Abstract
The ordered series of proliferation and differentiation from hematopoietic progenitor cells is disrupted in leukemia, resulting in arrest of differentiation at immature proliferative stages. Characterizing the molecular basis of hematopoietic differentiation is therefore important for understanding and treating disease. Retinoic acid induces expression of ankyrin repeat-containing protein with a suppressor of cytokine signaling box 2 (ASB2) in acute promyelocytic leukemia cells, and ASB2 expression inhibits growth and promotes commitment, recapitulating an early step critical for differentiation. ASB2 is the specificity subunit of an E3 ubiquitin ligase complex and is proposed to exert its effects by regulating the turnover of specific proteins; however, no ASB2 substrates had been identified. Here, we report that ASB2 targets the actin-binding proteins filamin A and B for proteasomal degradation. Knockdown of endogenous ASB2 in leukemia cells delays retinoic acid-induced differentiation and filamin degradation; conversely, ASB2 expression in leukemia cells induces filamin degradation. ASB2 expression inhibits cell spreading, and this effect is recapitulated by knocking down both filamin A and filamin B. Thus, we suggest that ASB2 may regulate hematopoietic cell differentiation by modulating cell spreading and actin remodeling through targeting of filamins for degradation.
Introduction
Hematopoiesis is organized as a hierarchy of events controlled by both genetic commitment and external regulatory factors. Whether a hematopoietic stem cell self-renews or differentiates down the myeloid, lymphoid, or erythromegakaryocytic lineages is determined by the pathways that regulate cell-cycle status and gene expression profile. In acute myeloid leukemia, cells are arrested at an immature step of differentiation leading to an accumulation of granulocyte and monocyte precursors in the bone marrow and blood. All-trans retinoic acid (RA) induces differentiation of acute promyelocytic leukemia (APL) cells; this serves as an effective therapy and provides an opportunity to investigate the differentiation process.1 We identified ASB2 (ankyrin repeat-containing protein with a suppressor of cytokine signaling box 2) as a gene activated during RA-induced maturation of APL cells.2,3 ASB2 is also a target gene of the promyelocytic leukemia/retinoic acid receptor alpha (PML–RAR-α) oncogenic transcription factor characteristic of APL.2,4 ASB2 expression inhibits growth and promotes commitment, recapitulating an early step critical for differentiation of myeloid leukemia cells.2 ASB2 encodes a protein that harbors ankyrin repeats and a BC motif located within a suppressor of cytokine signaling (SOCS) box. By interacting with the Elongin BC complex through its BC box ASB2 can assemble with a Cullin5/Rbx module to reconstitute an E3 ubiquitin ligase complex that stimulates polyubiquitylation by the E2 ubiquitin-conjugating enzyme Ubc5.5,6 Within this complex, ASB2 is thought to target proteins for proteosomal degradation. However, ASB2 targets remained unknown.
Filamins are actin cross-linking protein found on stress fibers, in lamellae and in filopodiae. In addition to organizing F-actin, filamins anchor transmembrane and cytoplasmic signaling proteins involved in motility, adhesion, and cell-shape modulation to the actin cytoskeleton, providing mechanical stability to the cell membrane and cell-cell or cell-extracellular matrix connections.7-9 Filamins also regulate the activity of several transcription factors.8,10 Filamins play essential roles throughout development and in the adult organism. Mutations in each of the human filamin genes have been linked to disease with phenotypes, including embryonic lethality, defective neuronal migration, valvular dystrophy, congenital malformations, and myofibrillar myopathy.11-13 This diversity reveals that filamins perform a variety of essential functions, particularly with respect to the skeletal and cardiovascular systems. Furthermore, although it appears that complete loss of filamin A (FLNa) is usually lethal during embryonic development, the similarities between some phenotypes associated with FLNa and filamin B (FLNb) missense mutations and the overlap in their tissue distribution and binding partners suggest the potential for functional redundancy between these isoforms.
Here we show that ASB2 ubiquitin ligase activity drives proteasome-mediated degradation of the actin-binding proteins FLNa and FLNb. This reveals a novel mechanism for controlling filamin levels through ubiquitin-mediated proteasomal degradation that has the potential to impact a wide range of filamin-dependent processes.
Methods
Cell lines, culture conditions, and measurements of differentiation
NB4 and PLB985 cells were used as described.2 HeLa and HEK293T cells were grown in Dulbecco modified Eagle medium (DMEM) containing 4.5 g/L glucose (Invitrogen, Carlsbad, CA) and 5% fetal bovine serum (PAA Laboratories, Coelbe, Germany). NIH3T3 cells were grown in DMEM containing 4.5 g/L glucose and 5% newborn calf serum (PAA Laboratories). HT1080 cells were grown in DMEM containing 4.5 g/L glucose and 10% fetal calf serum (Atlanta Biologicals, Norcross, GA).
Exponentially growing NB4 and PLB985 cells were seeded at 2 × 105 and 1 × 105 cells/mL 16 hours before all-trans RA treatment, respectively. Cell viability was evaluated by direct cell counting (trypan blue dye exclusion method). Differentiation was assessed by: (1) cell morphology on cytospin slides stained with May-Grünwald-Giemsa (Sigma-Aldrich, St Louis, MO), (2) the percentage of cells with nitro blue tetrazolium (Sigma-Aldrich) deposits, and (3) the percentage of CD11b-positive cells and fluorescence intensity by FACScan (BD Biosciences, San Jose, CA) using PC5-conjugated anti-CD11b antibodies (Beckman Coulter, Fullerton, CA). For proteasome inhibition, cells were incubated with 0.5 or 1 μM MG132 or 20 μM LLnL (Euromedex, Souffelweyersheim, France).
FLNa and FLNb double knockdown HT1080 cells (FLNabKD) were obtained by transfecting HT1080 wild type (WT) with short hairpin RNA (shRNA) against human FLNa (in pSM2c vector; Open Biosystems, Huntsville, AL). After 2 days, the transfected cells were selected using 4 μg/ml puromycin. After selection FLNa knockdown cells were transfected with shRNA against human FLNb (in pGIPZ vector; Open Biosystems) and 2 days later transfected cells were selected using 4 μg/mL puromycin and 1 mg/mL hygromycin.
For cell-spreading assays, 24 hours after transfection, NIH3T3 cells were trypsinized, washed in phosphate-buffered saline (PBS), and incubated in suspension for 1 hour at 37°C in serum-free MEM containing 0.2% bovine serum albumin (BSA). Cells were then plated on fibronectin-coated slides (BD Biosciences) for 45 minutes. Cells were considered spread if the cytoplasmic surface area was at least twice the nuclear surface area. HT1080 WT, HT1080 transfected with green fluorescent protein (GFP)–ASB2wt, and HT1080 FLNab KD were trypsinized, replated on glass coverslips coated with 5 μg/mL fibronectin (Sigma-Aldrich), and fixed with 4% paraformaldehyde in PBS at the indicated time. Images were then taken in differential interference contrast (DIC) using a 40× objective and cell area was measured using ImageJ software.
Plasmid constructs
Construction of an ASB2LA-mutated plasmid was achieved using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). For this, the forward mutated oligonucleotide sequence was (mutated bases in bold): 5′-CTCCAAGACCTGCGGCTCACCTTTGCCG-3′. Deletion of the SOCS box (amino acids 545-587) was generated by polymerase chain reaction (PCR) amplification. ASB2 WT, LA, and ΔSOCS fragments were subcloned into: (1) the pEGFP-C3 expression vector (Clontech, Mountain View, CA); (2) the pDsRed-monomerC1 expression vector (Clontech); (3) pMTCB6+-derived expression vectors to direct the expression of ASB2 fused to the FLAG epitope or to GFP at its amino terminus under the control of the zinc-inducible sheep metallothionein promoter2 ; and (4) the pBacPAK8 plasmid (Clontech). The pSG5FN-ASB2 expression vector was as described.5 The Rbx2 coding sequence (RZPD, Berlin, Germany) was subcloned into the pBacPAK8 vector. The human pcDNA3-FLNa-GFP14 and pCl-puro-FLNb-GFP15 expression constructs have been used previously. Human ubiquitin was subcloned into the pCMV3Tag7 vector (Stratagene) to direct the expression of ubiquitin fused to 3 Myc-tag at its amino terminus.
