Abstract
Herein we have analyzed chemokine involvement in the trafficking of developing and mature mouse natural killer (NK) cells in the bone marrow (BM). We observed drastic changes of CCR1, CXCR3, and CXCR4 expression and function during progression from precursor NK (pNK) cells to immature DX5− NK (iNK) and mature DX5+ NK (mNK) cells. pNK and mNK cells expressed the 3 receptors, while only CXCR4 was detected on iNK cells. Correspondingly, mNK cells migrated to CXCL12, CXCL10, and CCL3, and pNK and iNK cells to CXCL12, whereas pNK cells migrated to CCL3 and CXCL10 only after CXCL12 stimulation. Comparison of BM, peripheral blood, and spleen mNK cell populations revealed that CXCL12, CXCL10, and CCL3 preferentially affected BM mNK cell migration. Administration of the CXCR4 antagonist, AMD-3100, to C57BL/6 mice induced strong reduction of mNK and iNK cells in the BM and increased their number in blood and spleen. Conversely, CCL3 administration selectively mobilized mNK cells from the BM and this effect correlated with its ability to inhibit CXCL12-mediated mNK cell responses in vitro. Our results suggest that the combined action of chemokines selectively regulates localization of NK cell subsets in the BM and direct their maturation and migration to the periphery.
Introduction
Natural killer (NK) cells are a lymphocyte subset that contributes to the early host response to viral infection and cancer by promoting contact-dependent cytotoxicity and producing cytokines upon recognition of altered cells.1-3 These functions are acquired mainly during differentiation in the bone marrow (BM) that provides cytokine- and growth factor–mediated signals along with the adhesive support required for NK cell development.4,5 Sequential steps of mouse NK cell differentiation and maturation have been recently characterized6,7 : NK cells derive from a lymphoid precursor (pNK) expressing the common IL2R/IL15R beta chain (CD122) and the activating receptor NKG2D and lacking lineage markers. Immature NK (iNK) cells express NK1.1 and CD94 along with the alphav integrin chain (CD51) and low levels of CD11b expression, and have poor cytotoxic and cytokine secretion capacity. During maturation, an intimate contact with IL15-presenting cells drives Ly49 C-type lectin receptor expression and a phase of proliferation-dependent expansion.8,9 Mature NK (mNK) cells acquire CD49b (defined by the DX5 mAb clone) and lose CD51 expression; among this population, cells with increased expression of CD11b and CD43 are considered fully functional. The phenotypic and functional heterogeneity of NK cells in the peripheral organs, together with the observation that also pNK and iNK can be found outside BM, suggest that the BM is involved mainly in the initial steps of NK cell differentiation, and that final maturation also occurs in different organs.6,10-12
Leukocyte development in the BM is characterized by changes in their migration properties that enable coordinated relocation of individual precursors during their sequential stages of maturation.
Chemokines are a family of small polypeptides that, together with adhesion receptors, control leukocyte trafficking in homeostatic and inflammatory conditions by regulating migration from the blood into tissues and subsequent microenvironmental localization within the tissues.13-15 Changes in the migration properties during leukocyte development are reflected by switches in chemokine receptor profile and expression of selected chemokines in distinct microenvironments.16,17 In addition, chemokines such as CXCL8 and CCL3 that are secreted by BM stromal cells in response to basal or inflammatory stimuli are potent mobilizing factors for selected BM cell populations.18-21 Thus, chemokines can regulate both retention/mobilization of leukocytes from the BM to the blood and their recruitment into tissues.
NK cell subsets localize in several organs such as spleen, liver, lung, uterus, along with BM during steady state,10-12,22 and it is likely that NK cell trafficking in these organs is controlled by chemotactic receptors. Consistently, Walzer et al recently demonstrated that the lysophospholipid S1P receptor S1P5 provides an egress signal for both BM and lymph node NK cells.23 Upon stimulation, NK cells are recruited in a chemokine-dependent manner from peripheral blood to sites of viral infection and tumor to exert their functions. A critical role for CCL3-mediated NK cell recruitment in liver and lung has been demonstrated for the defense against cytomegalovirus, Klebsiella pneumoniae, or Mycobacterium bovis infection.24-26 In addition, knockout models have shown a critical role for CXCR3 in the recruitment of NK cells to injured lung or to lymph nodes draining inflamed tissues.27,28
To reach their target organ, NK cells must move from their storage places such as BM to peripheral blood. Thus, understanding how NK cells are mobilized from BM could assist the development of novel NK cell–based immunotherapy for cancer and chronic infections.29
While the role of chemokines has been addressed for B- and T-lymphocyte development within primary lymphoid organs, little is known about the role of these molecules in the regulation of the homing, retention, and migration of NK precursor and developing cells within BM. As NK cell trafficking is strongly influenced by the cells' ability to respond to chemokines, we investigated whether chemokines could play a role in regulating NK cell localization during development in the BM both under steady-state and inflammatory conditions.
Our study shows differences in the chemotaxis response profile of developing and mature mouse NK cells to selected chemokines, and the ability of endogenous CXCL12 and exogenous CCL3 to act as retention and mobilizing factors, respectively, for NK cell subsets in the BM.
Methods
Mice
Female C57BL/6 mice were purchased from Charles River (Calco, Italy) housed at the mouse facility Department of Histology and Medical Embryology, University of Rome La Sapienza under specific pathogen-free conditions. Sentinel mice were screened for seropositivity to Sendai virus, rodent coronavirus, and Mycoplasma pulmonis by murine Immunocomb test (Charles River) and were found negative. For all experiments, mice between 6 and 10 weeks of age were used.
Antibodies, chemokines, and reagents
Armenian hamster anti-CD3ϵ (145–2C11) and mAbs directly conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), peridinin chlorophyll protein 5.5 (PerCP), allophycocyanin (APC), or biotin, and specific for the following antigens (clone name in parentheses) were used in this study: NK1.1 (PK136), CD49b (DX5), CD11b (M1/70), CD122 (TM-beta1), CD19 (6D5), CXCR4 (2B11), CXCR3 (220803), and CCR5 (C34-3448). Abs and APC-conjugated streptavidin were purchased from Pharmingen (Becton Dickinson, San Diego, CA), R&D Systems (Minneapolis, MN), eBioscience (San Diego, CA), and Biolegend (San Diego, CA). Alexafluor 647–conjugated goat polyclonal anti-CCR1 and control IgG were from Santa Cruz Biotechnologies (Santa Cruz, CA).
