show that mutational deletion of the distal NFAT site but not the GATA-1 element nor the proximal NFAT site diminished transcriptional activity. These data indicate that unlike T cell–specific CD154 expression, only the distal NFAT element is required for megakaryocyte-specific promoter activity.
Megakaryocyte-specific promoter activity is dependent on the distal NFAT site. Untreated and 4-day PMA-differentiated CMK cells were transfected with the reporter constructs containing mutations to the putative transcription factor binding sites shown (represented by the schematics). Twenty-four hours after transfection, luciferase activity was assessed and expressed as percent of activity relative to the −391-bp wild-type construct. The bars represent means plus or minus the standard error from 3 independent experiments.
Megakaryocyte-specific promoter activity is dependent on the distal NFAT site. Untreated and 4-day PMA-differentiated CMK cells were transfected with the reporter constructs containing mutations to the putative transcription factor binding sites shown (represented by the schematics). Twenty-four hours after transfection, luciferase activity was assessed and expressed as percent of activity relative to the −391-bp wild-type construct. The bars represent means plus or minus the standard error from 3 independent experiments.
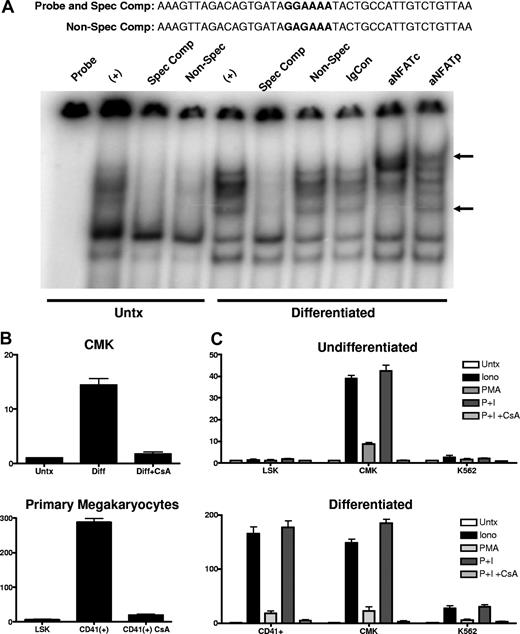
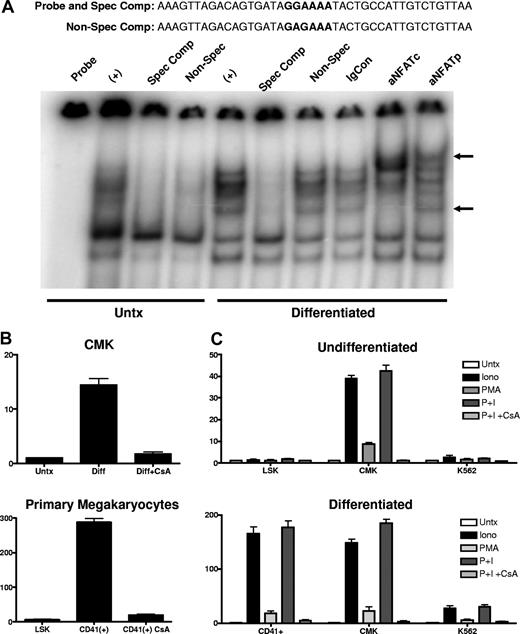
To verify NFAT protein occupancy of the distal NFAT site, EMSAs were used to examine the in vitro binding of nuclear protein, isolated from untreated and differentiated CMK cells, to an oligonucleotide probe containing the distal NFAT and the 20 flanking nucleotides. Figure 7A shows differentiation-independent, constitutive binding of a multiprotein complex to the oligonucleotide probe containing the distal NFAT site. The binding of the complex in the reaction containing the nuclear extract from untreated CMK cells was susceptible to competition by the inclusion of a 50-fold molar excess of the unlabeled specific probe (spec comp) as well as the unlabeled probe with a point mutation in the NFAT site (nonspec comp), suggesting nonspecific or low-affinity binding. In contrast, the complex bound to the probe using extracts from the differentiated CMK cells were susceptible to competition by the unlabeled specific probe but not to the probe containing the mutation to the NFAT site, suggesting specific, high-affinity interaction with the NFAT site. Verification of NFAT family binding was demonstrated by showing a “supershifted” band by the inclusion of antibodies to human NFATp and NFATc, but not nonimmune serum (IgCon).