Specific silencing of ASB2 was achieved using a shRNA-expressing vector. Nucleotides 96 to 114 (sh no. 1) and 1370 to 1388 (sh no. 2) of the hASB2 coding sequence were chosen as target for shRNA. The small interfering RNA (siRNA) sequences were used to construct 60-mer shRNA oligonucleotides, which were then synthesized (MWG Biotech, Ebersberg, Germany) and ligated into the pSUPER.neo.gfp expression vector (Oligoengine, Seattle, WA) under the control of the H1 promoter. The following oligonucleotides were used (underlined, sense and antisense sequences; bold, restriction enzyme sites; nonbold italics, polIII termination signals; bold italics, loop with linker): sh no. 1: 5′-GATCCCCCGAACATCGACGCCTATATTTCAAGAGAATATAGGCGTCGATGTTCGTTTTTGGAAA-3′, sh no. 2: 5′-GATCCCCGCACGAGGCCGCATACTATTTCAAGAGAATAGTATGCGGCCTCGTGCTTTTTGGAAA-3′.
All constructs were verified by DNA sequencing.
In vivo expression and protein extracts
PLB985 cells were transfected using the nufleofector T solution and the O17 program, as recommended by the manufacturer (Amaxa, Gaithersburg, MD). Cells were then cultured for 48 hours before selection with 0.4 mg/mL G418 (Invitrogen). HeLa and NIH3T3 cells were transfected using Jet PEI (PolyPlus Transfection, New York, NY). Cells were washed twice in PBS one time and resuspended in whole cell extract buffer (50 mM Tris-HCl, pH 7.9, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 0.1% Nonidet P40 [NP40], 10% glycerol, 1 mM dithiotreitol, 1 mM Na3VO4, 50 mM NaF, and 1% protease inhibitor cocktail (P8340; Sigma-Aldrich)). After 2 freeze-thaw cycles in liquid nitrogen, the resulting cell lysates were cleared by a 10-minute 20 000g centrifugation at 4°C. HEK293T and HT1080 WT cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 0.5 M NaCl, 10 mM MgCl2, protease inhibitors cocktail) and then cleared by a 10-minute 15 000g centrifugation at 4°C.
Antibodies
The serum raised against ASB2 (1PNA) has been described previously.2 Anti-Flag (2EL-1B11) was purchased from Euromedex. The mouse monoclonal antibody to polyubiquitinylated proteins (clone FK1), anti-FLNa (clones PM6/317 and TI10), and anti–non-muscle myosin II heavy chain A were from BIOMOL International (Plymouth Meeting, PA), Millipore (Billerica, MA), and Covance (Princeton, NJ), respectively. Polyclonal anti-FLNb was purchased from Chemicon International (Temecula, CA) and anti-GFP was from Rockland Immunochemicals (Gilbertsville, PA). The antihuman FLNa antiserum, which cross-reacts with mouse FLNa, was raised in rabbits.16
Immunofluorescence
Cells were fixed in 4% paraformaldehyde in PBS supplemented with 15 mM sucrose and permeabilized with 0.1% Triton X-100. After blocking with 3% BSA in PBS, immunostaining of cells was performed using appropriate dilutions of antibody solutions as follows: antibodies to polyubiquitinylated proteins in 1:10 000, human FLNa in 1:5000, mouse FLNa in 1:1000, nonmuscle myosin IIA in 1:5000, and FLNb 1:200. F-actin was visualized with Alexa 568- or rhodamine-phalloidin (Fisher Scientific, Pittsburgh, PA) in 1:400. Secondary antibodies used were Alexa Fluor 488 coupled to goat anti-rabbit, and Alexa 633 or Cy3 coupled to goat anti-mouse. Preparations were mounted in mowiol (Calbiochem, San Diego, CA). When indicated, the numbers of cells were counted among a randomly selected pool of at least 100 cells expressing GFP-tagged proteins.
For Figures 1A-C, 2C, 5C, 6B,C, and 7A, slides were viewed with a DM-RE Leica Microscope (Leica Microsystems, Rueil Malmaison, France). Images were acquired through an I2 filter-set and N2.1 filter set (Leica Microsystems) using 40×/1.00-0.5 PL or 100×/1.4-0.7 PL oil objectives and a CoolSnap HQ CCD camera (Photometrics, Roper Scientifics, Evry, France) driven by MetaView (Roper Scientifics) and were processed with the open source software ImageJ 1.37c (National Institutes of Health [NIH], Bethesda, MD) and Adobe Photshop CS2 version 9.0 software (Adobe Systems, San Jose, CA).
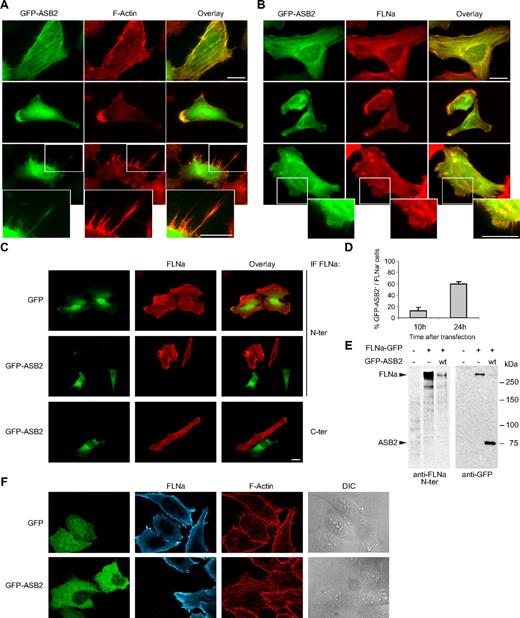
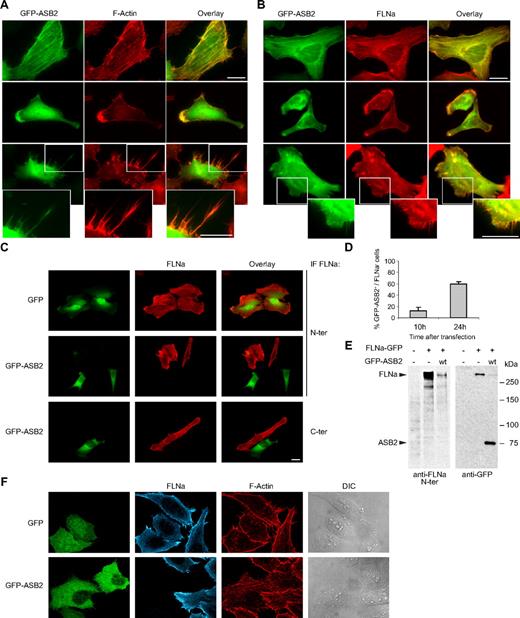
ASB2 induces FLNa degradation. GFP-tagged ASB2 colocalizes with F-actin (A) and FLNa (B) in stress fibers (top), lamellae (middle), and filopodiae (bottom). HeLa cells were imaged 10 hours after transfection with GFP-ASB2 expression vector. Actin was detected with Alexa 568-phalloidin and FLNa with an antibody against its N terminus. Higher magnification views of the lower panels are also shown (bottom). (C) HeLa cells were transfected with GFP or GFP-ASB2 expression vectors as indicated; 24 hours later, FLNa was localized by staining the cells with antibodies directed against the N terminus (N-ter) or C terminus (C-ter) of the protein. Scale bars represent 20 μm. (D) FLNa-negative, GFP-ASB2-positive HeLa cells were counted 10 and 24 hours after transfection among a randomly selected pool of at least 100 cells expressing GFP-ASB2. Results are mean plus or minus SEM from 3 independent experiments. (E) NIH3T3 cells were mock-transfected or transfected with FLNa-GFP and GFP-ASB2 expression vectors for 24 hours, as indicated; 20-μg aliquots of whole cell extracts were immunoblotted with antibodies directed against the N-ter of human FLNa and GFP. Antibodies to human FLNa do not recognize endogenous mouse FLNa. (F) Triple staining showing F-actin organization and FLNa expression in HeLa cells expressing GFP or GFP-ASB2.