Recombinant mouse CCL3 and CXCL10 and human CXCL12 were from Peprotech EC (London, United Kingdom), and were reconstituted in stock solution in water at 1 mg/mL. Bovine serum albumin (BSA), saponin, and AMD-3100 were from Sigma-Aldrich (St Louis, MO).
Cell preparation
BM cells were isolated by extensive flushing of femurs and tibias with PBS and passing the disrupted marrow through a 70-μm cell strainer (Falcon; Becton Dickinson). Spleens were pressed through a 70-μm cell strainer with a rubber syringe plunger in RPMI 10% fetal bovine serum (FBS) to obtain single-cell suspension. Blood samples were collected into heparin-containing tubes. For chemokine receptor staining and chemotaxis experiments, blood cells were loaded and separated using Lympholyte (CL5031; Cedarlane Lab, Hornby, ON), washed, and maintained in RPMI 10% FBS. Cells from BM, blood, and spleen were incubated for 1 hour at 37°C in a CO2 incubator to remove plastic-adherent cells, and for resensitization of potentially desensitized chemokine receptors. Cells were then washed and resuspended in staining buffer for chemokine receptor staining, or in migration or adhesion medium for functional studies.
Immunofluorescence and fluorescence-activated cell sorting analysis
One million cells from the indicated organs or 105 cells from blood were resuspended in 20 μL staining buffer (PBS without Ca++/Mg++, BSA 0.5%, EDTA 1 mM, and NaN3 0.05%) containing anti-CD16/32 (24G2) mAb for 10 minutes on ice. Cells were stained with either biotin-conjugated antimouse CD122 or with fluorochrome-conjugated DX5, antimouse CD3ϵ, and NK1.1 for 45 minutes, to detect pNK and i/mNK cells, respectively. Anti-CD122–stained cells were washed and incubated for 30 minutes with an antibody mixture containing fluorochrome-conjugated DX5, antimouse CD19, CD3ϵ, and NK1.1 plus streptavidin-APC.
Analysis of chemokine receptor expression was performed by cell staining with either biotinylated antimouse CCR5, antimouse CXCR4, APC-conjugated antimouse CXCR3, or control mAb for 45 minutes. After washing, cells were stained with the cocktail of fluorochrome-conjugated antibodies (plus streptavidin-APC when required) to identify pNK, iNK, and mNK cells. Intracellular staining with Alexafluor 647–conjugated antimouse CCR1 was performed by fixing and permeabilizing cells already stained with the antibody cocktails described in this paragraph, with 1.5% paraformaldehyde and 0.5% saponin. CCR1 staining specificity was controlled using F4/80+ macrophages and CD19+ B cells as positive and negative controls, respectively (data not shown).
Cells were analyzed on a modified 2-laser FACScalibur (Becton Dickinson) and flow cytometry data were elaborated using Cellquest software (Becton Dickinson). A representative gating of pNK, iNK, and mNK is shown in Figure 1A.
Migration assay
Chemotaxis assays with the concentrations of CCL3, CXCL12, and CXCL10 indicated in figure legends were performed in 5-μm pore Transwell insert as described.30 BM (5 × 105), spleen (5 × 105), and blood cells (2 × 105) from individual animals were loaded on the upper well. After 90 minutes, the contents of the lower chemotaxis well were transferred to a polypropylene tube after Transwell insert removal. Cells were centrifuged at 300g for 10 minutes, and then resuspended in 20 μL staining buffer containing anti-CD16/32 (24G2) mAb. Cells were then incubated on ice for 10 minutes and a mixture of the appropriate antibodies was added. Cells were further incubated on ice for 30 minutes, washed with 2 mL staining buffer, and resuspended in 200 μL of the same buffer. Cell migration was counted by fluorescence-activated cell sorting (FACS) as previously described and calculated as percentage of input cells.30 For chemokine pretreatment, BM cells were incubated with 5 μg/mL chemokine for 10 minutes at room temperature, washed, and resuspended in migration medium for chemotaxis.
Adhesion assay
Adhesion assay was performed on 96-well plates. Wells were coated as previously described with murine ICAM-1 (mICAM-1) or with BSA30 and blocked with 50μL FBS for 10 minutes at 37°C. After FBS removal, 1.5 × 105 BM cells in 50 μL were added to the wells and incubated for 10 minutes at 37°C. PBS or CXCL12 (10 μL) was then added to CCL3 pretreated or untreated cells, for 20 minutes. Thereafter, the assay was stopped by placing the plate on ice and washing each well 3 times with PBS-containing divalent cations. To detach adherent cells, 100 μL recovery medium (PBS without divalent cations, 5 mM EDTA) was added into each well for 20 minutes at 37°C. Cells were then resuspended by extensive pipetting and transferred to a polypropylene tube containing 1 mL staining buffer. The wells were washed with 100 μL recovery medium that was combined with previously collected cells. Triplicate wells were pooled to reach sufficient amounts of cells for staining. One hundred percent adhesion was made by staining 150 μL total input cells. Data were calculated as percentage of input cells.
In vivo AMD-3100 and CCL3 administration
For subcutaneous injection, CCL3 and AMD-3100 were diluted in PBS at the indicated concentration. Mice were given either control diluent, AMD-3100 (100 μg), or CCL3 (1 μg) in 50 μL. Timing and dosage were based on our own preliminary studies and were consistent with previous reports.31,32 At the end of treatment, mice were killed by cervical dislocation and cells were isolated as described above.
After removal of contaminating erythrocytes, cells were counted, and 106 BM or spleen cells, or 30 to 50 μL whole blood cells were stained with the Abs indicated in the figure legends and analyzed by flow cytometry. Total number of cells was obtained by assuming that total blood volume is 1.5 mL and that BM cells isolated from tibia and femur represent 9% of the total BM.33 The total number of NK and pNK cells in the organs was obtained taking into account the percentage of CD3−NK1.1+ and CD122+Lin− (CD3−CD19−DX5−NK1.1−) cells, respectively, within the total lymphocytes. Lymphocyte number was calculated by evaluating the percentage of lymphocytes (electronically gated according to side scatter and forward scatter) within the total population after excluding dead cells with a second gate. In some experiments, annexin and 7-AAD staining was performed on BM NK cell populations following both in vitro and in vivo treatment with AMD-3100 or CCL3, and no changes in NK cell viability were observed (data not shown).
