Abstract
Genome-wide analyses of the relationship between H3 K79 dimethylation and transcription have revealed contradictory results. To clarify this relationship at a single locus, we analyzed expression and H3 K79 modification levels of wild-type (WT) and transcriptionally impaired β-globin mutant genes during erythroid differentiation. Analysis of fractionated erythroid cells derived from WT/Δ locus control region (LCR) heterozygous mice reveals no significant H3 K79 dimethylation of the β-globin gene on either allele prior to activation of transcription. Upon transcriptional activation, H3 K79 di-methylation is observed along both WT and ΔLCR alleles, and both alleles are located in proximity to H3 K79 dimethylation nuclear foci. However, H3 K79 di-methylation is significantly increased along the ΔLCR allele compared with the WT allele. In addition, analysis of a partial LCR deletion mutant reveals that H3 K79 dimethylation is inversely correlated with β-globin gene expression levels. Thus, while our results support a link between H3 K79 dimethylation and gene expression, high levels of this mark are not essential for high level β-globin gene transcription. We propose that H3 K79 dimethylation is destabilized on a highly transcribed template.
Introduction
Posttranslational modifications of histones have been implicated in establishing and maintaining different transcriptional states, although the link of a subset of histone modifications to transcription state is controversial. For example, H3 K79 methylation was originally proposed to be a marker of a permissive transcription state in yeast and is excluded from telomeric regions.1,2 In addition, our genome-wide analysis in Drosophila Kc cells suggests that H3 K79 dimethylation is correlated with transcriptional activation, similar to H3 K4 dimethylation and H3 acetylation.3 However, a recent report using chromatin immunoprecipitation (ChIP)–microsequencing (ChIP-seq) suggests that H3 K79 dimethylation does not show preferential association with either active or silenced regions, and H3 K79 trimethylation is correlated with silencing.4 While these contradictory conclusions may stem from differences in experimental conditions (antibody specificities, cell types, crosslinking and immunoprecipitation conditions, etc), they may also be due to the complex nature of these modifications. H3 K79 dimethylation is induced by a transactivator5 and is a component of the elongation complex (for reviews, see Shilatifard,6 Zhu et al,7 and Osley8 ), suggesting that this mark is involved in the activation of transcription. In contrast, additional data suggests that H3 K79 methylation may not be involved in transcription per se: analyses of cells at different cell-cycle stages suggest that the timing of H3 K79 dimethylation is inversely correlated with pol II and other active marks, such as acetylated H3 and H3 K4 methylation.9 Finally, this mark has also been proposed to be involved in DNA repair pathways (for a review, see Karagiannis and El-Osta10 ). Given the complex nature of the H3 K79 methylation, we decided to investigate it through development and differentiation along wild-type (WT) and transcriptionally impaired mutant β-globin gene loci
The murine β-globin gene locus is a model for studying the molecular mechanisms of gene expression in higher eukaryotes during development and differentiation. The locus contains multiple embryonic and adult β-globin genes. The β-globin genes are expressed highly only in erythroid cells and their expression is regulated by the locus control region (LCR), which consists of several DNase I hypersensitive sites (HSs) spanning 30 to 60 kb upstream of the adult βmajor-globin gene. The LCR, which shows increased colocalization with β-globin promoters upon activation,11-13 is involved in preinitiation complex formation, initiation, and elongation.14,15 The locus often (40%-60%) colocalizes with other erythroid-specific genes at transcription factories upon induction,16 and the LCR plays a role in relocating the entire locus from the periphery to the nuclear interior during maturation, accompanying colocalization of the locus with foci of phosphorylated pol II.15
Here we examine H3 K79 dimethylation along the adult β-globin genes in primary erythroid cells. First, we analyzed unsorted primary cells derived from ΔLCR/WT heterozygous mice17 and found that H3 K79 dimethylation is dramatically increased in the ΔLCR allele. To examine the relationship between expression and modification levels further, we combined a novel method (graduated fractionation of labeled cells [GFLC]), with allele-specific chromatin immunoprecipitation (ChIP),14 and found that H3 K79 dimethylation is correlated with gene activation upon maturation, but the levels are reduced in an LCR-dependent manner. Despite the reduction in modification levels in the WT allele, immunofluorescence in situ hybridization (immunoFISH) analyses reveal that the WT and ΔLCR β-globin alleles colocalize with H3 K79 dimethylation foci at equal frequency. This suggests that although the WT template contains much lower levels of the modification, both alleles are located near nuclear subcompartments containing modified histones. Finally, analyses of primary erythroid cells from mice with a deletion of HS2 and HS3 of the LCR (ΔHS2-3) suggest that an increasing level of transcription results in a reduction of H3 K79 dimethylation. These results raise the possibility that H3 K79 dimethylation is a dynamically regulated mark induced upon transcriptional activation, but is destabilized on a highly transcribed chromatin template.