ASB2 induces FLNa degradation. GFP-tagged ASB2 colocalizes with F-actin (A) and FLNa (B) in stress fibers (top), lamellae (middle), and filopodiae (bottom). HeLa cells were imaged 10 hours after transfection with GFP-ASB2 expression vector. Actin was detected with Alexa 568-phalloidin and FLNa with an antibody against its N terminus. Higher magnification views of the lower panels are also shown (bottom). (C) HeLa cells were transfected with GFP or GFP-ASB2 expression vectors as indicated; 24 hours later, FLNa was localized by staining the cells with antibodies directed against the N terminus (N-ter) or C terminus (C-ter) of the protein. Scale bars represent 20 μm. (D) FLNa-negative, GFP-ASB2-positive HeLa cells were counted 10 and 24 hours after transfection among a randomly selected pool of at least 100 cells expressing GFP-ASB2. Results are mean plus or minus SEM from 3 independent experiments. (E) NIH3T3 cells were mock-transfected or transfected with FLNa-GFP and GFP-ASB2 expression vectors for 24 hours, as indicated; 20-μg aliquots of whole cell extracts were immunoblotted with antibodies directed against the N-ter of human FLNa and GFP. Antibodies to human FLNa do not recognize endogenous mouse FLNa. (F) Triple staining showing F-actin organization and FLNa expression in HeLa cells expressing GFP or GFP-ASB2.
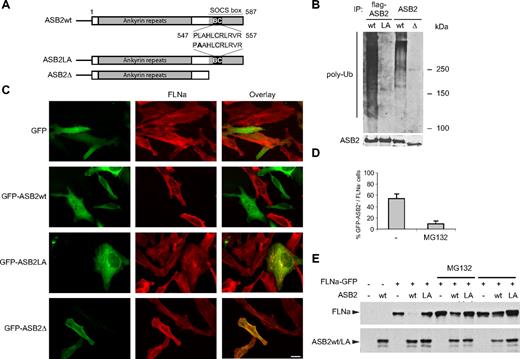
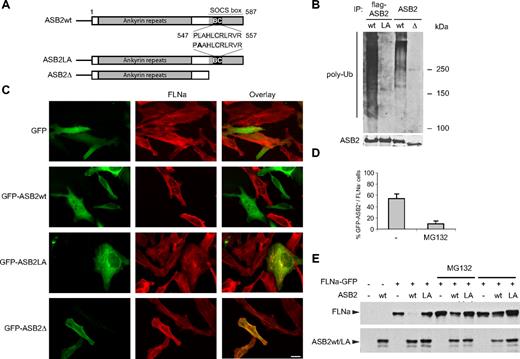
ASB2-induced FLNa degradation is dependent on ASB2 ubiquitin ligase activity and the proteasome. (A) Schematic representation of ASB2, ASB2LA, a BC box mutant, and ASB2-Δ, a deletion mutant lacking the SOCS box. The BC box mutation is indicated in bold. (B) ASB2-Δ and ASB2LA do not activate formation of polyubiquitin chains by the E2 enzyme. Elongin B, elongin C, cullin 5, and Rbx2 together with ASB2wt, ASB2LA, or ASB2-Δ were expressed in Sf21 cells. ASB2 complexes were immunoprecipitated (IP) with anti-flag or -ASB2 antibodies as indicated, and incubated with Uba1, Ubc5a, ubiquitin, and adenosine triphosphate. Their ability to stimulate polyubiquitination was assessed by Western blotting with antibodies to polyubiquitinylated proteins. ASB2 immunoprecipitation was assessed by blotting with anti-ASB2 antibodies. (C) ASB2 ubiquitin ligase activity is required for FLNa degradation. HeLa cells were transfected with GFP, GFP-ASB2wt, GFP-ASB2LA, or GFP-ASB2-Δ expression vectors and analyzed 24 hours after transfection using an antibody directed against the N-ter of FLNa. Scale bar represents 20 μm. (D) ASB2-induced FLNa degradation is dependent on proteasome activity. Ten hours after transfection with GFP-ASB2wt, HeLa cells were left untreated (−) or treated with 1 μM MG132 for 14 hours. Percentages of FLNa-negative GFP-ASB2-positive cells were counted among a randomly selected pool of at least 100 cells expressing GFP-ASB2. Results are mean plus or minus SEM from 3 independent experiments. (E) FLNa-GFP degradation by ASB2 is dependent on proteasome activity. NIH3T3 cells were transfected with FLNa-GFP, Myc-ubiquitin, and DsRed (−), DsRed-ASB2wt (wt), or DsRed-ASB2LA (LA) expression vectors. Seven hours after transfection, cells were left untreated or incubated with 1 μM MG132 or 20 μM LLnL for 17 hours, as indicated. A total of 20-μg aliquots of whole cell extracts were immunoblotted with antibodies directed against the N-ter of human FLNa and ASB2. Antibodies to human FLNa do not recognize endogenous mouse FLNa.
ASB2-induced FLNa degradation is dependent on ASB2 ubiquitin ligase activity and the proteasome. (A) Schematic representation of ASB2, ASB2LA, a BC box mutant, and ASB2-Δ, a deletion mutant lacking the SOCS box. The BC box mutation is indicated in bold. (B) ASB2-Δ and ASB2LA do not activate formation of polyubiquitin chains by the E2 enzyme. Elongin B, elongin C, cullin 5, and Rbx2 together with ASB2wt, ASB2LA, or ASB2-Δ were expressed in Sf21 cells. ASB2 complexes were immunoprecipitated (IP) with anti-flag or -ASB2 antibodies as indicated, and incubated with Uba1, Ubc5a, ubiquitin, and adenosine triphosphate. Their ability to stimulate polyubiquitination was assessed by Western blotting with antibodies to polyubiquitinylated proteins. ASB2 immunoprecipitation was assessed by blotting with anti-ASB2 antibodies. (C) ASB2 ubiquitin ligase activity is required for FLNa degradation. HeLa cells were transfected with GFP, GFP-ASB2wt, GFP-ASB2LA, or GFP-ASB2-Δ expression vectors and analyzed 24 hours after transfection using an antibody directed against the N-ter of FLNa. Scale bar represents 20 μm. (D) ASB2-induced FLNa degradation is dependent on proteasome activity. Ten hours after transfection with GFP-ASB2wt, HeLa cells were left untreated (−) or treated with 1 μM MG132 for 14 hours. Percentages of FLNa-negative GFP-ASB2-positive cells were counted among a randomly selected pool of at least 100 cells expressing GFP-ASB2. Results are mean plus or minus SEM from 3 independent experiments. (E) FLNa-GFP degradation by ASB2 is dependent on proteasome activity. NIH3T3 cells were transfected with FLNa-GFP, Myc-ubiquitin, and DsRed (−), DsRed-ASB2wt (wt), or DsRed-ASB2LA (LA) expression vectors. Seven hours after transfection, cells were left untreated or incubated with 1 μM MG132 or 20 μM LLnL for 17 hours, as indicated. A total of 20-μg aliquots of whole cell extracts were immunoblotted with antibodies directed against the N-ter of human FLNa and ASB2. Antibodies to human FLNa do not recognize endogenous mouse FLNa.
For Figures 4A and 7D,E, slides were viewed with a Nikon Eclipse TE2000-E inverted microscope (Nikon, Melville, NY) using a Nikon Plan Fluo objective at 40×/0.60 and ProLong Gold antifade reagent (Molecular Probes, Eugene, OR). Images were acquired using IPLab software (Scanalytics, Vienna, VA) and Hamamatsu (Hamamatsu Photonics, Hamamatsu City, Japan) camera model C4742-95-12ER. Images were processed with ImageJ version 1.39 (NIH).
Triple-immunofluorescence staining was performed using biotinylated goat anti-mouse IgG1 (Southern Biotechnology Associates, Birmingham, AL) as the secondary antibody, cells were incubated for 20 minutes with Cy5-conjugated streptavidin (Jackson ImmunoResearch Laboratories, West Grove, PA) and analyzed with a laser scanning confocal microscope (LSM 510; Carl Zeiss, Thornwood, NY) using a 63× oil objective and light source wavelengths of 488, 543, and 633 nm.
Quantification of FLNa expression by fluorescence-activated cell sorting
Cells were fixed in 3% paraformaldehyde in PBS and permeabilized in PMZ-T (1× PBS, 0.2% BSA, 0.3% Triton X-100, and 50 mM NH4Cl). To assess FLNa levels, cells were incubated at room temperature for 1 hour with 2 μg/mL mouse anti-FLNa (clone PM6/317) or anti-mouse IgG1 (BD Biosciences) as isotypic control, and then for 1 hour with Alexa Fluor 647–conjugated anti-mouse antibodies (Invitrogen). Saturating concentrations of anti-FLNa antibodies were used. Samples were analyzed by flow cytometry using a FACScan.
