Abstract
Hemochromatosis is predominantly associated with the HFE p.C282Y homozygous genotype, which is carried by approximately 1 person in 200 in Northern European populations. However, p.C282Y homozygosity is often characterized by incomplete penetrance. Here, we describe the case of a woman who had a major structural alteration in the HFE gene. Molecular characterization revealed an Alu-mediated recombination leading to the loss of the entire HFE gene sequence. Although homozygous for the HFE deleted allele, the woman had a phenotype similar to that seen in most women homozygous for the common p.C282Y mutation. Contrasting with previously reported results in Hfe knockout and Hfe knockin mice, our report gives further evidence that progression of the disease depends on modifying factors.
Introduction
Hemochromatosis (HC) refers to a group of inherited disorders of iron metabolism characterized by progressive iron accumulation in parenchymal cells. If not recognized and treated, this progressive iron loading can lead to tissue damage and severe clinical complications, including cirrhosis, hepatocellular carcinoma, and cardiomyopathy.1
The best-known and predominant form of HC is an adult-onset autosomal recessive condition usually associated with the HFE p.C282Y/p.C282Y genotype. In Northern European populations this genotype is carried by approximately 1 person in 200.2 However, its phenotypic expression is heterogeneous and clearly depends on a balance between accentuating and reducing factors.3-5
Apart from the founder p.C282Y mutation, 21 non-neutral variations have been identified in HFE. Of these, 52% are missense mutations and the remainder includes nonsense mutations (2/21), splicing mutations (2/21), and small deletions (4/21). To date, no large deletions have been reported.2,6
In the present study, we report the case of a woman of Sardinian descent who had a major structural alteration in the HFE gene. Molecular characterization revealed an Alu-mediated recombination causing the loss of the complete HFE gene sequence. Although homozygous for the HFE deleted allele, the woman displayed only a moderate iron overload. This observation does not support previously reported results in Hfe mouse models suggesting that the common p.C282Y mutation does not produces a null allele.7 Rather, it indicates that HFE has a limited role in maintenance of iron homeostasis and emphasizes the importance of modifying factors.8
Methods
Family members
Blood samples from the reported case, her parents, and her children were collected after informed consent was obtained in accordance with the Declaration of Helsinki. The institutional review board of the Centre Hospitalier Universitaire de Brest approved this study.
Reference sequence
DNA from the proband gave an informative result for the 2 HFE flanking genes, namely HIST1H1C and HIST1H4C. This showed that the extent of the deletion was no longer than the chromosomal region bordering the HFE gene. The sequence encompassing these 2 genes, from nucleotide 26163000 to 26213000 of chromosome 6, was extracted from the University of California Santa Cruz (UCSC) human genome browser (http://genome.ucsc.edu; March 2006 assembly), and this contig was used to design primers to map the deletion. The sequence was filtered for regions with homology repetitive DNA sequences using RepeatMasker (Institute for Systems Biology, Seattle, WA).
PCRs and sequencing
Primer sequences and positions (regarding Figure 1B) are presented in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Polymerase chain reaction (PCR) conditions for each experiment (mapping, gap, and rearrangement specific PCR) are available on request. Amplicons were purified and sequenced using the BigDye Terminator version 1.1 chemistry according to the manufacturer's recommendations (Applied Biosystems, Foster City, CA).
Semiquantitative fluorescent multiplex PCR assay
Semiquantitative fluorescent multiplex PCR (QFM-PCR) assay was performed as previously described.9 Conditions specific to HFE are available on request.
Results and discussion
The reported case was a 47-year-old woman of Sardinian descent (Italy). She was referred to us because of a slight but chronic increase in serum ferritin. During a follow-up period of 15 months, the serum ferritin concentration fluctuated between 524 and 554 μg/L (N < 250 μg/L) and the transferrin saturation level ranged from 52% to 55% (N < 40%). The woman suffered no clinical features except for persistent fatigue. Common causes of acquired hyperferritinemia were excluded. Laboratory testing showed normal hematologic constants (hemoglobin, 13.4 g/dL; red blood cells, 4.32 million/mL; white blood cells, 5.7 million/mL; platelets, 228 000/mL; mean corpuscular volume, 93 fL), liver enzymes (alanine aminotransferase, 18 IU/L; aspartate aminotransferase, 23 IU/L; gamma-glutamyltransferase, 20 IU/L), and normal blood sugar parameters (glucose, 0.96 g/L). In addition, the woman denied any history of thalassemia, alcohol abuse, viral hepatitis, blood transfusion, or chronic iron supplementation. She gave birth to 3 children. Her menstrual periods began at the age of 16 and were irregular. She is not yet menopausal. She has never been a blood donor. At present, the woman is included in a phlebotomy program that involves removal of 300 mL of blood every 2 weeks. Under those conditions, the removal of 2.4 g of iron has lowered serum ferritin to below 100 μg/L.