Statistics
The Student t test was used to analyze data. P value of .05 was chosen as the limit of statistical significance.
Results
BM NK cells modulate chemokine receptor profile and function during the development
Changes in migration properties during leukocyte development are reflected by switches in chemokine receptor profile that allow the cell to direct toward new chemokine-expressing microenvironments.16,34 NK cells develop in the BM through a series of stages, namely pNK, iNK, and finally mNK cells.9,35 To investigate the contribution of chemokines in NK cell development, we initially analyzed the surface expression of chemokine receptors on BM NK cells at different stages of maturation. We focused on CXCR4, CCR1, CCR5, and CXCR3 receptors because their ligands, CXCL12, CCL3, and CXCL10, regulate peripheral NK cell trafficking during inflammation, and modulate the BM localization of immature cells of other lineages.16,24-26
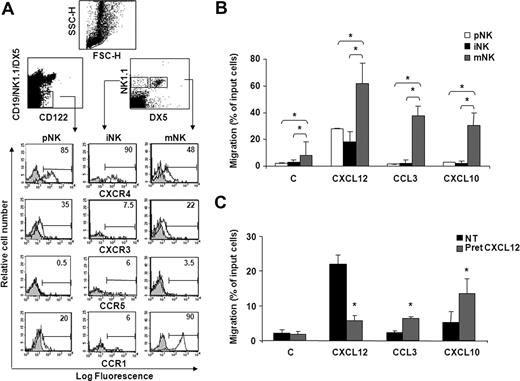
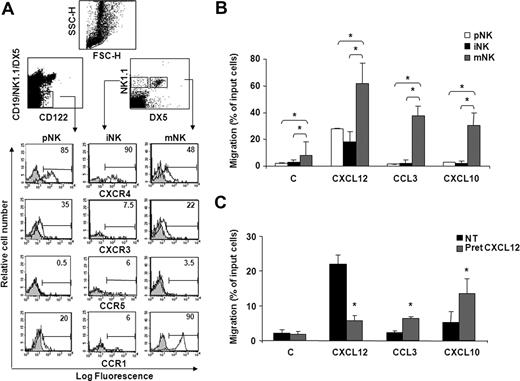
Immunofluorescence and FACS analysis of cells stained with specific Abs shows that the majority of pNK and iNK cells express high levels of CXCR4, while lower levels were detected on mNK cells (Figure 1A; Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article). In addition, we observed a biphasic profile of CXCR3 and CCR1 expression during development, as pNK and mNK cell subsets were significantly positive for these receptors, whereas low levels were detectable on iNK cells. CCR5 expression was barely detectable on all BM NK cell subsets.
BM NK cell chemokine receptor expression and chemotactic response profile to CXCL12, CXCL10, and CCL3 during development. (A) Cell surface expression of chemokine receptors on developing NK cells. BM cells from individual C57BL/6 mice were stained with anti-CXCR4, anti-CXCR3, or anti-CCR5 mAb in combination with antibodies that identify pNK (anti-CD122, anti-CD3ϵ, anti-CD19, anti-NK1.1, and DX5), iNK, and mNK (anti-CD3ϵ, anti-NK1.1, and DX5) cells. CD3− lymphocytes were gated on the CD122+lin− population (top left dot plot) and on the NK1.1+DX5− or NK1.1+DX5+ population (top right dot plot). Shown are the histogram plot overlays of chemokine receptor antibody staining (empty histogram) against isotype control Ab (gray histogram). To analyze CCR1 expression, intracellular staining was performed on permeabilized cells using Alexafluor 647–conjugated goat polyclonal anti-CCR1 (empty histogram) or normal IgG (gray histogram). Numbers in the histograms indicate the percentage of positive cells of a representative experiment of at least 3 performed. Control staining was always less than 2%. (B) The chemotactic response of NK cells changes during development in the BM. In vitro chemotaxis assay of cells isolated from BM of individual animals was performed as described in “Methods.” One million cells were allowed to migrate in response to recombinant murine CXCL10 and CCL3 and human CXCL12 (used at 200 ng/mL) for 90 minutes. At the end of the experiment, duplicate wells were pooled, and cells were centrifuged and stained with the antibody cocktail used to identify pNK, iNK, and mNK cells (C indicates medium alone). Data are expressed as percentage of input cells and represent the means plus or minus SD of percentage migration of cells from a total of 8 to 12 animals analyzed in independent experiments. Student t test was performed by comparing migration of different BM NK cell subsets with each other; *P < .05. Migration of pNK versus iNK is not significantly different and thus omitted for sake of simplicity. (C) CXCL12 induces migration of pNK cells and promotes their chemotaxis to CCL3 and CXCL10. BM pNK cell migration was performed after cell pretreatment with 5 μg/mL CXCL12. Cells were washed and allowed to migrate in response to the indicated chemokines for 90 minutes. Migrated cells were then collected and stained with anti-CD122, anti-CD3ϵ, anti-CD19, anti-NK1.1, and DX5. Migrating pNK cells were identified as CD122+/lin− cells and quantified as positive events in 150 seconds acquisitions. C indicates control. Data are expressed as percentage of input cells and represent the means plus or minus SD of 6 independent experiments. Student t test was performed by comparing migration of untreated versus CXCL12-pretreated pNK cells. *P < .05.