In vitro ubiquitylation assays
Recombinant baculoviruses encoding ASB2LA, ASB2-ΔSOCS, and Rbx2 were generated with the BacPAK baculovirus expression system (Clontech). Baculoviruses encoding ASB2, elongin B, elongin C, and cullin 5 were as described.5 Expression of recombinant proteins in Sf21 insect cells was as previously described.5 Cell extracts were immunoprecipitated with anti-Flag antibodies coupled to agarose beads (Sigma-Aldrich). After 3 washes with binding buffer and 2 washes with a buffer containing 40 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid-NaOH, pH 7.9, 60 mM potassium acetate, 1 mM MgCl2, 0.5 mM ethylenediaminetetraacetic acid, 10% glycerol, 2 mM dithiothreitol, the beads were mixed with 133 nM Uba1, 666 nM UbcH5a, 333 μM ubiquitin (Boston Biochem, Cambridge, MA) in a 20-μL reaction containing 4 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid NaOH, pH 7.9, 6 mM potassium acetate, 5 mM MgCl2, 1 mM dithiothreitol, and 1.5 mM adenosine triphosphate. Reactions were incubated for 1 hour at 30°C. When indicated, 133 nM APP-BP1/Uba3, 1 μM Ubc12, and 25 μM NEDD8 (Boston Biochem) were added. FLNa ubiquitylation assays were performed using immunopurified FLNa as a substrate in the reaction described above in this section. Briefly, 150 μg NB4 cell extracts was precleared by incubating 30 minutes in the presence of protein A-Sepharose beads (GE Healthcare, Little Chalfont, United Kingdom) at 4°C. Anti-FLNa (clone PM6/317) was added to the cell protein extract in a binding buffer adjusted to 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% NP40. After 2 hours of incubation, immunocomplexes were recovered with protein A-Sepharose. After 3 washes with binding buffer, proteins were eluted with 100 mM phosphate buffer, pH 12.5, and buffered to pH 8.5 for in vitro ubiquitylation assays. Reaction products were fractionated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by immunoblotting with anti-FLNa and anti-ASB2 antibodies.
Results
ASB2 expression induces loss of FLNa
As a first step toward identifying ASB2 targets, the subcellular localization of GFP-tagged ASB2 transiently expressed in HeLa cells was assessed by fluorescence microscopy. Ten hours after transfection, ASB2 colocalized with F-actin in stress fibers, lamellae, and filopodiae (Figure 1A). Similar observations were made using DsRed or Flag-tagged ASB2 (data not shown). Thus, actin-associated proteins may be targets for ASB2-mediated degradation.
The subcellular localization of ASB2 is reminiscent of that of the actin-binding protein FLNa; 10 hours after transfection of HeLa cells, endogenous FLNa colocalized with GFP-ASB2 in most cells (Figure 1B). However, 24 hours after transfection, FLNa was undetectable in 59% plus or minus 5% of GFP-ASB2-expressing cells (Figure 1C,D), suggesting that ASB2 may induce FLNa degradation. No loss of FLNa was observed in cells expressing GFP (Figure 1C). Calpain proteolysis of FLNa generates major N- and C-terminal fragments of approximately 190 and 100 kDa, respectively,17 and this is implicated in FLNa-interacting protein (FILIP)–mediated FLNa degradation.18 However, ASB2-induced loss of FLNa staining was observed using antibodies directed against either the N or the C terminus of FLNa, suggesting that the absence of FLNa in ASB2-expressing cells is doubtful to be simply the result of proteolytic cleavage of FLNa (Figure 1C). To confirm these results in another cell type and using biochemical assays, NIH3T3 cells were cotransfected with vectors expressing FLNa-GFP without or with GFP-ASB2. Twenty-four hours after transfection, Western blotting revealed that GFP-ASB2 expression resulted in a loss of FLNa-GFP (Figure 1E). Furthermore, no proteolytic fragments of FLNa-GFP were detected using antibodies directed against the N terminus of FLNa or against the GFP. Although filamins act as actin cross-linkers, no drastic alteration of the actin cytoskeleton was observed in ASB2-expressing cells that have lost FLNa expression (Figure 1F).
ASB2 ubiquitin ligase activity drives proteasome-mediated FLNa degradation
To determine whether E3 ubiquitin ligase activity is required in ASB2-mediated FLNa degradation, we generated ASB2 mutants defective in complex formation. Within the ASB2 SOCS box, the BC box defines a binding site for the elongin BC complex, whereas the Cul5 box determines the binding specificity for the cullin5/Rbx2 module.5,6,19 A deletion mutant lacking the entire SOCS box (ASB2-Δ) and a BC box mutant (ASB2LA) was constructed (Figure 2A). When expressed in Sf21 cells, immunopurified WT ASB2/elongin BC/cullin5/Rbx2 complex stimulated formation of ubiquitin conjugates by UbcH5a (Figure 2B), as previously demonstrated for the ASB2/elongin BC/cullin5/Rbx1 complex.5 However, despite comparable levels in the immunoprecipitations, ASB2-Δ and ASB2LA mutants failed to activate formation of polyubiquitin chains by the E2 enzyme (Figure 2B). The effect of these ASB2 mutants on FLNa levels was then tested. Twenty-four hours after transfection of HeLa cells with GFP-ASB2LA or GFP-ASB2-Δ, levels of endogenous FLNa were unaffected, whereas FLNa could not be detected in 55% plus or minus 7% of cells expressing GFP-ASB2wt (Figure 2C). This suggests that formation of an active E3 ubiquitin ligase complex is required for ASB2-mediated FLNa degradation. Loss of FLNa is mediated via the proteasome as treatment of HeLa cells with the proteasome inhibitor MG132 abolished ASB2-induced FLNa degradation (Figure 2D). To confirm these results, NIH3T3 cells were cotransfected with vectors expressing DsRed, DsRed-ASB2wt, or DsRed-ASB2LA together with an FLNa-GFP expression vector. Twenty-four hours after transfection, Western blotting revealed that DsRed-ASB2wt expression resulted in a loss of FLNa-GFP (Figure 2E), whereas FLNa-GFP levels were not altered in cells expressing ASB2LA. Consistent with results in HeLa cells, proteasome inhibitors blocked FLNa-GFP degradation induced by DsRed-ASB2wt (Figure 2E).
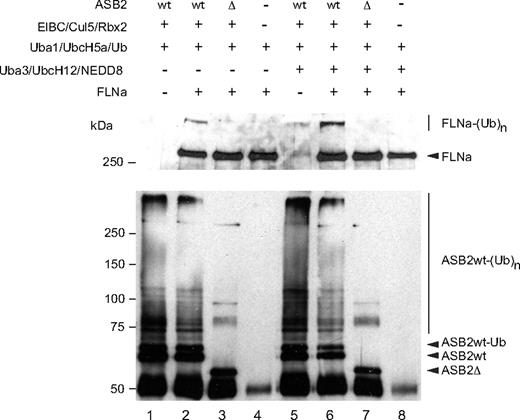
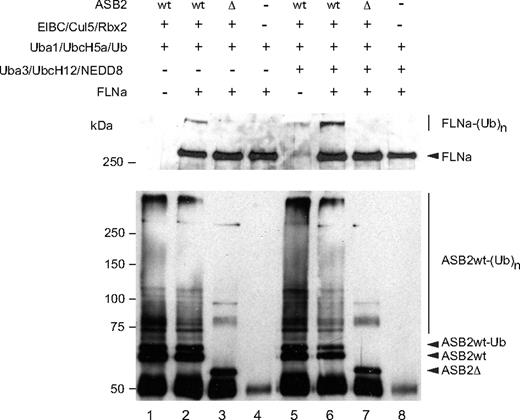
To determine whether ASB2 can promote FLNa ubiquitylation, in vitro, substrate ubiquitylation assays were performed using purified FLNa. Even in the absence of specific substrate, the immunopurified WT ASB2/elongin BC/cullin5/Rbx2 complex stimulated formation of a ladder of ubiquitin conjugates, showing its intrinsic E3 ligase activity. Among the proteins that were polyubiquitylated in this reaction, ASB2 was found to be polyubiquitylated (Figure 3 bottom panel, lanes 1 and 2). However, when FLNa was used as a substrate, ubiquitylation of FLNa by UbcH5a in the presence of the ASB2/Elongin BC/Cullin5/Rbx2 complex, but not in the presence of ASB2-Δ, was observed (Figure 3 top panel, lanes 2 and 3). Because NEDD8 modification of cullins activates the ubiquitin ligase activity of cullin-based E3 ligases20-22 and because cullins are not neddylated after expression in insect cells, we wanted to determine whether ASB2-induced FLNa ubiquitylation was enhanced in the presence of the 3 components of the neddylation machinery, APP-BP1/Uba3, Ubc12, and NEDD8. As can be seen in Figure 3, addition of the NEDD8 machinery stimulated ubiquitin conjugation significantly and resulted in increased conversion of FLNa into high molecular mass ubiquitin adducts (Figure 3 top panel, lane 6). Thus, ASB2 ubiquitin ligase activity drives ubiquitin-mediated proteasomal degradation of FLNa.