Methods
Antibodies
This information is described in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
GFLC-AutoMacs
Primary cells from mice heterozygous for a deletion of the β-globin LCR17 were used in this assay. Livers from day-13.5 ΔLCR (HbbD)/WT (HbbS)–H heterozygous fetuses17 were dissociated by pipetting in RPMI media containing 10% fetal calf serum (FCS). In general, one 13.5-day fetal liver contains 10 to 14 million cells, and 50 to 70 livers are used. Cells were filtered, centrifuged, and resuspended in modified AutoMacs buffer (phosphate-buffered saline [PBS] containing 2% FCS and 2 mM EDTA) at 20 million cells/mL. Cells were mixed with an equal volume of modified AutoMacs buffer containing anti–TER-119–PE, anti-CD71 (transferrin receptor)–FITC, and anti–c-kit (CD117)–APC (BD Bioscience, San Jose, CA) simultaneously.18 It is important to use sufficient amounts of the TER-119–PE antibody to achieve efficient fractionation as well as to increase purity. Typically, 1 μL (BD Bioscience; lot no. 21115), 2 μL (lot no. 45705; BD Pharmingen, San Jose, CA), or 2.5 μL (lot no. 5061102070; Miltenyi Biotec, Bergisch Gladbach, Germany) of anti–TER-119–PE per 1 million cells was used. After incubating for 20 minutes on ice, stained cells were washed and resuspended in modified AutoMacs buffer at a final cell concentration of 10 million cells/50 μL. An equal volume of modified AutoMacs buffer containing anti-PE magnetic beads (Miltenyi Biotec) was added, so that the mixture contained 1 μL of beads per 10 million cells. Following bead incubation at 4°C for 15 minutes and washing, cell separation was performed with an AutoMacs Separator (Miltenyi Biotec) using the “possel” mode. Less than 200 million cells were loaded in each run. The positive fraction (fraction 1) was collected and resuspended in 1 mL StemPro media (Life Technologies, Gaithersburg, MD) containing nutrient and cytokines (2 U/mL erythropoietin, 100 ng/mL stem cell factor, and 40 ng/mL insulinlike growth factor 1),19 and stored on ice. The negative fraction was collected and resuspended in modified AutoMacs buffer (10 million cells/50 μL). An equal volume of modified AutoMacs buffer containing magnetic beads was added so that the mixture contained 1 μL of beads per 10 million cells, and processed as described. The positive fraction (fraction 2) was collected, resuspended in 1 mL of StemPro media containing nutrient and cytokines, and stored on ice. The negative fraction was collected and resuspended in modified AutoMacs buffer (10 million cells/50 μL). An equal volume of modified AutoMacs buffer-containing beads was added so that the mixture contained 3 μL of beads per 10 million cells, and processed as described. The positive fraction (fraction 3) was collected, resuspended in 1 mL of StemPro media containing nutrient and cytokines, and stored on ice. The negative fraction was collected and resuspended in modified AutoMacs buffer (10 million cells/50 μL). An equal volume of modified AutoMacs buffer containing beads was added so that the mixture contained 10 μL of beads per 10 million cells, and processed as described. The positive fraction (fraction 4) was collected, resuspended in 1 mL of StemPro media containing nutrient and cytokines, and stored on ice. The negative fraction was collected and resuspended in modified AutoMacs buffer (10 million cells/50 μL). The final fractionation again used 10 μL of beads per 10 million cells, but was done using the “possel-S” mode (slower flow rate). The positive (fraction 5) and negative (fraction 6) fractions were collected, resuspended in 1 mL of StemPro media containing nutrient and cytokines, and stored on ice. All bead binding reactions and washes were performed at 4°C. Cell numbers were determined at each step using a Z2 coulter counter (Beckman Coulter, Fullerton, CA). Isolated fractions were analyzed by flow cytometry (Figure 3; Table S1). Repeat separations yield reproducible profiles of fractionated cells but depend on optimization to assure the same intensity of PE staining to normalize different batches of TER-119–PE antibodies. It should be noted that both temperature and incubation time must be strictly controlled throughout the experiment to obtain reproducible fractionation profiles.
Of note, although cells were fractionated via their ability to bind TER-119 antibody, we found increasing numbers of TER-119− cells in the immature cell fractions (eg, Figure 1A fraction 5). This is likely nonspecific binding to the column due to the high bead-to-cell ratio used for recovering TER-119dim cells in fractions 4 and 5. Regardless, thisTER-119− population is small (approximately 5%) in those populations and thus does not have a significant effect on results.
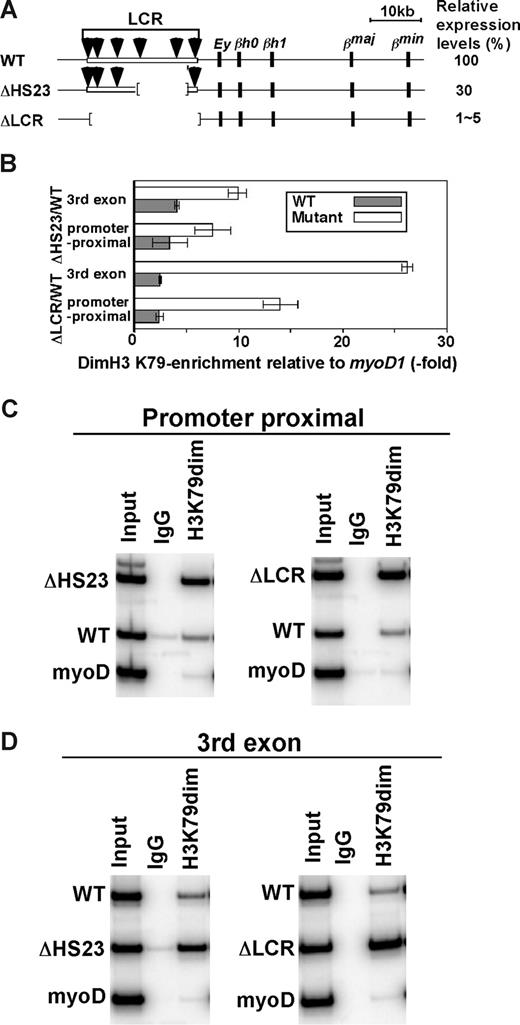
Allele-specific analyses of histone modifications. The promoter proximal region (A) and third exon (B) were analyzed in ΔLCR (HbbD)/WT (HbbS) heterozygous mouse spleen cells. Enrichments of the WT (▩) and ΔLCR (□) adult β-globin gene relative to myoD1 in sample materials relative to those in input materials are plotted. Each bar represents the average of multiple experiments. Error bars represent SD. (C) Allele-specific ChIP in primitive erythroid (EryP) cells derived from ΔLCR (HbbD)/WT (HbbS) ES cells. Enrichment of the WT (▩) and ΔLCR (□) βh1-globin gene relative to amylase in sample materials normalized to input DNA is shown. The regions analyzed were the promoter proximal and third exon of the βh1-globin gene. Each bar represents the average of multiple experiments. Error bars represent SD.