BM NK cell chemokine receptor expression and chemotactic response profile to CXCL12, CXCL10, and CCL3 during development. (A) Cell surface expression of chemokine receptors on developing NK cells. BM cells from individual C57BL/6 mice were stained with anti-CXCR4, anti-CXCR3, or anti-CCR5 mAb in combination with antibodies that identify pNK (anti-CD122, anti-CD3ϵ, anti-CD19, anti-NK1.1, and DX5), iNK, and mNK (anti-CD3ϵ, anti-NK1.1, and DX5) cells. CD3− lymphocytes were gated on the CD122+lin− population (top left dot plot) and on the NK1.1+DX5− or NK1.1+DX5+ population (top right dot plot). Shown are the histogram plot overlays of chemokine receptor antibody staining (empty histogram) against isotype control Ab (gray histogram). To analyze CCR1 expression, intracellular staining was performed on permeabilized cells using Alexafluor 647–conjugated goat polyclonal anti-CCR1 (empty histogram) or normal IgG (gray histogram). Numbers in the histograms indicate the percentage of positive cells of a representative experiment of at least 3 performed. Control staining was always less than 2%. (B) The chemotactic response of NK cells changes during development in the BM. In vitro chemotaxis assay of cells isolated from BM of individual animals was performed as described in “Methods.” One million cells were allowed to migrate in response to recombinant murine CXCL10 and CCL3 and human CXCL12 (used at 200 ng/mL) for 90 minutes. At the end of the experiment, duplicate wells were pooled, and cells were centrifuged and stained with the antibody cocktail used to identify pNK, iNK, and mNK cells (C indicates medium alone). Data are expressed as percentage of input cells and represent the means plus or minus SD of percentage migration of cells from a total of 8 to 12 animals analyzed in independent experiments. Student t test was performed by comparing migration of different BM NK cell subsets with each other; *P < .05. Migration of pNK versus iNK is not significantly different and thus omitted for sake of simplicity. (C) CXCL12 induces migration of pNK cells and promotes their chemotaxis to CCL3 and CXCL10. BM pNK cell migration was performed after cell pretreatment with 5 μg/mL CXCL12. Cells were washed and allowed to migrate in response to the indicated chemokines for 90 minutes. Migrated cells were then collected and stained with anti-CD122, anti-CD3ϵ, anti-CD19, anti-NK1.1, and DX5. Migrating pNK cells were identified as CD122+/lin− cells and quantified as positive events in 150 seconds acquisitions. C indicates control. Data are expressed as percentage of input cells and represent the means plus or minus SD of 6 independent experiments. Student t test was performed by comparing migration of untreated versus CXCL12-pretreated pNK cells. *P < .05.
Our findings that CXCR4, CXCR3, and CCR1 are expressed and modulated during NK cell development prompted us to analyze the chemotaxis of NK cells at each stage of development to CXCL12, CXCL10, and CCL3. In preliminary experiments, chemokines were titered from 2 to 500 ng/mL, and optimal concentrations were subsequently used in all the experiments. mNK cells migrated in response to all chemokines tested, while iNK and pNK cells markedly responded only to CXCL12. Notably, pNK cells displayed a lower ability to migrate in response to CXCL12 than mNK cells (Figure 1B), irrespective of their higher CXCR4 expression levels. Similarly, pNK cells poorly migrated to CXCL10 (2.3-fold over basal migration) and did not respond to CCL3 in spite of considerable expression of CXCR3 and CCR1 (Figure 1C). In addition, the migration in the absence of chemokine (basal migration) was higher in mNK cells than in pNK and iNK cells, suggesting that the migratory capability of BM NK cells improves during maturation. Nevertheless, a short pretreatment (5 minutes) of BM cells with CXCL12, a chemokine that is highly expressed in the BM microenvironment, significantly enhanced pNK cell chemotactic response to CCL3 and CXCL10, suggesting that the chemokine milieu of this organ can influence cell migration (Figure 1C). This effect was blocked in the presence of AMD3100, a selective CXCR4 antagonist, indicating the specificity of CXCR4 triggering (data not shown). Furthermore, CXCL12 pretreatment did not alter CXCR3 and CCR5 expression (up to 90 minutes) indicating that the increased migration is not due to regulation of receptor expression.
Collectively, these results indicate that BM NK cells express functional CXCR4, CXCR3, and CCR1, the only receptor for CCL3 we could detect on BM NK cells.
iNK and mNK cells from different anatomic compartments exhibit different expression levels of chemokine receptors, and differently migrate in response to chemokines
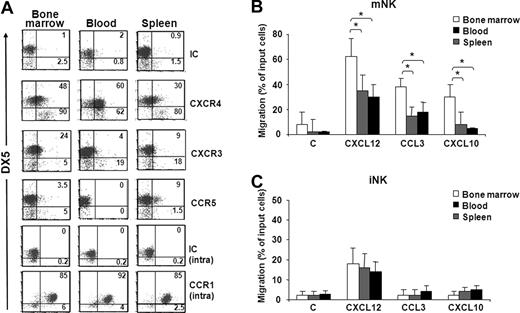
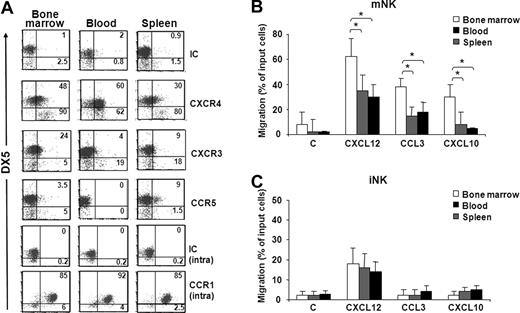
To study the involvement of chemokines in the trafficking of NK cell subsets from BM to other anatomic compartments, we initially analyzed chemokine receptor expression and responsiveness of iNK and mNK cells from BM, blood, and spleen. Our results show that CXCR4 is highly expressed by mNK cells obtained from BM and blood compared with spleen mNK cells (Figure 2A; Figure S1C). BM mNK cells also expressed higher levels of CXCR3 than pNK and iNK cells. Conversely, CCR5 expression was detected only on a minute subset of spleen mNK cells. High intracellular levels of CCR1 were found on mNK cells independent of their anatomic origin, but the relative membrane expression is currently unknown.
Chemokine receptor expression profile and chemotaxis of iNK and mNK cells from different anatomic compartments. (A) Freshly isolated cells from BM, spleen, and blood of individual C57BL/6 mice were stained with specific antibodies against CXCR4, CXCR3, CCR5, and CCR1 in combination with anti-NK1.1–, anti-CD3ϵ–, and DX5-specific antibodies. Expression of chemokine receptors was analyzed on DX5− and DX5+ NK cells (NK1.1+CD3−) of the electronically gated lymphocytes. Numbers in the dot plots indicate the percentage of positive cells within the iNK (DX5−, bottom left and right) and mNK (DX5+, top left and right) cell populations. A representative experiment of 6 performed is shown. (B,C) In vitro chemotaxis assay of cells isolated from different organs was performed as described in “Methods” and in Figure 1B legend. mNK and iNK cell migration was quantified by enumerating the positive events contained in the NK1.1+/CD3−/DX5+ gate (mNK, B) and in the NK1.1+/CD3−/DX5− gate (iNK, C) after 150-second acquisition for BM and blood cells and 75-second acquisition for spleen cells. Results presented are the means plus or minus SD of 6 independent experiments. Student t test was performed by comparing migration of mNK cells (B) or iNK cells (C) from different anatomic compartments from each other; *P < .05. Differences that are not statistically significant are omitted for sake of simplicity.