ASB2, but not ASB2LA, induces polyubiquitylation of FLNa. Recombinant ASB2/elongin BC/cullin5/Rbx2 complexes were purified as in Figure 2B. All samples contained purified Uba1, UbcH5a, and ubiquitin. The 3 components of the NEDD8 pathway were added as indicated. Purified FLNa was also provided as indicated and subjected to ubiquitylation. Aliquots of the reaction mixture were analyzed by Western blotting using anti-FLNa (upper panel) and anti-ASB2 antibodies (lower panel).
ASB2, but not ASB2LA, induces polyubiquitylation of FLNa. Recombinant ASB2/elongin BC/cullin5/Rbx2 complexes were purified as in Figure 2B. All samples contained purified Uba1, UbcH5a, and ubiquitin. The 3 components of the NEDD8 pathway were added as indicated. Purified FLNa was also provided as indicated and subjected to ubiquitylation. Aliquots of the reaction mixture were analyzed by Western blotting using anti-FLNa (upper panel) and anti-ASB2 antibodies (lower panel).
ASB2 expression induces proteasomal degradation of FLNb
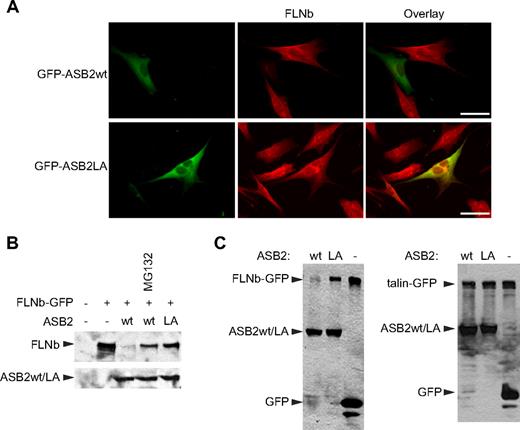
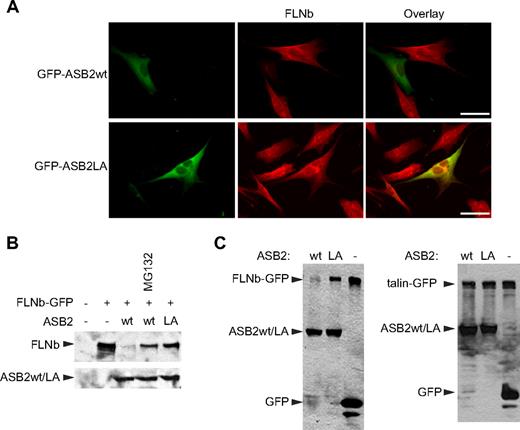
To test whether ASB2 can also induce proteasomal degradation of the other widely expressed mammalian filamin, FLNb, NIH3T3 cells were transfected with vectors expressing GFP-ASB2wt or GFP-ASB2LA. Twenty hours after transfection, FLNb was undetectable in 76% plus or minus 5% of GFP-ASB2wt–expressing cells, whereas no loss of FLNb was observed in cells expressing GFP-ASB2LA (Figure 4A). Furthermore, coexpression of GFP-ASB2wt, but not GFP-ASB2LA, resulted in the reduction of FLNb-GFP levels (Figure 4B). As observed for FLNa, ASB2-induced loss of FLNb-GFP was prevented by proteasome inhibition (Figure 4B). Thus, ASB2 targets both FLNa and FLNb for degradation. ASB2 does, however, show specificity as no degradation of another coexpressed actin-binding protein talin-GFP was observed (Figure 4C), nor were levels of the endogenous actin-binding proteins vinculin or non-muscle myosin heavy chain IIA affected (data not shown).
ASB2-induced FLNb degradation is dependent on ASB2 ubiquitin ligase activity and the proteasome. (A) ASB2-induced FLNb degradation depends on ASB2 ubiquitin ligase activity. NIH3T3 cells were transfected with GFP-ASB2wt or GFP-ASB2LA expression vectors, replated on coverslips coated with 5 μg/mL fibronectin 5 hours after transfection, fixed 15 hours after replating, and analyzed using an antibody directed against FLNb. Scale bar represents 50 μm. (B) FLNb-GFP degradation by ASB2 is dependent on proteasome activity. NIH3T3 cells were transfected for 24 hours with FLNb-GFP together with the GFP (−), GFP-ASB2wt (wt), or GFP-ASB2LA (LA) expression vectors in the absence or presence of 1 μM MG132 for 18 hours, as indicated. (C) HEK293T cells were transfected for 24 hours with FLNb-GFP (left) or GFP-talin (right) together with the GFP (−), GFP-ASB2wt (wt), or GFP-ASB2LA (LA) expression vectors, as indicated. In panels B and C, 20-μg aliquots of whole cell extracts were immunoblotted with antibodies to GFP.
ASB2-induced FLNb degradation is dependent on ASB2 ubiquitin ligase activity and the proteasome. (A) ASB2-induced FLNb degradation depends on ASB2 ubiquitin ligase activity. NIH3T3 cells were transfected with GFP-ASB2wt or GFP-ASB2LA expression vectors, replated on coverslips coated with 5 μg/mL fibronectin 5 hours after transfection, fixed 15 hours after replating, and analyzed using an antibody directed against FLNb. Scale bar represents 50 μm. (B) FLNb-GFP degradation by ASB2 is dependent on proteasome activity. NIH3T3 cells were transfected for 24 hours with FLNb-GFP together with the GFP (−), GFP-ASB2wt (wt), or GFP-ASB2LA (LA) expression vectors in the absence or presence of 1 μM MG132 for 18 hours, as indicated. (C) HEK293T cells were transfected for 24 hours with FLNb-GFP (left) or GFP-talin (right) together with the GFP (−), GFP-ASB2wt (wt), or GFP-ASB2LA (LA) expression vectors, as indicated. In panels B and C, 20-μg aliquots of whole cell extracts were immunoblotted with antibodies to GFP.