Allele-specific analyses of histone modifications. The promoter proximal region (A) and third exon (B) were analyzed in ΔLCR (HbbD)/WT (HbbS) heterozygous mouse spleen cells. Enrichments of the WT (▩) and ΔLCR (□) adult β-globin gene relative to myoD1 in sample materials relative to those in input materials are plotted. Each bar represents the average of multiple experiments. Error bars represent SD. (C) Allele-specific ChIP in primitive erythroid (EryP) cells derived from ΔLCR (HbbD)/WT (HbbS) ES cells. Enrichment of the WT (▩) and ΔLCR (□) βh1-globin gene relative to amylase in sample materials normalized to input DNA is shown. The regions analyzed were the promoter proximal and third exon of the βh1-globin gene. Each bar represents the average of multiple experiments. Error bars represent SD.
Cytospin preparation and Wright-Giemsa staining
Microscopes and software used in the procedures are described in Document S1.
Allele-specific ChIP
Two alleles of the murine β-globin gene, HbbD and HbbS, were analyzed. We have shown previously that expression levels of the adult β-globin gene from HbbD and HbbS alleles in WT (HbbD)/WT (HbbS) mice are similar.20 In this study, the HbbD allele contains either the ΔLCR or ΔHS23 mutation21 (M.A.B., manuscript in preparation), while the WT (HbbS) allele served as a control. As reported previously, ΔLCR (HbbD)/WT (HbbS) heterozygous fetuses contained one copy of the YAC (H) expressing the human β-globin gene (H).
For studies using adult spleen cells, ΔLCR (HbbD)/WT (HbbS)-H mice were treated with phenyl hydrazine (PHZ) for 3 days to increase the percentage of erythroid cells; spleens were harvested on day 6, as in our previous analyses.14 To compare ΔLCR and ΔHS23 alleles, fetal livers were obtained from 13.5-day embryos. For GFLC allele–specific ChIP, fetal liver cells from each GFLC fraction were cross-linked and processed as described previously.14 For experiments using fetal livers, we did not treat mice with PHZ, since most fetal liver cells are erythroid. These protocols were approved by the Fred Hutchinson Cancer Research Center (FHCRC) Institutional Review Board.
Duplex polymerase chain reaction (PCR) reactions and restriction digests were performed as previously described,14 except that the primers described in Document S1 were used. In allele-specific ChIP assays, it is important to use high-performance liquid chromatography (HPLC)–purified oligos to decrease nonspecific priming. Semiquantitative PCR procedures were as described previously.14,22 Enrichment of the β-globin gene relative to myoD1 or amylase in WT or mutant alleles in sample materials relative to those in control IgG bound materials (or input DNA) from each stage was calculated as described previously.14
RT-PCR analysis of GFLC fraction
This procedure is described in Document S1.
RNA/DNA immunoFISH
Slides for RNA/DNA, immunoFISH were largely prepared as published for primary transcript RNA FISH23 with some modifications. Prior to hybridization, the slides were equilibrated in 50% formamide/2X SSC (pH 7.0). Probe preparation is described in Document S1. Biotinylated (for RNA) and digoxigenin (DNA)–labeled in vitro–transcribed or nick-translated probes were hybridized (20 ng in 50 μL hybridization solution) to the cells in 50% formamide/10% dextran sulfate/2X SSC/5 mM ribonucleotide vanadate complex/0.05% BSA/0.1 mg/mL Cot-1DNA/1 μg/μL Escherichia coli tRNA. Following the addition of the hybridization cocktail to the slides, the coverslips were sealed with rubber cement and cellular and probe DNA denatured by placing the slides on a slidewarmer at 80°C for 5 to 7 minutes. Hybridization proceeded overnight at 37°C in a humid chamber. Slides were washed in 50% formamide/2X SSC (pH 7) at 37°C, rinsed in 2X SSC, and blocked in 145 mM NaCl/0.1M Tris (pH 7.5)/2% BSA/2 mM ribonucleotide vanadate complex. Detection by indirect immunofluorescence including 2 or 3 layers of signal amplification was performed as described.24 For primary transcripts, streptavidin–Alexa Fluor 594 conjugate (S-32356; Molecular Probes/Invitrogen, Carlsbad, CA) and biotinylated antistreptavidin (BA-0500, 1:200; Vector Laboratories, Burlingame, CA) were used. Antidigoxin-FITC (F3523, 1:250; Sigma-Aldrich, St Louis, MO) and anti-rabbit–Cy3 (1:200; Jackson Laboratories, West Grove, PA) were used to detect alleles and H3 K79 dimethylation, respectively.
Microscopy and data processing for FISH analyses
The procedures, microscopes, and software that were used are described in Document S1.