Chemokine receptor expression profile and chemotaxis of iNK and mNK cells from different anatomic compartments. (A) Freshly isolated cells from BM, spleen, and blood of individual C57BL/6 mice were stained with specific antibodies against CXCR4, CXCR3, CCR5, and CCR1 in combination with anti-NK1.1–, anti-CD3ϵ–, and DX5-specific antibodies. Expression of chemokine receptors was analyzed on DX5− and DX5+ NK cells (NK1.1+CD3−) of the electronically gated lymphocytes. Numbers in the dot plots indicate the percentage of positive cells within the iNK (DX5−, bottom left and right) and mNK (DX5+, top left and right) cell populations. A representative experiment of 6 performed is shown. (B,C) In vitro chemotaxis assay of cells isolated from different organs was performed as described in “Methods” and in Figure 1B legend. mNK and iNK cell migration was quantified by enumerating the positive events contained in the NK1.1+/CD3−/DX5+ gate (mNK, B) and in the NK1.1+/CD3−/DX5− gate (iNK, C) after 150-second acquisition for BM and blood cells and 75-second acquisition for spleen cells. Results presented are the means plus or minus SD of 6 independent experiments. Student t test was performed by comparing migration of mNK cells (B) or iNK cells (C) from different anatomic compartments from each other; *P < .05. Differences that are not statistically significant are omitted for sake of simplicity.
mNK cells from all 3 compartments migrated in response to CXCL12, CCL3, and CXCL10, with BM mNK cells migrating significantly more than the peripheral (Figure 2B). In fact, compared with BM, the percentage of migrated cells was lower in blood and spleen (1.8- and 2-fold for CXCL12, 2.2- and 2.4-fold for CCL3, and 3.6- and 4.4-fold for CXCL10, respectively).
The analysis of iNK cells revealed that only CXCR4 is expressed in all 3 compartments, and accordingly CXCL12 promoted iNK cell migration (Figure 2C). Unlike in BM, a small fraction of iNK cells from spleen also expressed CXCR3, although they did not significantly respond to CXCL10.
These results indicate that BM mNK cells have an higher propensity to respond to CXCL12, CCL3, and CXCL10, and this may affect their distribution under steady-state and inflammatory conditions.
The CXCR4 antagonist AMD-3100 and the CCR1/5 agonist CCL3 selectively promote mobilization of BM NK cell subsets
Next we tested the function of CXCL12/CXCR4 axis and CCL3 on NK cell trafficking in vivo. In addition to its role in driving developing hematopoietic cells to specialized niches within BM, CXCR4/CXCL12 axis can potently regulate the homing and retention of cells within this organ.16,18,36-38 Conversely, CCL3 is known for its ability to promote the release of BM hematopoietic precursors (HPCs) and stem cells (HSCs) in a CCR1-dependent manner.32
The role of CXCR4 and CCR1 in NK cell distribution in vivo was investigated using AMD3100, a selective CXCR4 antagonist and CCL3. AMD-3100 (100 μg) or CCL3 (1 μg) was given subcutaneously by one single injection, and animals were killed at indicated time periods. The different subsets of NK cells in BM, spleen, and blood were identified by immunofluorescence and FACS analysis as described above (Figures 3,4). Within CD3−NK1.1+ NKcells, mNK cell subpopulations were further defined as DX5+CD11blow and DX5+CD11bhigh, the latter being the most mature subset.9
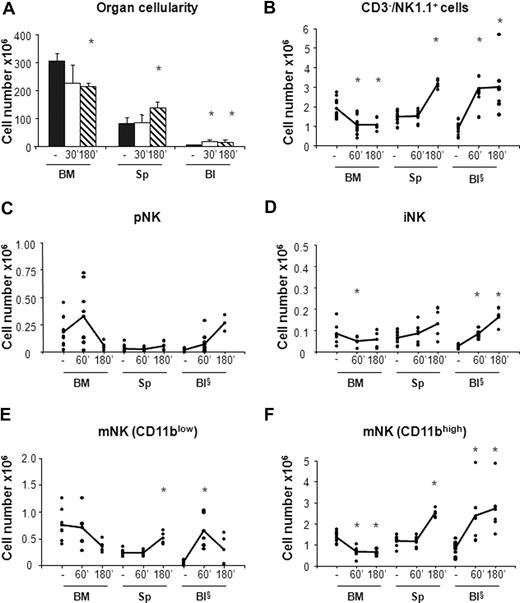
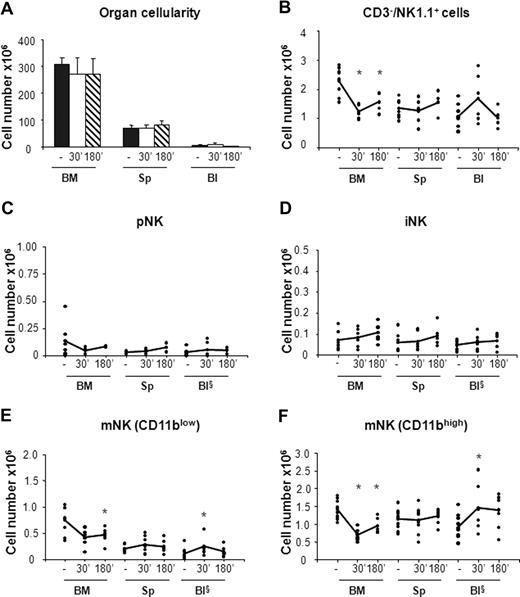
In vivo administration of the CXCR4 antagonist AMD-3100 promotes mobilization of iNK and mNK cells from the BM. Six- to 10-week-old C57BL/6 mice were subcutaneously injected with 50 μL PBS alone (−) or containing 2 mg/mL AMD-3100 and then killed at 60 minutes and at 60 or 180 minutes, respectively, after treatment. Cells from BM, spleen (Sp), and blood (Bl) cells were counted and stained either with anti-CD122, anti-CD19, anti-CD3ϵ, anti-NK1.1, and DX5 or with anti-CD3ϵ, anti-NK1.1, anti-CD11b, and DX5 mAbs to determine the percentage of pNK (C) and iNK/mNK (D-F) cells, respectively. Each dot represents the total cell number in the indicated organ of individual mice from at least 4 independent experiments performed and was calculated as described in “Methods.” Student t test was performed to compare the tissue distribution of NK cell subsets in mice treated with vehicle control with that of mice treated with AMD-3100. *P < .05. Differences that are not statistically significant are omitted for sake of simplicity. §Cell number × 105.