Knockdown of ASB2 in myeloid leukemia cells delays RA-induced differentiation and FLNa and FLNb degradation
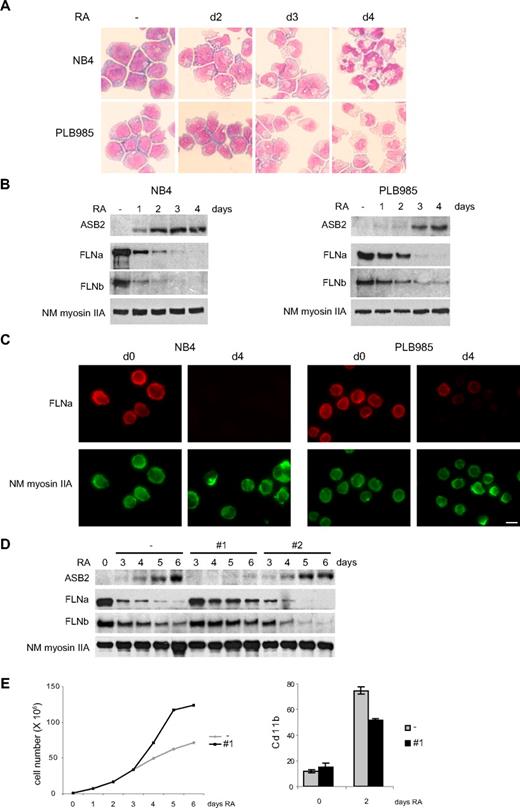
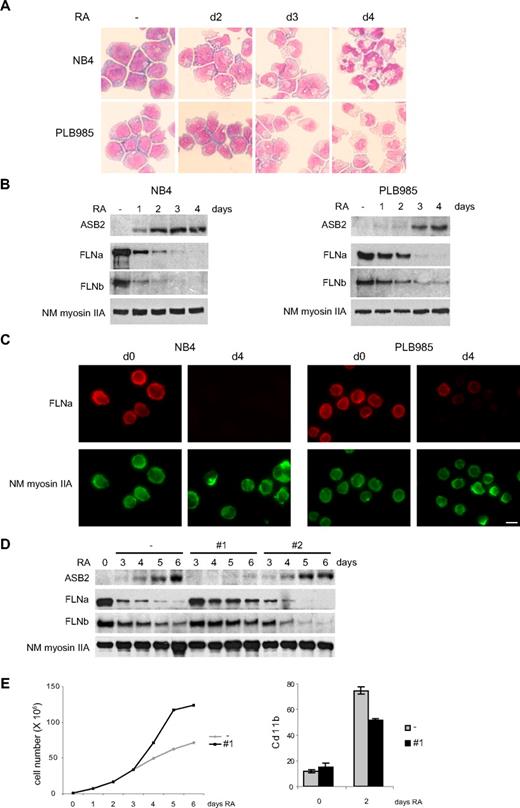
ASB2 was originally identified as an RA-induced gene in APL. We therefore investigated FLNa and FLNb expression in promyelocytic NB4 and myeloblastic PLB985 cells. When cultured with RA, these cells differentiated toward the granulocytic pathway (Figure 5A). Differentiation of PLB985 cells was slower and, accordingly, RA-induced ASB2 expression was delayed in these cells (Figure 5B). In both cell types, ASB2 up-regulation correlated with loss of FLNa and FLNb (Figure 5B), and FLNa was not or barely detectable in the differentiated cells (Figure 5C). To examine the role of ASB2 in filamin degradation during RA-induced differentiation of PLB985 cells, we established stable PLB985 cell lines expressing shRNAs directed against ASB2. In the absence of RA, these lines grow at the same rate as control cells (data not shown). ASB2 knockdown in PLB985 cells cultured in the presence of RA was obtained with sequence 1 (Figure 5D). In these cells, FLNa and FLNb degradation was delayed (Figure 5D). Cells expressing the shRNA no. 2, which did not affect ASB2 expression, showed a kinetic of filamin degradation identical to cells transfected with the empty vector (Figure 5D). Furthermore, ASB2 knockdown delayed the RA-induced growth arrest of PLB985 cells (Figure 5E) and reduced the number of cells expressing the differentiation marker CD11b (Figure 5E). However, the morphology and nitro blue tetrazolium reduction of control and ASB2 knockdown PLB985 cells were similar after 6 days of RA treatment (data not shown). Thus, ASB2 knockdown delays growth arrest and differentiation of RA-treated PLB985 cells and ASB2 controls filamin degradation during RA-induced differentiation of myeloid leukemia cells.
ASB2 induces degradation of FLNa and FLNb in differentiating myeloid leukemia cells. (A-C) Down-regulation of FLNa and FLNb in myeloid leukemia cells induced to differentiate correlates with ASB2 induction. NB4 and PLB985 cells were treated with 10−6 M of all-trans RA. Differentiation was assessed by cell morphology on May-Grünwald-Giemsa-stained cytospin slides (A). Expression of ASB2, FLNa, FLNb, and non-muscle myosin IIA (NM myosin IIA) was analyzed by Western blot using 20-μg aliquots of whole cell extracts (B) and by immunofluorescence (C). (D) ASB2 knockdown delayed RA-induced FLNa and FLNb degradation. Stable PLB985 cell populations expressing shRNAs directed against ASB2 (no. 1 and no. 2) or transfected with the empty vector (−) were treated with 10−6 M RA for different times as indicated; 10-μg aliquots of the protein extracts were analyzed by Western blotting. (E) PLB985 cell populations expressing shRNA no. 1 or transfected with the empty vector (−) were untreated and treated with RA. (Top panel) Representative experiment of 3 showing numbers of viable cells. (Bottom panel) Percentages of CD11b-positive cells. Error bars represent SDs from the results of 3 independent experiments.
ASB2 induces degradation of FLNa and FLNb in differentiating myeloid leukemia cells. (A-C) Down-regulation of FLNa and FLNb in myeloid leukemia cells induced to differentiate correlates with ASB2 induction. NB4 and PLB985 cells were treated with 10−6 M of all-trans RA. Differentiation was assessed by cell morphology on May-Grünwald-Giemsa-stained cytospin slides (A). Expression of ASB2, FLNa, FLNb, and non-muscle myosin IIA (NM myosin IIA) was analyzed by Western blot using 20-μg aliquots of whole cell extracts (B) and by immunofluorescence (C). (D) ASB2 knockdown delayed RA-induced FLNa and FLNb degradation. Stable PLB985 cell populations expressing shRNAs directed against ASB2 (no. 1 and no. 2) or transfected with the empty vector (−) were treated with 10−6 M RA for different times as indicated; 10-μg aliquots of the protein extracts were analyzed by Western blotting. (E) PLB985 cell populations expressing shRNA no. 1 or transfected with the empty vector (−) were untreated and treated with RA. (Top panel) Representative experiment of 3 showing numbers of viable cells. (Bottom panel) Percentages of CD11b-positive cells. Error bars represent SDs from the results of 3 independent experiments.
ASB2 ubiquitin ligase activity drives FLNa and FLNb degradation in myeloid leukemia cells
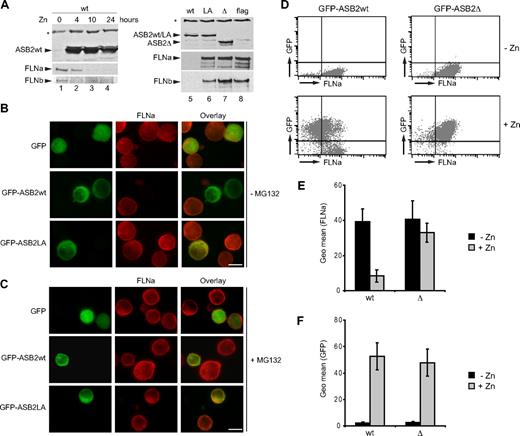
We next investigated PLB985 cells stably expressing empty vector, WT, or mutant ASB2 under the control of the zinc-inducible metallothionein promoter. When zinc was added to the media, ASB2wt increased and endogenous FLNa and FLNb decreased (Figure 6A lanes 1-4). However, although equivalent amounts of ASB2wt, ASB2LA, and ASB2-Δ proteins were expressed, only WT ASB2 induced filamin degradation (Figure 6A lanes 5-8). To visualize ASB2-induced FLNa degradation at the single-cell level, GFP, GFP-ASB2wt, or GFP-ASB2-Δ was transiently expressed in PLB985 cells (Figure 6B). FLNa was undetectable in 90% of the cells expressing GFP-ASB2wt but was unaffected by the ASB2 mutant. FLNa degradation was abolished by MG132 treatment of PLB985 cells (Figure 6C).
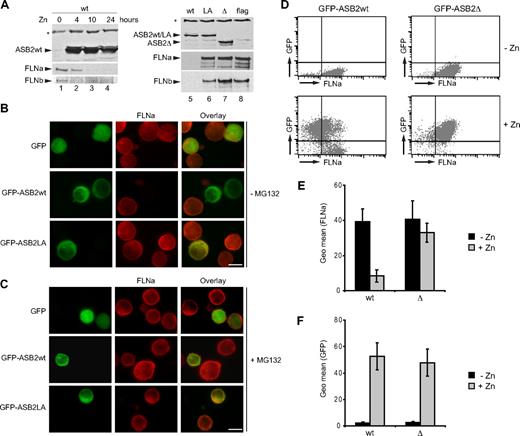
ASB2-induced degradation of FLNa is dependent on ASB2 ubiquitin ligase activity and proteasome in myeloid leukemia cells. (A) ASB2wt was induced in PLB985/MT-ASB2 treated with 65 μM ZnSO4 for the indicated times (lanes 1-4). ASB2wt was induced in PLB985/MT-ASB2 cells treated with 10 μM ZnSO4 (lane 5). ASB2LA (lane 6) and ASB2-Δ (lane 7) were induced in PLB985/MT-ASB2LA and PLB985/MT-ASB2-Δ cells treated with 100 μM ZnSO4. PLB985/MT-Flag cells were used as controls and treated with 100 μM ZnSO4 (lane 8). Protein extracts (20 μg) were separated by SDS-PAGE and immunoblotted for ASB2, FLNa (N-ter), and FLNb. * indicate a nonspecific band. PLB985 cells were nucleofected with the GFP, GFP-ASB2wt, and GFP-ASB2LA expression vectors for 8 hours and treated without (B) or with 0.5 μM MG132 (C). Expression of FLNa was analyzed by immunocytochemistry using an antibody directed against the N-ter of FLNa. Colocalization of ASB2 and FLNa is indicated in the merged image (yellow). Scale bar represents 20 μm. (D-F) Whereas FLNa expression is not influenced by GFP or GFP-ASB2-Δ, FLNa expression is dramatically reduced in GFP-ASB2wt–expressing leukemia cells. GFP, GFP-ASB2wt, and GFP-ASB2-Δ were induced in PLB985/MT-GFP, PLB985/MT-GFP-ASB2wt, and PLB985/MT-GFP-ASB2-Δ cells with 100 μM ZnSO4 for 48 hours, respectively. After permeabilization and fixation, cells were stained with anti-FLNa or anti-IgG1 antibodies and Alexa Fluor 647–conjugated anti-mouse antibodies. A total of 10 000 cells were analyzed by flow cytometry. (D) Dot plots of a representative experiment showing FLNa expression before and after ZnSO4 treatment. (E) Expression of FLNa in GFP–ASB2wt- and GFP–ASB2-Δ–expressing cells. (F) Expression of GFP-ASB2wt and GFP–ASB2-Δ after ZnSO4 treatment. Results are mean plus or minus SEM from 3 independent experiments.