NFAT protein occupies the distal NFAT site, and calcium mobilization is sufficient to induce endogenous CD154 expression. (A) Nuclear extracts prepared from untreated and PMA differentiated CMK cells were incubated with an oligonucleotide probe representing the −340 to −300 nt region of the CD154 promoter (the distal NFAT site in bold). Specificity of the protein complex formation was demonstrated by inclusion of cold oligonucleotides in the binding reaction. Specific competitor (spec com) contained a 50-fold molar excess of the identical probe. The nonspecific competitor reaction contains 50-fold molar excess of the identical probe with 2 point mutations in the distal NF-AT site (GGAAA to GAGAA). Supershifting is observed in reactions containing anti-NFATc and anti-NFATp but not immunoglobulin control (IgCon). (B) Relative CD154 mRNA expression in PMA-differentiated CMK cells and primary murine megakaryocytes (CD41+) derived from 12-day cytokine-differentiated LSK cells in the presence or absence of CsA. (C) CD154 mRNA expression in undifferentiated and differentiated mouse primary cells (LSK and CD41+), CMK, and K562 cells left untreated or treated with PMA, ionomycin, or both (or both PMA and ionomycin in the presence of CsA) for 2 hours. The bars represent means plus or minus the standard error from 2 independent experiments.
NFAT protein occupies the distal NFAT site, and calcium mobilization is sufficient to induce endogenous CD154 expression. (A) Nuclear extracts prepared from untreated and PMA differentiated CMK cells were incubated with an oligonucleotide probe representing the −340 to −300 nt region of the CD154 promoter (the distal NFAT site in bold). Specificity of the protein complex formation was demonstrated by inclusion of cold oligonucleotides in the binding reaction. Specific competitor (spec com) contained a 50-fold molar excess of the identical probe. The nonspecific competitor reaction contains 50-fold molar excess of the identical probe with 2 point mutations in the distal NF-AT site (GGAAA to GAGAA). Supershifting is observed in reactions containing anti-NFATc and anti-NFATp but not immunoglobulin control (IgCon). (B) Relative CD154 mRNA expression in PMA-differentiated CMK cells and primary murine megakaryocytes (CD41+) derived from 12-day cytokine-differentiated LSK cells in the presence or absence of CsA. (C) CD154 mRNA expression in undifferentiated and differentiated mouse primary cells (LSK and CD41+), CMK, and K562 cells left untreated or treated with PMA, ionomycin, or both (or both PMA and ionomycin in the presence of CsA) for 2 hours. The bars represent means plus or minus the standard error from 2 independent experiments.
The apparent role of NFAT prompted us to assess if CD154 expression was calcineurin dependent. For this, CMK cells were differentiated in the presence or absence of the calcineurin inhibitor CsA, then assessed for CD154 expression by RT-PCR. To control for nonspecific toxic effects of prolonged CsA exposure, we also assessed viability, percent of polyploidy, and alteration in CD41 mRNA levels. The data in Figure 7B shows that CsA suppressed CD154 expression in PMA-differentiated CMK cells. No alteration in cell viability, levels of CD41, or numbers of polyploid cells were observed (data not shown). To confirm similar effects in primary megakaryocytes, LSK cells were differentiated to the megakaryocyte lineage over 12 days, then cultured in the absence or presence of CsA for 48 hours. Similar to CMK cells, CsA also suppresses endogenous CD154 expression in primary murine megakaryocytes. The current studies confirm a role for NFAT in differentiation-dependent CD154 expression. We next wanted to assess the effect of transient NFAT activation on CD154 expression before and after megakaryocyte differentiation. CD154 mRNA expression was assessed in primary murine stem cells (LSK), undifferentiated CMK, and K562 cells after 2 hours of exposure to ionomycin, PMA, both, or both in the presence of CsA. Likewise, CD154 expression was also assessed in CD41+ megakaryocytes derived from LSK cells, as well as PMA-differentiated CMK and K562 after identical treatment. The data in Figure 7C show that short-term ionomycin and PMA treatment strongly induced CD154 in the undifferentiated CMK, but not the LSK or K562. In contrast, PMA and ionomycin are strong CD154 inducers in all 3 differentiated cell types. Also noted was that CsA completely inhibited induced CD154 expression in all cell types. These results show that CD154 is a target gene of NFAT in megakaryocytes and appears to be lineage specific.
Discussion
CD154 is established as a key player in both the initiation and progression of the adaptive immune response based on greatly diminished immunologic responses as a result of CD154 deficiency.5 CD40 ligation by CD154 is crucial for the regulation of cytokine production, cell proliferation, cell adhesion, programmed cell death in diverse cell types, particularly B cells, and dendritic cells. Emerging evidence implicate platelet-derived, as opposed to lymphoid cell–derived, CD154 as a key modulator of the pathology of several chronic inflammatory diseases, including autoimmune disorders and atherosclerosis, due primarily to the observed CD154-mediated, proinflammatory effects on nonlymphoid cells such as monocytes and macrophages, as well as vascular endothelial and smooth muscle cells.35-41
Based on the abundant expression of CD154 expression by activated platelets, the current study characterized the CD154 regulation in platelet precursor cells, megakaryocytes.