In vivo administration of the CXCR4 antagonist AMD-3100 promotes mobilization of iNK and mNK cells from the BM. Six- to 10-week-old C57BL/6 mice were subcutaneously injected with 50 μL PBS alone (−) or containing 2 mg/mL AMD-3100 and then killed at 60 minutes and at 60 or 180 minutes, respectively, after treatment. Cells from BM, spleen (Sp), and blood (Bl) cells were counted and stained either with anti-CD122, anti-CD19, anti-CD3ϵ, anti-NK1.1, and DX5 or with anti-CD3ϵ, anti-NK1.1, anti-CD11b, and DX5 mAbs to determine the percentage of pNK (C) and iNK/mNK (D-F) cells, respectively. Each dot represents the total cell number in the indicated organ of individual mice from at least 4 independent experiments performed and was calculated as described in “Methods.” Student t test was performed to compare the tissue distribution of NK cell subsets in mice treated with vehicle control with that of mice treated with AMD-3100. *P < .05. Differences that are not statistically significant are omitted for sake of simplicity. §Cell number × 105.
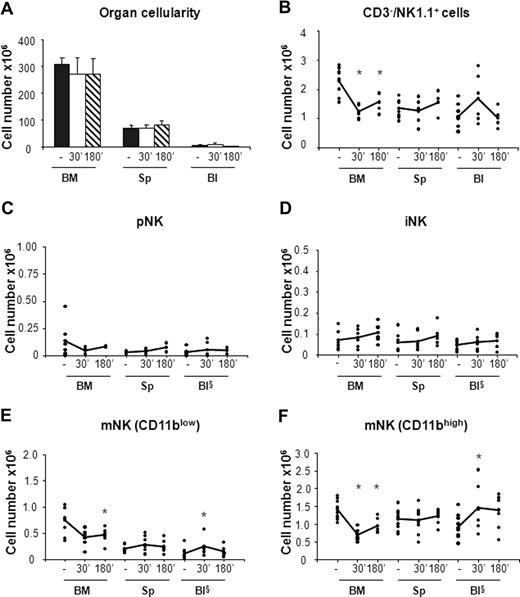
In vivo administration of CCL3 selectively promotes mobilization of mNK cells from the BM. Six- to 10-week-old C57BL/6 mice were subcutaneously injected with 50 μL PBS alone (−) or containing 20 μg/mL CCL3. Mice treated with PBS or with CCL3 were killed at 30 minutes and at 30 or 180 minutes, respectively, after treatment. Cells from BM, spleen (Sp), and blood (Bl) were counted and stained either with anti-CD122, anti-CD19, anti-CD3ϵ, anti-NK1.1, and DX5 antibodies or with anti-CD3ϵ, anti-NK1.1, anti-CD11b, and DX5 mAbs. Each dot represents the total cell number in the indicated organ of individual mice from at least 4 independent experiments performed and was calculated as described in “Methods.” Student t test was performed to compare the tissue distribution of NK cell subsets in mice treated with vehicle control with that of mice treated with CCL3. *P < .05. Differences that are not statistically significant are omitted for sake of simplicity. §Cell number × 105.
In vivo administration of CCL3 selectively promotes mobilization of mNK cells from the BM. Six- to 10-week-old C57BL/6 mice were subcutaneously injected with 50 μL PBS alone (−) or containing 20 μg/mL CCL3. Mice treated with PBS or with CCL3 were killed at 30 minutes and at 30 or 180 minutes, respectively, after treatment. Cells from BM, spleen (Sp), and blood (Bl) were counted and stained either with anti-CD122, anti-CD19, anti-CD3ϵ, anti-NK1.1, and DX5 antibodies or with anti-CD3ϵ, anti-NK1.1, anti-CD11b, and DX5 mAbs. Each dot represents the total cell number in the indicated organ of individual mice from at least 4 independent experiments performed and was calculated as described in “Methods.” Student t test was performed to compare the tissue distribution of NK cell subsets in mice treated with vehicle control with that of mice treated with CCL3. *P < .05. Differences that are not statistically significant are omitted for sake of simplicity. §Cell number × 105.
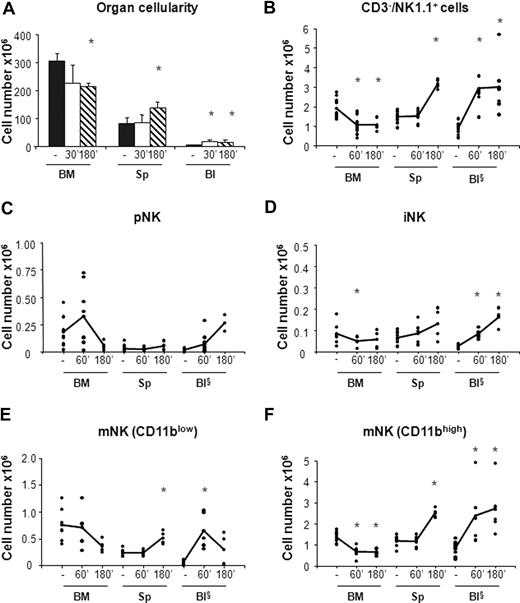
In agreement with previous observations,31 we found that CXCR4 blockade resulted in 20% reduction of total BM cellularity that was accompanied by a rapid increase in the number of white blood cells (3- to 4-fold) and by a 2-fold increase in the number of splenocytes at 3 hours after AMD-3100 administration (Figure 3A). By contrast, the total number of cells in the organs analyzed did not vary in mice treated with CCL3 (Figure 4A).
AMD-3100 administration led to 1.9-fold decrease in the number of total BM NK cells (CD3−/NK1.1+) that was associated with a parallel increase in the blood and in the spleen (Figure 3B). As BM is the only organ exhibiting decreased numbers of NK cells following AMD-3100 administration, the increase in circulating and spleen NK cells likely reflects mobilization from the BM.