ASB2-induced degradation of FLNa is dependent on ASB2 ubiquitin ligase activity and proteasome in myeloid leukemia cells. (A) ASB2wt was induced in PLB985/MT-ASB2 treated with 65 μM ZnSO4 for the indicated times (lanes 1-4). ASB2wt was induced in PLB985/MT-ASB2 cells treated with 10 μM ZnSO4 (lane 5). ASB2LA (lane 6) and ASB2-Δ (lane 7) were induced in PLB985/MT-ASB2LA and PLB985/MT-ASB2-Δ cells treated with 100 μM ZnSO4. PLB985/MT-Flag cells were used as controls and treated with 100 μM ZnSO4 (lane 8). Protein extracts (20 μg) were separated by SDS-PAGE and immunoblotted for ASB2, FLNa (N-ter), and FLNb. * indicate a nonspecific band. PLB985 cells were nucleofected with the GFP, GFP-ASB2wt, and GFP-ASB2LA expression vectors for 8 hours and treated without (B) or with 0.5 μM MG132 (C). Expression of FLNa was analyzed by immunocytochemistry using an antibody directed against the N-ter of FLNa. Colocalization of ASB2 and FLNa is indicated in the merged image (yellow). Scale bar represents 20 μm. (D-F) Whereas FLNa expression is not influenced by GFP or GFP-ASB2-Δ, FLNa expression is dramatically reduced in GFP-ASB2wt–expressing leukemia cells. GFP, GFP-ASB2wt, and GFP-ASB2-Δ were induced in PLB985/MT-GFP, PLB985/MT-GFP-ASB2wt, and PLB985/MT-GFP-ASB2-Δ cells with 100 μM ZnSO4 for 48 hours, respectively. After permeabilization and fixation, cells were stained with anti-FLNa or anti-IgG1 antibodies and Alexa Fluor 647–conjugated anti-mouse antibodies. A total of 10 000 cells were analyzed by flow cytometry. (D) Dot plots of a representative experiment showing FLNa expression before and after ZnSO4 treatment. (E) Expression of FLNa in GFP–ASB2wt- and GFP–ASB2-Δ–expressing cells. (F) Expression of GFP-ASB2wt and GFP–ASB2-Δ after ZnSO4 treatment. Results are mean plus or minus SEM from 3 independent experiments.
To additionally confirm these data and to provide more detailed information regarding the efficiency of ASB2-mediated FLNa down-regulation in myeloid leukemia cells, the expression of FLNa was determined together with the respective GFP expression by FACS analysis. Figure 6D shows the FLNa expression profile of PLB985/MT-GFP-ASB2 and PLB985/MT-GFP-ASB2-Δ cells cultured with or without ZnSO4. The geometric mean FLNa expression in GFP-ASB2wt–expressing cells was dramatically reduced to 18% plus or minus 2% of that seen in nonexpressing cells, whereas FLNa expression was not influenced by GFP-ASB2-Δ, although similar quantities of ASB2 and ASB2-Δ could be detected by flow cytometry (Figure 6D). Hence, ASB2 ubiquitin ligase activity mediates proteasomal degradation of FLNa in myeloid leukemia cells.
ASB2 regulates integrin-mediated cell spreading through degradation of FLNs
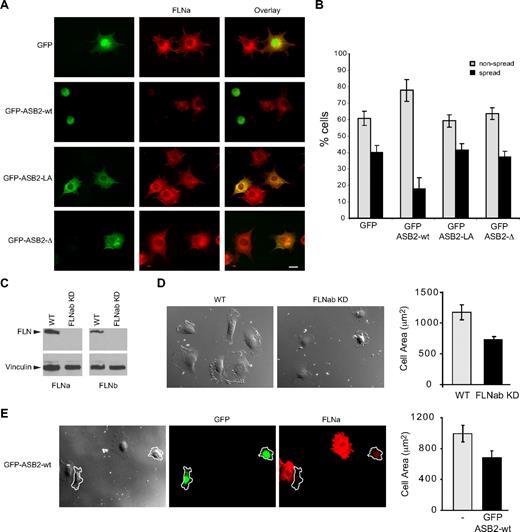
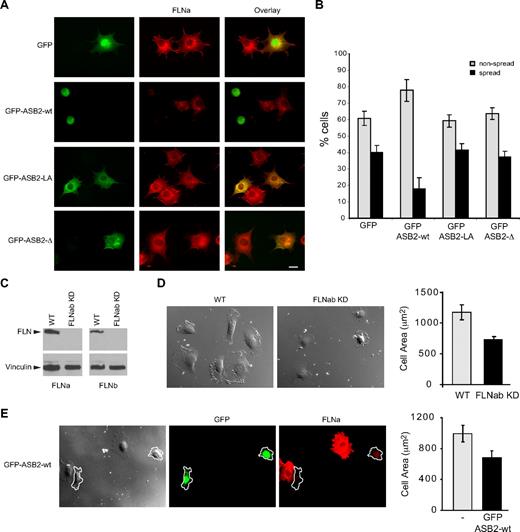
To characterize further the effects of ASB2, NIH3T3 cells were transfected with GFP, GFP-ASB2wt, GFP-ASB2LA, and GFP-ASB2-Δ expression vectors, 24 hours later cells were trypsinized, and then plated on fibronectin-coated slides. Expression of ASB2, but not ASB2 mutants inhibited spreading of NIH3T3 cells 45 minutes after plating on fibronectin-coated slides (Figure 7A,B). Similar results were obtained in HeLa cells (data not shown). To demonstrate the involvement of degradation of FLNs in ASB2-mediated effects on cell spreading, we tested stable FLNa and FLNb knockdown HT1080 cells (Figure 7C). When plated on fibro-nectin, the area of these cells was significantly decreased compared with that of control cells (Figure 7D). Similar observations were made in HT1080 cells expressing WT ASB2 (Figure 7E). Thus, ASB2 regulates integrin-mediated cell spreading through FLNa and FLNb degradation.
ASB2-induced degradation of FLNs inhibits cell spreading on fibronectin. (A,B) NIH3T3 cells were transfected with GFP, GFP-ASB2wt, GFP-ASB2LA, or GFP–ASB2-Δ expression vectors for 24 hours, trypsinized, and serum arrested for 1 hour in suspension. Cells were plated on fibronectin-coated coverslips and fixed after 45 minutes. (A) Cells were stained with anti-FLNa antibodies as indicated. Scale bar represents 20 μm. (B) Percentages of nonspread (▩) and spread (■) cells are plotted as mean plus or minus SEM from 3 independent experiments. (C) FLNa and FLNb double knockdown in HT1080. (D) Spreading quantification. HT1080 WT or HT1080 FLNabKD cells were replated on fibronectin-coated coverslips and fixed after 40 minutes. Cell areas of at least 100 cells were measured and plotted as mean plus or minus SEM. (E) HT1080 WT were transfected with GFP-ASB2wt and 24 hours later replated on fibronectin-coated coverslips, fixed after 40 minutes, and stained for FLNa. Cell areas were measured and cells expressing GFP-ASB2wt (outlined in the image) were compared with untransfected cells on the same coverslip. Scale bar represents 20 μm. The plot shows the mean area plus or minus SEM.