Results
Histone modifications along ΔLCR and WT alleles in primary erythroid cells
We performed an allele-specific ChIP analysis of several “active” histone marks associated with the adult β-globin gene in ΔLCR/WT heterozygous mouse spleen cells during recovery from a PHZ-induced hemolytic anemia in which more than 80% of cells are erythroid.14 This allele-specific assay exploits nucleotide polymorphisms between two β-globin locus haplotypes, HbbS and HbbD, and permits the simultaneous analysis of a WT HbbS allele and a ΔLCR HbbD allele in heterozyotic mice.14,20,25 After ChIP, PCR was performed on DNA recovered from antibody-bound material, and the resulting products were digested with restriction enzymes that distinguish amplification products from the HbbD and HbbS alleles. As both WT and ΔLCR alleles are in the same nuclear environment, this approach allows accurate comparison of subtle differences in histone modification states between the alleles by assuring that observed differences in histone/histone modification enrichment are not due to differences in modifying enzyme concentrations in nuclei, sample preparation, or the maturational and cell-cycle stages of cells.14 In addition, comparison of the WT HbbS allele product to an external myoD control allows comparison between cell fractions.
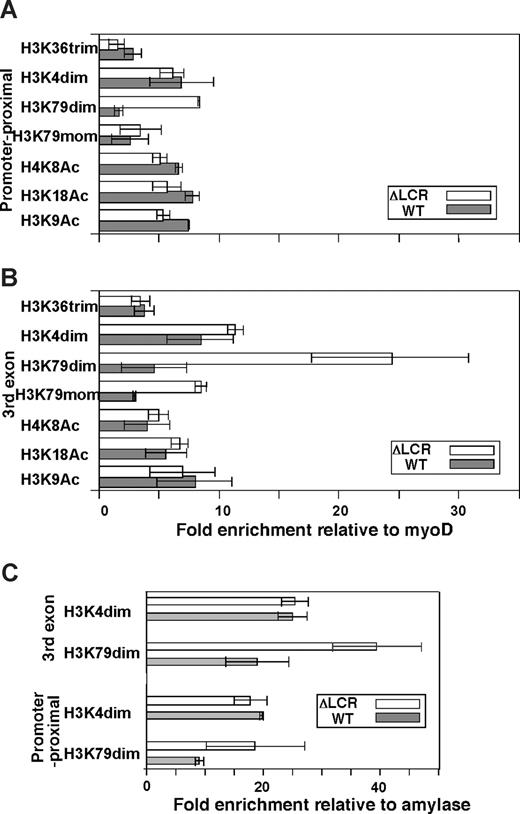
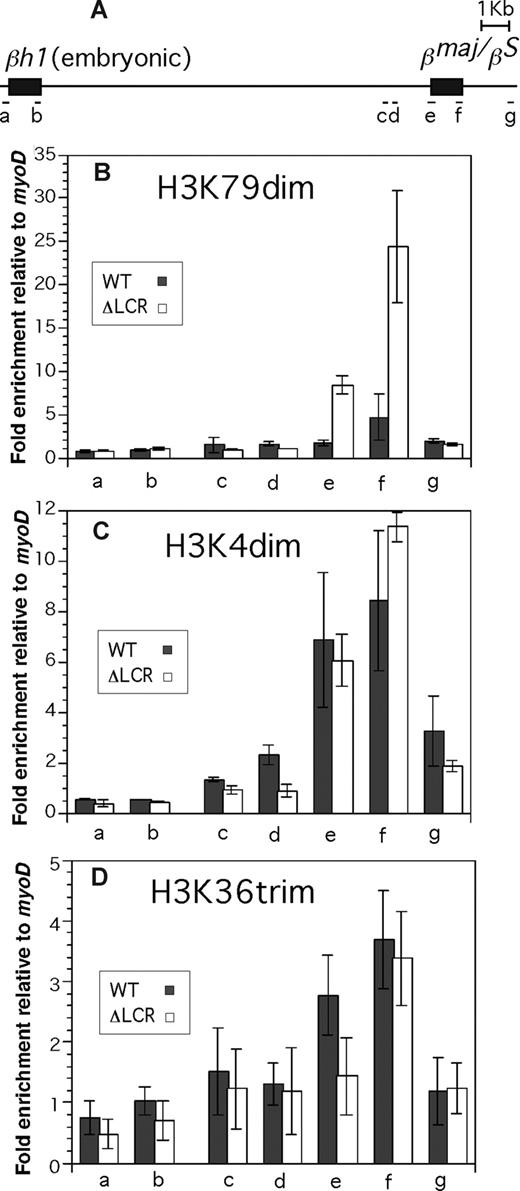
We first analyzed histone acetylation along the βmajor (ΔLCR allele) and βS (WT allele)–globin genes using antibodies against specific single residue modifications of H3 and H4. As shown in Figure 1, acetylated H3 K9 and K18, and acetylated H4 K8 are highly and equally enriched in WT and ΔLCR alleles at both the promoter proximal region (Figure 1A) and the third exon (Figure 1B). These results are consistent with our previous analysis of βmajor-globin genes using antibodies against K9 and/or K14 acetylated H3 and panacetylated H4.25 Further analysis of H3 K4 dimethylation and H3 K79 mono- and dimethylation revealed significant enrichment in both WT and ΔLCR β-globin genes (Figure 1). H3 K36 trimethylation, which is linked to transcriptional elongation, was detected in the third exon of the WT and ΔLCR alleles at similar levels (Figure 1B), but not in the silenced βh1-globin gene (Figure 2D). The modification was modestly but significantly (P < .01) enriched at the promoter proximal region of the WT compared with ΔLCR alleles, consistent with the higher elongation efficiency in WT versus ΔLCR alleles.14,15 Interestingly, while H3 K4 dimethylation enrichment was similar along WT and ΔLCR genes, a 5.4-fold reduction in H3 K79 dimethylation was observed at the promoter-proximal region and third exons of the WT βmajor gene relative to the ΔLCR allele (Figure 1A,B). H3 K79 trimethylation was not detected at either allele (data not shown). Analyses of the embryonic βh1-globin gene and surrounding regions of the adult β-globin gene (Figure 2A) suggest that the peak enrichment of H3 K79 dimethylation is located in the coding region of the active globin gene in the locus (Figure 2B). In contrast, as suggested in previous genome-wide analyses,4,26 H3 K4 dimethylation appears to spread 5′ and 3′ of the active gene (Figure 2C). While neither H3 K79 nor K4 dimethylation was detected in the embryonic βh1-globin gene in adult spleens, both marks were detected in the βh1-globin gene in embryonic stem (ES) cells differentiated to the primitive erythroid lineage where βh1-globin is expressed (Figure 1C). This provides further evidence that both marks are correlated with active transcription. Similar to the adult globin gene in spleens, we found a reduction in H3 K79 dimethylation in the WT compared with ΔLCR