CXCR4 blockade induced the mobilization from the BM of both iNK cells, and the CD11blow and CD11bhigh mNK cell subsets (Figure 3D-F). CD11blow circulating mNK cell number increased 6-fold after 1 hour and decreased thereafter, while iNK as well as CD11bhigh mNK cells increased after 1 hour and reached the maximal values 3 hours after injection (4-fold and 3-fold increase over vehicle control mice, respectively). Interestingly, while the number of each NK cell subset in the blood increased rapidly during the time period analyzed, the increase in the spleen was slower, suggesting that NK cell entry in this organ is delayed by arrest in other compartments.
When we analyzed the effect of CCL3 administration, we observed 1.5- to 1.9-fold decrease of the total NK cell number in the BM associated with increased levels of circulating NK cells (Figure 4B). Nevertheless, the CCL3-induced increase of NK cells in the blood does not completely reflect the reduction of NK cells in BM, suggesting that mobilized BM NK cells rapidly distribute in other organs during the time course analyzed. This hypothesis is supported by the increased number of cells detected in liver after treatment, whereas no increase was observed in lung NK cells (data not shown).
The analysis of CCL3 treatment on the different NK cell subsets indicates that, unlike CXCR4 antagonist, this chemokine strongly affected the distribution of mNK cells but not of pNK and iNK cells. mNK cell number (both CD11blow and CD11bhigh) increased in the blood (1.5- and 2.5-fold, respectively), while their BM counterparts underwent a rapid decrease (1.5- and 2-fold, respectively; Figure 4E,F).
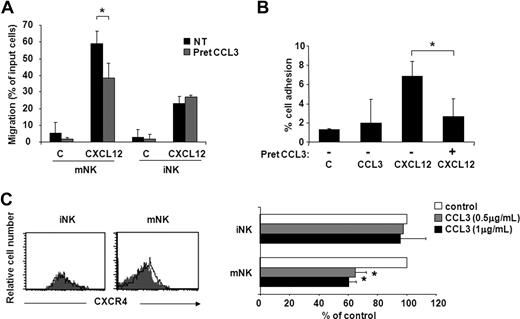
Based on recent observations showing that mouse CXC chemokine KC stimulates neutrophil release from BM by CXCR4 heterologous desensitization,39 we evaluated whether CCL3 could affect CXCR4 function in vitro. As shown in Figure 5, pretreatment of BM cells with 5 μg/mL CCL3 led to a marked decrease of CXCL12-induced chemotaxis (30%) and adhesion to ICAM-1 (80%-100%) of mNK cells. By contrast, CXCL12-mediated migration of iNK cells was not affected by CCL3 (Figure 5A), thus suggesting that CXCR4 blockade by this chemokine contributes to the observed CCL3-mobilizing effect on the mNK cell population (Figure 4E,F). The molecular mechanisms responsible for CCL3 action on CXCR4 are not completely known but involve receptor down-regulation (35%-40% reduction at 45-minute incubation) on mNK cells. As expected based on the results described above, CCL3 did not induce any change in CXCR4 expression in iNK cells (Figure 5C).
CCL3 selectively inhibits CXCL12-induced chemotaxis and adhesion to ICAM-1 of mNK cells. iNK and mNK cell migration (A) and mNK cell adhesion (B) were performed after BM cell pretreatment with or without 5 μg/mL CCL3. Cells were washed and allowed to migrate or to adhere to ICAM-1 in response to CXCL12 (used at 200 ng/mL and 5 μg/mL, respectively). iNK and mNK cell migration and mNK cell adhesion were quantified by enumerating the events contained in the NK1.1+/CD3−/DX5− gate (iNK) and NK1.1+/CD3−/DX5+ gate (mNK) after 150-second acquisition. Data are expressed as percentage of input cells and are the means plus or minus SD of 4 independent experiments performed. (C) CXCR4 cell surface expression on iNK and mNK cells was evaluated following 45-minute BM cell incubation with CCL3 (0.5 and 1 μg/mL) or control vehicle as described in “Methods.” Left panels show histogram plot overlays of CXCR4 antibody staining of untreated (control: empty histogram) or CCL3-treated (0.5 μg/mL, gray histogram) cells. Right panel shows the geometric mean fluorescence intensity (MFI) of CCL3-treated cells expressed as percentage of control (untreated) cells. MFI of isotype control staining did not vary after treatment with CCL3 (not shown). Student t test was performed by comparing CXCL12-supported migration (A) and adhesion (B), and CXCR4 cell surface expression (C) of untreated versus CCL3-pretreated BM NK cells. *P < .05.
CCL3 selectively inhibits CXCL12-induced chemotaxis and adhesion to ICAM-1 of mNK cells. iNK and mNK cell migration (A) and mNK cell adhesion (B) were performed after BM cell pretreatment with or without 5 μg/mL CCL3. Cells were washed and allowed to migrate or to adhere to ICAM-1 in response to CXCL12 (used at 200 ng/mL and 5 μg/mL, respectively). iNK and mNK cell migration and mNK cell adhesion were quantified by enumerating the events contained in the NK1.1+/CD3−/DX5− gate (iNK) and NK1.1+/CD3−/DX5+ gate (mNK) after 150-second acquisition. Data are expressed as percentage of input cells and are the means plus or minus SD of 4 independent experiments performed. (C) CXCR4 cell surface expression on iNK and mNK cells was evaluated following 45-minute BM cell incubation with CCL3 (0.5 and 1 μg/mL) or control vehicle as described in “Methods.” Left panels show histogram plot overlays of CXCR4 antibody staining of untreated (control: empty histogram) or CCL3-treated (0.5 μg/mL, gray histogram) cells. Right panel shows the geometric mean fluorescence intensity (MFI) of CCL3-treated cells expressed as percentage of control (untreated) cells. MFI of isotype control staining did not vary after treatment with CCL3 (not shown). Student t test was performed by comparing CXCL12-supported migration (A) and adhesion (B), and CXCR4 cell surface expression (C) of untreated versus CCL3-pretreated BM NK cells. *P < .05.