We show that CD154 is differentially expressed in various in vitro hematopoietic cell lines and primary human and mouse hematopoietic precursor cells. Specifically, CD154 mRNA is highly expressed in primary mouse bone marrow–derived CD41+ cells versus CD34+ precursors and CD41+-depleted bone marrow cells. Further, CD154 expression is enhanced from undetectable levels in both mouse and human primary CD34+ stem cell precursors, to high-level expression following cytokine-induced megakaryocyte-specific differentiation. Our observations in primary megakaryocytes were recapitulated in various hematopoietic cell lines by demonstrating that CD154 expression is enhanced in cell lines either representative of the megakaryocyte lineage (CMK and Meg-01) or after megakaryocyte-specific differentiation of a multipotient precursor cell line (K562). We conclude from these data that CD154 appears to be a member of megakaryocyte-associated gene repertoire. Our data showing megakaryocyte-specific CD154 expression in the in vitro cell lines also provided a model system for the preliminary investigation into the mechanisms of CD154 regulation in the unique cellular context of megakaryocytes.
Our observations that enhanced CD154 expression and promoter activity appears associated specifically with a mature megakaryocyte phenotype originally led us to hypothesize that CD154 was potentially regulated via GATA and Ets-1, families of transcriptional regulators shown to be key mediators of megakaryocyte-restricted genes including gpIIb, gpIX, and PF-4.42,43 Although the proximal 391-bp promoter region of CD154 contained GATA and Ets-1 binding sites, promoter truncation analysis demonstrated that megakaryocyte-specific CD154 expression was instead dependent on functional consensus NFAT sites in the proximal promoter region. EMSA and supershifting analysis indeed confirmed NFATp and NFATc protein occupancy of the distal NFAT site (Figure 7A). When these data were considered, along with the observations that (1) CsA abrogates differentiation-induced CD154 expression, and (2) CD154 is rapidly induced following short-term PKC and intracellular calcium mobilization (also inhibited by CsA) in CMK and primary CD41+ cells, but not in K562 and CD34+ precursor cells, a megakaryocyte-specific role for NFAT is suggested.
A novel role for the nonlymphoid tissue-specific regulation of genes by NFAT was originally implied by Kiani et al, where extensive expression analysis in primary bone marrow–derived megakaryocytes, culture-derived megakaryocytes, and various hematopoietic cells lines, including CMK, showed that the pattern of NFAT family protein expression is lineage specific among the hematopoietic cell compartment.44,45 Primary culture-derived megakaryocytes and CMK cells selectively and abundantly express several NFAT isoforms, including NFATp. In contrast, NFAT expression was profoundly suppressed in other myeloid lineages. Implied from this previous study as well as the current data is that NFAT plays a role in lineage-specific gene expression in megakaryocytes. Although no traditional megakaryocyte-associated target genes have been as yet identified, the same group showed specific inhibition of NFAT in megakaryocytes suppressed both constitutive and ionomycin-inducible expression of Fas ligand, another TNF family protein important for thrombogenesis.45 Based on both our observations as well as previous studies by Kiani et al, megakaryocyte differentiation is associated with NFAT expression and activation, but it is not clear if NFAT is required for the expression of megakaryocyte-restricted genes. Although preliminary, it was observed that the 4-day PMA-induced differentiation of CMK or in vitro–derived primary megakayocytes in the presence of CsA had no observable effects on viability, cellular morphology, or gpIIb expression, suggesting that inhibition of NFAT did not alter megakaryocyte differentiation. Nonetheless, our observations reported herein may have clinical implications in that several widely used immunosuppressive drugs may impact platelet-associated CD154 expression without altering platelet numbers or function.
In summary, we present evidence that suggests platelet-associated CD154 is derived from megakaryocytes through the novel observation that megakaryocytes express full-length, membrane-bound CD154. Our regulation studies reveal that constitutive CD154 appears to be regulated, at least in part, by inducible pathways traditionally associated with activated T cells. The involvement of NFAT, coupled with the ability to biochemically modulate megakaryocyte CD154 levels with widely used calcineurin inhibitors, imply a potential therapy to modulate platelet CD154 and potentially impact progression of systemic inflammatory diseases.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health (NIH) Career Development Award K01 DK067338-02 (S.A.C.) and NIH R01-AI60924 (T.L.R).
National Institutes of Health
Authorship
Contribution: S.A.C. designed the research; S.A.C. and D.L.S. performed the research and analyzed data; and S.A.C. and T.L.R. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Scott A. Crist, Purdue University, Department of Comparative Pathobiology and the Purdue University Cancer Center, Rm 401 Hansen Bldg, 201 S University St, West Lafayette, IN 47907-2064; e-mail:scrist@purdue.edu.