allele in the βh1-globin gene in primitive erythroid cells (Figure 1C). As the trimethyl form of H3 K79 was not detected in either allele (data not shown), and the monomethyl form of H3 K79 was also reduced along the WT allele (Figure 1A,B), it is unlikely that the reduction in H3 K79 dimethylation is caused by a conversion from di- to trimethyl or di- to monomethyl forms. Despite the dramatic reduction in this modification along the WT allele, total H3 content was similar in WT and ΔLCR alleles, suggesting that reduction of H3 K79 dimethylation is not simply due to a depletion of histones from the WT allele upon high-level transcription (Table S1). It is possible, however, that active histone replacement27-32 from modified to unmodified H3 may play a role in reducing this mark at the WT allele (“Discussion”).
Enrichment profiles of histone modifications in the βh1 and βmajor/βS globin gene. Allele-specific analyses were conducted in ΔLCR (HbbD)/WT (HbbS) heterozygous mouse spleen cells. (A) The βh1 and the βmajor/βS globin genes are shown. Positions of PCR amplicons (a-g) are shown as bars. Primers and restriction enzymes used in this analysis are described in Document S1. Enrichment of modifications relative to myoD1 in the WT (▩) and ΔLCR (□) alleles is shown. (B) H3 K79 dimethylation. (C) H3 K4 dimethylation. (D) H3 K36 trimethylation. Each bar represents the average of multiple experiments. Error bars represent SD.
Enrichment profiles of histone modifications in the βh1 and βmajor/βS globin gene. Allele-specific analyses were conducted in ΔLCR (HbbD)/WT (HbbS) heterozygous mouse spleen cells. (A) The βh1 and the βmajor/βS globin genes are shown. Positions of PCR amplicons (a-g) are shown as bars. Primers and restriction enzymes used in this analysis are described in Document S1. Enrichment of modifications relative to myoD1 in the WT (▩) and ΔLCR (□) alleles is shown. (B) H3 K79 dimethylation. (C) H3 K4 dimethylation. (D) H3 K36 trimethylation. Each bar represents the average of multiple experiments. Error bars represent SD.
GFLC separates fetal liver into populations varying in their degree of erythroid maturation
Spleen cells derived from PHZ-treated mice are highly enriched for erythroid cells, but they are heterogeneous, containing nonerythroid cells (eg, B-lymphocytes) as well as erythroid cells at different levels of maturation33-35 (data not shown). Erythroid cells display dramatic changes in nuclear morphology, chromatin organization, and expression during differentiation. To determine whether this is associated with alterations in histone modifications, and if so, whether the LCR plays a role, we wanted to re-examine the WT and ΔLCR alleles at defined stages of erythroid maturation. Therefore, we developed GFLC, a novel large-scale surface antigen-based fractionation method allowing for stage-specific ChIP analysis.
Our approach used TER-119, an erythroid-specific antibody that demonstrates increased surface binding during maturation.36 In contrast, surface expression of c-kit (CD117), a marker of immature hematopoietic cells, falls during erythroid maturation, and CD71 (transferrin receptor) increases in erythroid progenitors and then falls with terminal maturation. Flow cytometry using combinations of these markers has led to the small-scale isolation and analysis of erythroid cells at defined stages of maturation,15,18,37 but is impractical for large-scale isolation sufficient for multiple ChIP analyses. Our approach was to stain fetal liver with anti-CD117–APC, anti-CD71–FITC, and TER-119–PE to facilitate characterization of fractions, followed by successive rounds of positive selection for TER-119–staining cells using anti-PE–labeled magnetic beads.