Discussion
We show here that developing BM NK cells undergo changes in the expression levels of selected chemokine receptors, namely CXCR4, CXCR3, and CCR1 and in the chemotactic responses to their respective ligands. In addition, we present in vivo evidence that the CXCR4/CXCL12 axis is critically involved in the retention of iNK and mNK cells in the BM, whereas CCL3, a ligand of CCR1, regulates mNK cell mobilization through a direct chemotactic action and the ability to inhibit CXCR4 expression and function.
We first addressed the chemokine receptor expression and functional responses of BM NK cell subsets. Our findings demonstrate that CXCR4 is expressed at higher levels on more immature stages of development (pNK and iNK cells) and substantially decreases during differentiation on mNK cells. Within mNK cells, the more mature CD11bhigh subset expresses lower levels of the receptor than CD11blow cells, an intermediate stage of development (Figure S2).
BM NK cells at all stages of development migrate to CXCL12, and mNK cells migrate much better than more immature subsets, although they express lower receptor levels. This finding is not unique to this cell type as it has been previously shown that BM plasma blasts as well as germinal center B cells can express this receptor in the absence of relevant chemotaxis to CXCL12.40,41 It is possible that pNK and iNK cells intrinsically have a lower migratory phenotype than mNK cells related to their different maturation/activation state. Higher CXCR4 expression levels may be associated to and required for other specialized functions independent of migration, such as promotion of cell survival and/or adhesion to BM ligands or to membrane-bound CXCL12 itself.
Surprisingly, we also found that a significant fraction of pNK cells also express receptors for inflammatory chemokines, CXCR3 and CCR1, and readily migrate in response to their respective ligands, CXCL10 and CCL3, but only upon CXCL12 stimulation.
The activation of CCR1 and CXCR3 function in CXCL12 preactivated pNK cells is consistent with previous findings obtained in other cell types.42 As we did not observe any variation of CXCR3 and CCR5 expression upon CXCL12 treatment (data not shown), we suggest that this chemokine regulates signaling events downstream to these receptors, as also previously reported for plasmacytoid dendritic cells.43 The physiologic role of CXCR3 and CCR1 on pNK cells is unclear: it is possible that as for other BM immature hematopoietic cells, they contribute to the regulation of pNK cell survival, proliferation, and mobilization.32,44-47
The majority of mature and functional NK cells found in the BM may constitute a reservoir, which can be mobilized quickly upon infection or stress. In addition, it is likely that all BM NK cell subsets exit into circulation during steady state as they are found in peripheral compartments.10-12 In this context, it is interesting to note that BM NK cells migrate significantly better than blood-circulating and spleen mNK cells in response to CXCL12, CCL3, and CXCL10 (Figure 2B). The higher expression of CXCL10 receptor, CXCR3, in the former population is consistent with this result. Unlike CXCR3, we found that CXCR4 is expressed at comparable levels on mNK cells from BM and blood in spite of the lower CXCL12 responsiveness of the latter population. This suggests that the strength of CXCL12-induced migration of blood mNK cells is regulated by factors decreasing CXCR4 function rather than its expression.36,37 In addition, the finding that BM mNK cells migrate better than their peripheral counterpart as well as the more immature NK cell subsets may depend on stimuli derived from BM microenvironment, such as IL15, capable of promoting their preferential activation and modulating chemokine receptor expression and function.6,48
Because of the different profile of chemotactic responses we observed during NK cell development, we also determined to what extent the CXCR4/CXCL12 interaction and CCL3 regulate the release of NK cell subsets from marrow.
We have demonstrated that in vivo administration of the CXCR4 antagonist AMD-3100 induces iNK and mNK cell mobilization and accumulation in the spleen showing that activation of CXCR4 by CXCL12 is important for the retention of different NK cell subsets in the BM, and thus controls their release from BM during steady state (Figure 3). In addition, our findings indicate that CCL3 is required for mNK cell egress from BM and can act at least in part by interfering with the CXCR4/CXCL12 axis. Indeed, we found that in vitro exposure to CCL3 selectively and efficiently reduced CXCR4-mediated chemotaxis and almost completely blocked the adhesion to ICAM-1 of mNK cells, likely downmodulating cell surface CXCR4 (Figure 5). As iNK cells do not express any detectable CCL3 receptor, the selectivity of the CCL3-inhibitory effect on CXCR4-mediated expression and function of mNK cells is likely to be attributable to the expression of a functional CCR1 on this latter population.
Wald et al49 observed increased mobilization of NK cells from BM and spleen after ConA-induced hepatitis, and showed that mobilization from BM was CXCR3 and CCR1 independent, even if NK cells were recruited to liver in a CCL3-dependent manner. Although their results apparently contrast with our demonstration that CCL3 mobilizes mNK cells from BM, it should be noted that in an inflammatory context also the other CCL3 receptor, CCR5, may be up-regulated on NK cells and mediate CCL3 response.
Collectively, our evidence indicates that in vivo CXCR4 inhibition is sufficient per se to promote the exit of NK cells at different developmental stages from the BM, and suggests that CCL3-mediated inhibition of CXCR4 function strongly contributes to the CCL3-mobilizing effect on mNK cells.
These findings together with the observation that CXCL12 promotes pNK cell responsiveness to CCL3 and CXCL10 support the hypothesis that the integration of chemokine signals received by developing and mature NK cells regulate their selective localization in BM and direct their maturation and migration to the periphery.
Moreover, our study on the mechanisms underlying the chemokine-mediated NK cell mobilization from BM would be of great relevance for a more rational use of NK cells in the immunotherapy against cancer and infectious disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Francesca Di Rosa, Elisabetta Parretta, and Angela Gismondi for precious advice.
This work was supported by grants from the Italian Association for Cancer Research (AIRC), the Italian Ministry of University and Research (MIUR), and the Center of Excellence (BEMM).
Authorship
Contribution: G.B. designed research, performed experiments, and wrote the paper; G.S. designed and performed experiments, analyzed and discussed results, and revised the paper; D.B. performed experiments and revised the paper; S.M. contributed to flow cytometric analysis; S.S. discussed results and revised the paper; A.S. designed research and contributed to writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Giovanni Bernardini, Department of Experimental Medicine, University of Rome La Sapienza, Viale Regina Elena 324, Rome, 00161, Italy; e-mail: giovanni.bernardini@uniroma1.it; or Angela Santoni, Department of Experimental Medicine, University of Rome La Sapienza, Viale Regina Elena 324, Rome, 00161, Italy; e-mail: angela.santoni@uniroma1.it.