We searched for the p.C282Y (exon 4) and p.H63D (exon 2) HFE variations using the 5′-nuclease allelic discrimination method, or TaqMan (Applied Biosystems, Foster City, CA) method. Because no signal was detected from the case DNA, we tested other PCR reactions. The inability to amplify the HFE exons was confirmed, and positive results were systematically obtained from PCR reactions targeting other loci (data not shown). The existence of a large deletion involving the entire HFE sequence was finally proved by QFM-PCR (Figure S1).
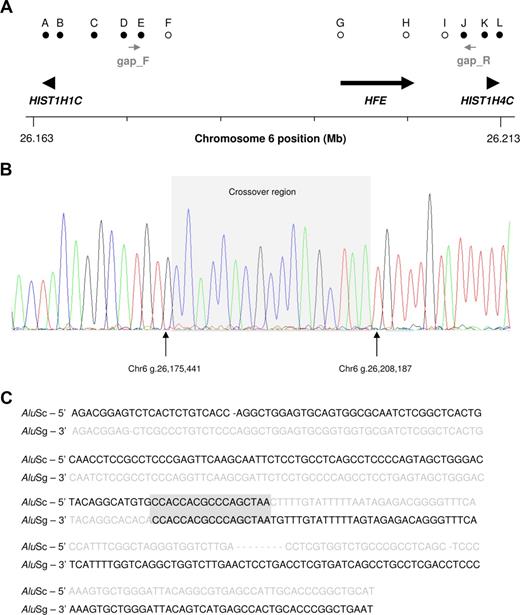
Mapping PCRs were therefore designed to further assess the size of the deletion (Figure 1A). Because primer pair E amplified a product from the case DNA but primer pair F did not, the telomeric breakpoint was presumed to be located within the regions defined by these 2 primer pairs. Likewise, the centromeric end of the deleted segment was localized between primer pairs I and J. These mapping PCRs were consistent with a deletion between 29.3 and 33.7 kb.
Detection and molecular characterization of a large deletion at the HFE locus. (A) Summary of mapping PCRs showing location of primer pairs on the chromosome band 6p21.3. indicate a region deleted in the reported case and indicate an unmodified sequence. Positions and directions of the gap PCR primers are shown in grey. Numbering is based on the UCSC human genome assembly. (B,C) Sequencing electrophoretogram of the recombined allele and sequence alignment of the Alu elements (AluSc in 5′, and AluSg in 3′) responsible for the chromosomal alteration. The gray box highlights a 17 bp track of perfect homology that marks the crossover region. The sequence deleted in the new allele is shown in gray. Interestingly, the intervening sequence contains 2 sequence motifs, CCACCA and CCAGC. Both motifs represent truncated versions of the Chi hotspots (consensus sequence: 5′-GCTGGTGG-3′ or its complement, 5′-CCACCAGC) of generalized recombination.12,13 In particular, CCACCA has been previously reported as a mutational “super-hotspot” common to micro-deletions, micro-insertions, and indels.14
Detection and molecular characterization of a large deletion at the HFE locus. (A) Summary of mapping PCRs showing location of primer pairs on the chromosome band 6p21.3. indicate a region deleted in the reported case and indicate an unmodified sequence. Positions and directions of the gap PCR primers are shown in grey. Numbering is based on the UCSC human genome assembly. (B,C) Sequencing electrophoretogram of the recombined allele and sequence alignment of the Alu elements (AluSc in 5′, and AluSg in 3′) responsible for the chromosomal alteration. The gray box highlights a 17 bp track of perfect homology that marks the crossover region. The sequence deleted in the new allele is shown in gray. Interestingly, the intervening sequence contains 2 sequence motifs, CCACCA and CCAGC. Both motifs represent truncated versions of the Chi hotspots (consensus sequence: 5′-GCTGGTGG-3′ or its complement, 5′-CCACCAGC) of generalized recombination.12,13 In particular, CCACCA has been previously reported as a mutational “super-hotspot” common to micro-deletions, micro-insertions, and indels.14
A gap PCR was subsequently used in an attempt to amplify a DNA fragment containing the breakpoint junction (gap_F and gap_R primers in Figure 1B). An approximately 500 bp fragment was amplified in the reported case that was absent in a control case (as the primer binding sites were too distant for amplification to occur; data not shown). Sequencing of this fragment showed that it contained a gap between the telomeric position chr6 g.26175441 and the centromeric position chr6 g.26208187, signifying a deletion of 32744bp (Chr6 g.(26175442)_g.(26208186) del), which was consistent with the mapping PCR data (Figure 1B). Study of the junction sequence revealed that the deletion originated from an unequal recombination between 2 Alu repeats, leaving a crossover region of 17 bp (Figure 1C).