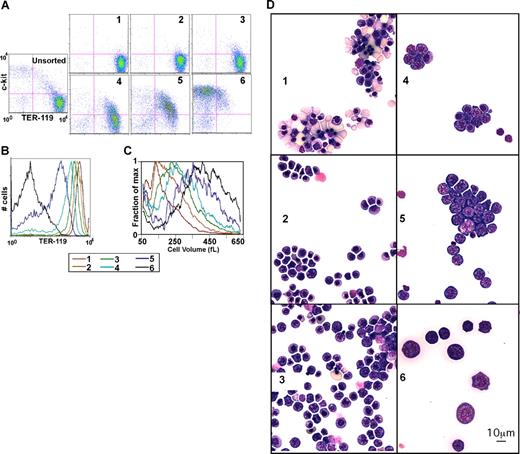
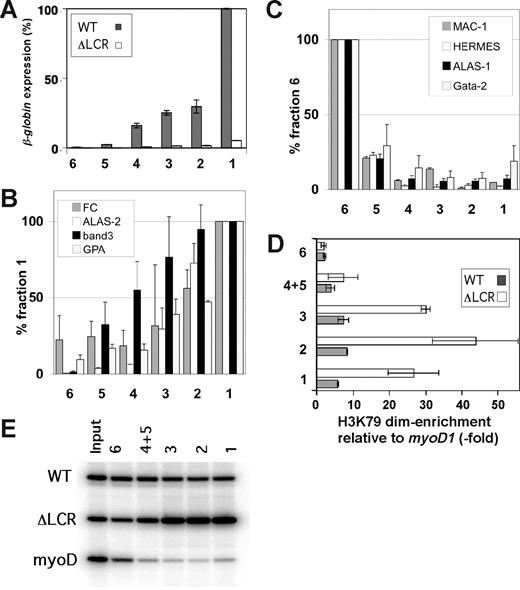
Day-13.5 fetal liver cells were used, as they are primarily erythroid. After triple-staining as discussed, the total fetal liver cell suspension was mixed with limiting amounts of anti-PE magnetic beads (1 μL/10 million cells), resulting in the preferential binding of cells with the highest TER-119 stain (ie, the most mature, erythroid cells). The first fraction obtained is comprised of red blood cells (RBCs), nucleated red cells, and a few orthochromatophilic erythroblasts (Figure 3). After addition of beads, fraction 2 is comprised of more immature cells, primarily orthochromatophilic and polychromatophilic erythroblasts. Similarly, successive rounds with increasing amounts of beads (fractions 3, 4, and 5) yielded increasingly immature cells (Figure 1D). As shown in Figure 1 and consistent with prior descriptions of erythroid maturation, progression from the most immature fraction (fraction 6) to the most mature fraction (fraction 1) is associated with a decrease in the percent of c-kit+ cells, an increase in the percentage of TER-119+ cells, an increase in TER-119 staining intensity (Figure 3A,B; Table S2), a decrease in cell size (Figure 3C), and a 200-fold increase in the level of globin expression (Figure 4A). Similarly, we found that expression levels of erythroid genes such as ferrochelatase (FC), band 3, aminolevulinic acid synthase 2 (ALAS-2), and glycophorin A (GPA) increase with maturation (Figure 4B). In contrast, levels of leukocyte-specific Mac-1, T-lymphoid marker CD44 (HERMES), and ubiquitously expressed aminolevulinic acid synthase 1 (ALAS-1) fall with maturation (Figure 3C). Finally as described previously, we find GATA-2 levels fall with differentiation (Figure 3C; for a review, see Bresnick et al38 ). Thus, morphologic, cell size, surface markers, and expression studies are consistent with GFLC fractionating fetal liver cells into populations varying in their degree of maturation.
GFLC fractionation. (A) Flow cytometric analysis of GFLC-sorted cells. Cells were first gated based on forward and side scatter to eliminate debris. The plots show intensities of c-kit–APC levels on the y-axis and TER-119–PE levels on the x-axis. Pink lines separate the quadrants (eg, c-kit–APC+ plus TER-119–PE+ on the top right, etc). Lines were positioned so that 95% would be in the bottom left (negative) quadrant in the unstained control. (B) Histograms show the TER-119–PE intensities of GFLC fractions. The plots show cell numbers on the y-axis and TER-119–PE levels on the x-axis. (C) Distribution of cell size in GFLC fractions. Fraction of max (y-axis) was estimated as follows: cell numbers at each cell volume were counted and normalized by peak cell numbers in each fraction. Data smoothing was performed (n = 7). (D) Wright-Giemsa–stained cytospin preparation of GFLC fractions. Fraction 1 consists predominantly of enriched enucleated reticulocytes and late orthochromatophilic erythroblasts. Early orthochromatophilic and polychromatophilic erythroblasts become more prominent in fraction 2. Fractions 3 and 4 are enriched in late polychromatophilic and basophilic erythroblasts. Fractions 5 and 6 are enriched in larger cells containing basophilic erythroblasts. Image acquisition information can be found in Document S1.
GFLC fractionation. (A) Flow cytometric analysis of GFLC-sorted cells. Cells were first gated based on forward and side scatter to eliminate debris. The plots show intensities of c-kit–APC levels on the y-axis and TER-119–PE levels on the x-axis. Pink lines separate the quadrants (eg, c-kit–APC+ plus TER-119–PE+ on the top right, etc). Lines were positioned so that 95% would be in the bottom left (negative) quadrant in the unstained control. (B) Histograms show the TER-119–PE intensities of GFLC fractions. The plots show cell numbers on the y-axis and TER-119–PE levels on the x-axis. (C) Distribution of cell size in GFLC fractions. Fraction of max (y-axis) was estimated as follows: cell numbers at each cell volume were counted and normalized by peak cell numbers in each fraction. Data smoothing was performed (n = 7). (D) Wright-Giemsa–stained cytospin preparation of GFLC fractions. Fraction 1 consists predominantly of enriched enucleated reticulocytes and late orthochromatophilic erythroblasts. Early orthochromatophilic and polychromatophilic erythroblasts become more prominent in fraction 2. Fractions 3 and 4 are enriched in late polychromatophilic and basophilic erythroblasts. Fractions 5 and 6 are enriched in larger cells containing basophilic erythroblasts. Image acquisition information can be found in Document S1.