ASB2-induced degradation of FLNs inhibits cell spreading on fibronectin. (A,B) NIH3T3 cells were transfected with GFP, GFP-ASB2wt, GFP-ASB2LA, or GFP–ASB2-Δ expression vectors for 24 hours, trypsinized, and serum arrested for 1 hour in suspension. Cells were plated on fibronectin-coated coverslips and fixed after 45 minutes. (A) Cells were stained with anti-FLNa antibodies as indicated. Scale bar represents 20 μm. (B) Percentages of nonspread (▩) and spread (■) cells are plotted as mean plus or minus SEM from 3 independent experiments. (C) FLNa and FLNb double knockdown in HT1080. (D) Spreading quantification. HT1080 WT or HT1080 FLNabKD cells were replated on fibronectin-coated coverslips and fixed after 40 minutes. Cell areas of at least 100 cells were measured and plotted as mean plus or minus SEM. (E) HT1080 WT were transfected with GFP-ASB2wt and 24 hours later replated on fibronectin-coated coverslips, fixed after 40 minutes, and stained for FLNa. Cell areas were measured and cells expressing GFP-ASB2wt (outlined in the image) were compared with untransfected cells on the same coverslip. Scale bar represents 20 μm. The plot shows the mean area plus or minus SEM.
Discussion
We have identified FLNa and FLNb as the first ASB2 targets and shown that ASB2 triggers ubiquitylation and proteasome-mediated degradation of these proteins in physiologically relevant settings. To our knowledge, this is the first example of FLNa and FLNb regulation via proteosomal degradation pathways and provides a mechanism by which ASB2 may modulate the many cytoskeletal and signaling pathways downstream of filamins. This further suggests that filamin degradation may play a role in ASB2-mediated hematopoietic cell differentiation.
As target cells for transformation in APL are probably a committed myeloid progenitor,23 it is probable that genes involved in myeloid differentiation are repressed in APL cells and de novo expressed when these cells are induced to differentiate by RA. We identified ASB2 as a gene activated during RA-induced maturation of APL cells.2,3 ASB2 is also a target gene of the PML–RAR-α oncogenic transcription factor characteristic of APL, and ASB2 expression inhibits growth and promotes commitment, recapitulating an early step critical for differentiation of myeloid leukemia cells.2 Although ASB2 is not sufficient on its own to induce terminal differentiation of myeloid leukemia cells,2 we show that knockdown of endogenous myeloid leukemia cell ASB2 delays RA-induced differentiation. Thus, ASB2 is a gene involved in cell differentiation whose function may be altered in APL.2,3 ASB2 forms part of an E3 ubiquitin ligase complex5,6 and was thought to target proteins for proteosomal degradation. Identification of ASB2 targets and verification that they are indeed targeted for degradation are therefore key to understanding ASB2 function. Here we show that ASB2 targets FLNa and FLNb to proteasomal degradation and that knockdown of endogenous ASB2 in myeloid leukemia cells delays FLNa and FLNb degradation. These suggest that ASB2-induced FLNa and FLNb degradation may regulate signaling pathways that are disrupted during transformation by PML–RAR-α. Indeed, alteration of receptors and subsequent signaling plays a critical role in the pathogenesis of acute myeloid leukemia.24,25 Accordingly, a number of signal transduction pathways are known to be activated during induced maturation of myeloid leukemia cells.25,26 Which pathways downstream of filamin are perturbed and the mechanisms by which ASB2-induced loss of filamin impacts cell differentiation are the subject of ongoing investigations.
We note that we cannot exclude the possibility that other ASB2 targets are also important during ASB2-induced differentiation, although we did not detect effects on talin, vinculin, or myosin heavy chain, and the cytoskeletal organization of ASB2-expressing cells was not grossly perturbed, suggesting that ASB2 does not induce widespread degradation of cytoskeletal proteins. It will nonetheless be important to identify the sites of ubiquitinylation within FLNa and FLNb and so to generate degradation-resistant filamins. However, the number of lysines in these large proteins (156 in FLNa and 173 in FLNb) combined with the possibility that multiple lysines may be modified makes this a major challenge to be addressed in future studies.
As WT ASB2 transiently colocalized with F-actin, ASB2 probably induces degradation of filamins that are associated with F-actin. When filamin is completely degraded, ASB2 is diffuse throughout the cytoplasm, suggesting that the colocalization of ASB2 with F-actin may be the result of ASB2 association with FLNa and/or FLNb. Accordingly, ASB2 mutants that are unable to stimulate filamin degradation accumulate on stress fibers. Surprisingly, despite the proposed roles for filamins in organization of F-actin, no drastic alteration of the actin cytoskeleton was observed in ASB2-transfected cells. However, expression of ASB2, but not ASB2, mutants did inhibit spreading of NIH3T3, HeLa, and HT1080 cells on fibronectin-coated slides. Thus, ASB2 regulates integrin-mediated cell spreading. Furthermore, knockdown of FLNa and FLNb also inhibited cell spreading on fibronectin, demonstrating that the cell-spreading defect of ASB2-expressing cells is the result of its effect on FLNa and/or FLNb degradation. Certain FLN-ligand interactions are regulated by receptor occupancy, phosphorylation, alternative splicing, and autoinhibition,14,15,27-29 but ASB2 represents a novel mechanism for controlling both FLNa and FLNb levels that has the potential to impact a wide range of filamin-dependent processes. Indeed, FLNa acts as a scaffold for signaling molecules involved in actin remodeling, including GTPases such as Rac1, Cdc42, RhoA, and RalA,30,31 Rho guanine nucleotide-exchange factors such as Trio32 and Lbc,33 Rho GTPase-activating proteins such as FilGAP and p190RhoGAP,34,35 p21-activated kinase 1,36 and the Rho kinase.37 Through FLNa degradation, ASB2 may regulate these pathways, thereby leading to the regulation of actin remodelling.
The ASB2 gene was identified as an RA-induced gene in APL cells.2,3 However, ASB2 is specifically expressed in immature normal hematopoietic cells, and its expression is lost in peripheral blood leukocytes2 ; so ASB2 is probably relevant during early hematopoiesis. The absence of ASB2 expression in mature leukocytes is consistent with the expression of FLNs in these cells.38 Cytoskeletal reorganization and response to mechanical stimuli are involved in the control of cell growth, differentiation, and stem cell lineage switching.39 Furthermore, integrin adhesion molecules, which bind directly to filamins, play a major role in anchoring hematopoietic stem cells to the hematopoietic niche during development40 and after transplantation in mice.41,42 In this regard, it is noteworthy that ASB2 can regulate integrin-mediated spreading, and it is tempting to speculate that, by targeting FLNa and/or FLNb to proteasomal degradation, ASB2 may modulate bone marrow homing and/or engraftment of hematopoietic stem cells.
In conclusion, our identification of filamins as targets for ASB2-induced degradation should stimulate future detailed mechanistic studies of the roles of filamins as actin cross-linkers or signaling scaffolds during hematopoiesis and reinforces the view that targeting proteins to proteasomal degradation may be an important step in the control of hematopoiesis.43
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank D. Heard for the design of shRNAs directed against ASB2, J. O'Bryan and A. Sonnenberg for ubiquitin and FLNb-GFP expression vectors, and D. Hudrisier for his generous help in assessing FLNa expression by FACS.
This work was supported by Centre National de la Recherche Scientifique, Université Paul Sabatier, Université Pierre et Marie Curie, National Institutes of Health (grant GM068600-01; D.A.C.), and by grants from the Agence Nationale de la Recherche (Program Jeunes Chercheuses, Jeunes Chercheurs), the Association pour la Recherche sur le Cancer, the Fondation de France, and the Fondation pour la Recherche Médicale (P.G.L.). M.L.H. was a fellow of Ministère de la Recherche et de la Technologie and was supported by the Association pour la Recherche sur le Cancer.
National Institutes of Health
Authorship
Contribution: M.L.H., I.L., M.B., Y.L., Z.R., and P.G.L. designed and performed the research and analyzed and interpreted the data; S.L. performed the research; C.M.-L. and D.A.C. designed the research and analyzed and interpreted the data; and D.A.C. and P.G.L. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pierre G. Lutz, Centre National de la Recherche Scientifique, UMR5089, Institut de Pharmacologie et de Biologie Structurale, 205 Route de Narbonne, F-31077 Toulouse, France; e-mail: Lutz: Pierre.Lutz@ipbs.fr.
References
Author notes
*M.L.H. and I.L. contributed equally to this work.