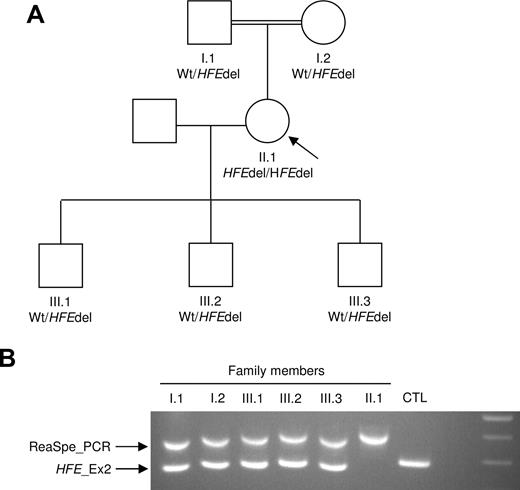
Finally, a rearrangement-specific PCR was designed to analyze segregation of the HFE deletion in family members and to facilitate future investigations. The patient was born to consanguineous parents and has 3 children (Figure 2A). Five relatives in the pedigree were found to be heterozygous for the HFE knocked-down allele (Figure 2B). The remaining copy of the HFE gene in each of the relatives was proved to be free of mutations. The rearrangement-specific PCR data were confirmed by QFM-PCR (Figure S1), while molecular characteristics of the reported Alu-mediated deletion were corroborated by array-comparative genomic hybridization (Figure S2).
Family study and rearrangement specific PCR design. (A) Family tree. The index case is indicated by ↖. Wt indicates wild-type allele; HFEdel, HFE knocked-down allele. (B) Rearrangement-specific PCR of the patient and her relatives. Primers were designed to amplify across the junctions and give a PCR product of 499 bp (ReaSpe_PCR). Also, a PCR was designed to amplify the wild-type sequence; this targets the HFE exon 2 (HFE_Ex2) and produces a 377 bp DNA fragment. Primer pairs were mixed to create a duplex PCR assay.
Family study and rearrangement specific PCR design. (A) Family tree. The index case is indicated by ↖. Wt indicates wild-type allele; HFEdel, HFE knocked-down allele. (B) Rearrangement-specific PCR of the patient and her relatives. Primers were designed to amplify across the junctions and give a PCR product of 499 bp (ReaSpe_PCR). Also, a PCR was designed to amplify the wild-type sequence; this targets the HFE exon 2 (HFE_Ex2) and produces a 377 bp DNA fragment. Primer pairs were mixed to create a duplex PCR assay.
This is the first report of a gross deletion in the context of hemochromatosis. The deletion was not detected in a cohort of 73 iron overload patients (negative or heterozygous for the p.C282Y mutation) of Northern European descent (data not shown). However, future investigations will help to determine whether the deletion is present at the population level, especially in Sardinia, and if it can explain other hemochromatosis cases. Using both Hfe knockout mice and mice homozygous for the p.C282Y mutation (knockin), Levy et al showed that the p.C282Y mutation produced a significant hepatic iron accumulation without resulting in a total lost of function.7 The woman diagnosed in the present study had a moderate iron overload similar to that seen in most women homozygous for the common p.C282Y mutation who do not suffer organ damage due to iron overload.5 This allowed us to argue that, whatever the nature and deleterious effects of the considered HFE mutation, progression of the disease depends on a complex interplay of modifying factors. The polygenic pattern of hepatic iron-loading inheritance has been demonstrated in Hfe knockout mice.10,11 Modifier genes clearly exist in humans but development of iron overload is also influenced by environmental factors,3,4 placing HFE-related hemochromatosis in the complex category of multifactorial traits.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Centre Hospitalier de Brest (Programme Hospitalier de Recherche Clinique) and the Conseil Scientifique de l'Etablissement Français Sang (project 2003.19).
Authorship
Contribution: G.L.G. designed the study and wrote the paper; J.-M.C. executed study of the junction sequence; I.G. was involved in the molecular analyses; S.Q. and C.L.M. assisted in execution of the molecular analyses; C.R., V.G., A.B., and N.P. provided data; and C.F. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Gerald Le Gac, Inserm U613, EFS – Bretagne, 46 rue Félix Le Dantec, 29200 Brest, France; e-mail: gerald.legac@univ-brest.fr.