H3 K79 dimethylation levels in the WT/ΔLCR adult globin gene during erythroid maturation. (A) Expression analysis of the WT (▩) and ΔLCR (□) adult β-globin genes in fractionated ΔLCR (HbbD)/WT (HbbS) primary cells. Bars represent the percentages of expression compared with the level of the WT allele in fraction 1. Fraction names are indicated at the bottom. Each bar represents the average of multiple RT-PCRs. Error bars represent SD. (B,C) Real-time RT-PCR analyses for erythroid (B) and non-erythroid (C) genes of GFLC fractions. Experimental values are standardized by HPRT expression levels for each fraction. Standardized values are normalized to fraction 1 (B) and fraction 6 (C). Error bars are SD. (D) Allele-specific ChIP analysis of GFLC-sorted fractions. Enrichment of H3 K79 dimethylation in the third exon of the WT (▩) and ΔLCR (gray circles) adult β-globin genes relative to myoD1 is plotted. Each point represents the average of multiple ChIPs. Error bars represent SD. The gel image is shown in panel E.
H3 K79 dimethylation levels in the WT/ΔLCR adult globin gene during erythroid maturation. (A) Expression analysis of the WT (▩) and ΔLCR (□) adult β-globin genes in fractionated ΔLCR (HbbD)/WT (HbbS) primary cells. Bars represent the percentages of expression compared with the level of the WT allele in fraction 1. Fraction names are indicated at the bottom. Each bar represents the average of multiple RT-PCRs. Error bars represent SD. (B,C) Real-time RT-PCR analyses for erythroid (B) and non-erythroid (C) genes of GFLC fractions. Experimental values are standardized by HPRT expression levels for each fraction. Standardized values are normalized to fraction 1 (B) and fraction 6 (C). Error bars are SD. (D) Allele-specific ChIP analysis of GFLC-sorted fractions. Enrichment of H3 K79 dimethylation in the third exon of the WT (▩) and ΔLCR (gray circles) adult β-globin genes relative to myoD1 is plotted. Each point represents the average of multiple ChIPs. Error bars represent SD. The gel image is shown in panel E.
As expected from the abundance of cell types in fetal liver, the yield of immature cells is greatly reduced compared with that of mature cells. Starting with 50 to 70 fetal livers, recovery ranges from more than 108 cells (fraction 1) to 1 to 2 × 107 cells (fraction 5; data not shown). Fractionations are reproducible as judged by comparison of surface marker profiles (2 independent GFLC profiles are presented in Table S1), although each lot of TER-119 antibody must be titrated (“Methods”).
H3 K79 dimethylation is a marker of gene activation, but does not correlate with expression levels
Allele-specific ChIP assays were performed on the GFLC fractions. With the exception of the most immature cell population (fraction 6), H3 K79 dimethylation is significantly enriched along the adult β-globin genes in both WT and ΔLCR alleles throughout maturation (Figure 4D), and the levels of this modificaiton correlate with the up-regulation of expression. However, the ChIP profile also reveals that the ΔLCR allele exhibits much higher levels of H3 K79 dimethylation than WT at each maturation stage except for the most immature fraction (Figure 3D).
Several studies have indicated that differences in histone modification can be associated with the localization of genes in different nuclear compartments enriched or depleted in those modifications. Thus, we performed 3D DNA-RNA immunoFISH analyses of the localization of the β-globin loci and H3 K79 dimethlyation foci in ΔLCR/WT heterozygous cells in fraction 2, in which the largest difference in K79 enrichment between ΔLCR and WT alleles is observed (Figure 5). Primary transcript signals were used to distinuguish the WT and ΔLCR alleles in the same cell (Figure 5E,F), and only clearly overlapping K79 and globin loci signals were scored as positive. Approximately 55% of WT and 57% of ΔLCR alleles clearly overlap H3 K79 dimethylation foci in these cells (Figure 5G). These results reinforce the conclusion from the ChIP analyses that H3 K79 dimethylation marks the active state of both WT and ΔLCR alleles. While ChIP analyses revealed a decrease of H3 K79 dimethylation in the WT allele, no significant differences in colocalization frequency between the WT and ΔLCR alleles with H3 K79 dimethylation foci were observed by DNA-RNA immunoFISH. This indicates that both WT and ΔLCR alleles are located near foci of modified histones, even though the modifications are not incorporated or maintained to the same extent in the 2 alleles. In combination, while our results are consistent with models in which H3 K79 dimethylation is associated with transcription, the modifi-cation levels do not always correlate with actual expression levels (“Discussion”).
DNA-RNA immunoFISH analyses of GFLC fraction 2 from ΔLCR/WT heterozygous fetal liver cells. 2 Z-sections, which contain allelic signals derived from the same cell, are shown (panels A and B, and panels C and D). Cells were stained by DAPI (A,C), anti–H3 K79 dimethylation (blue in panels B and D). The cells are also hybridized with a β-globin BAC (green in panels B and D) and the second intron of the adult β-globin (red in panels B and D) probes for detecting the loci and β-globin primary transcripts, respectively. Merged images of the alleles (green), primary transcripts (red), and H3 K79 dimethylation foci (blue) are shown. A part of each color channel from merged images in panels B and D is magnified and shown in panels E and F, respectively. Both color (top row) and gray (bottom) images are shown. The WT (green; panels B and E) and ΔLCR (green; panels D and F) alleles in heterozygous cells were identified based on primary transcript signals. Frequencies of colocalization of the WT and ΔLCR alleles are plotted (G). Each bar represents the average of 2 separate data sets (N = 20 + 26 [WT]; N = 28 + 29 [ΔLCR]). Error bars represent SD. Scale bars equal 10 μm. Image acquisition information can be found in Document S1.
DNA-RNA immunoFISH analyses of GFLC fraction 2 from ΔLCR/WT heterozygous fetal liver cells. 2 Z-sections, which contain allelic signals derived from the same cell, are shown (panels A and B, and panels C and D). Cells were stained by DAPI (A,C), anti–H3 K79 dimethylation (blue in panels B and D). The cells are also hybridized with a β-globin BAC (green in panels B and D) and the second intron of the adult β-globin (red in panels B and D) probes for detecting the loci and β-globin primary transcripts, respectively. Merged images of the alleles (green), primary transcripts (red), and H3 K79 dimethylation foci (blue) are shown. A part of each color channel from merged images in panels B and D is magnified and shown in panels E and F, respectively. Both color (top row) and gray (bottom) images are shown. The WT (green; panels B and E) and ΔLCR (green; panels D and F) alleles in heterozygous cells were identified based on primary transcript signals. Frequencies of colocalization of the WT and ΔLCR alleles are plotted (G). Each bar represents the average of 2 separate data sets (N = 20 + 26 [WT]; N = 28 + 29 [ΔLCR]). Error bars represent SD. Scale bars equal 10 μm. Image acquisition information can be found in Document S1.
Inverse correlation between levels of transcription and H3 K79 dimethylation
Our results suggest that H3 K79 dimethylation marks active genes, but, as revealed in the analysis of WT/Δ LCR herozygotes, there is an inverse correlation between this mark and expression. To further examine whether elevating levels of transcription reduce modification levels, we compared histone enrichment in mice heterozygotic for a targeted deletion of LCR HS2 and HS3 (Δ HS23) to the WT and ΔLCR mice. The ΔHS23 mutation results in a 70% reduction in β-globin gene transcription compared with the 95% to 99% reduction seen with the ΔLCR mutation (Figure 6A; M.A.B., unpublished results, May 2002). H3 K79 dimethylation enrichment on the ΔHS23 allele is intermediate to that on the WT and ΔLCR alleles (Figure 6B-D), reinforcing the inverse correlation between β-globin gene expression and H3 K79 dimethylation levels. Modification levels of WT HbbD and HbbS alleles were similar, as expected (data not shown). In combination, our results suggest that H3 K79 dimethylation is destabilized in highly transcribed templates.
H3 K79 dimethylation levels in the WT/ΔHS23 adult globin gene. (A) WT and mutant β-globin gene loci. The HbbD allele is shown. Arrows are DNaseI hypersensitive sites in the LCR. Black boxes represent β-like globin genes. Numbers to the right indicate the levels of expression in primary cells as the percentages of the WT level. (B) Allele-specific analyses of H3 K79 dimethylation enrichment in ΔLCR (HbbD)/WT (HbbS) and ΔHS23 (HbbD)/WT (HbbS) heterozygous mouse fetal liver cells. Enrichment of the WT type (▩) and ΔLCR or ΔHS23 (□) adult β-globin gene relative to myoD is shown. The promoter proximal and third exon of the adult β-globin gene were analyzed. Each bar represents the average of multiple experiments. Error bars represent SD. The gel image is shown in panels C and D.
H3 K79 dimethylation levels in the WT/ΔHS23 adult globin gene. (A) WT and mutant β-globin gene loci. The HbbD allele is shown. Arrows are DNaseI hypersensitive sites in the LCR. Black boxes represent β-like globin genes. Numbers to the right indicate the levels of expression in primary cells as the percentages of the WT level. (B) Allele-specific analyses of H3 K79 dimethylation enrichment in ΔLCR (HbbD)/WT (HbbS) and ΔHS23 (HbbD)/WT (HbbS) heterozygous mouse fetal liver cells. Enrichment of the WT type (▩) and ΔLCR or ΔHS23 (□) adult β-globin gene relative to myoD is shown. The promoter proximal and third exon of the adult β-globin gene were analyzed. Each bar represents the average of multiple experiments. Error bars represent SD. The gel image is shown in panels C and D.
Discussion
GFLC for large-scale fractionation of primary tissue cells
GFLC has several advantages over other techniques for the large-scale separation of subpopulations of cells. First, although it is impractical to use cell sorters to isolate adequate numbers of immature cells for conventional ChIP, sufficient cells from several stages of erythroid maturation are easily obtained by GFLC. In addition, this technique is easily scaled up with no or minimal increase in time. Although there is some overlap in the cell types present in sequential fractions, comparison of alternate fractions reveals distinct populations of cells. Importantly, more than 107 cells were obtained from rare subpopulations (approximately 5%), and this number is sufficient for molecular biology/biochemical analyses, including ChIP, ChIP-microarray, PCR-based chromatin analyses, and mass spectrometry analyses of proteins expressed at more than 1000 molecules per cell.39 Second, although we have separated cells into 6 different maturation stages, this approach could be modified to separate cells at a higher resolution simply by adding smaller amounts of magnetic beads to the negative fraction at each step. While elutriation can provide large-scale separation of primary cells, this may be complicated by the alterations in cell-cycle status, cell size, and cell density that occur during erythroid maturation. Third, by limiting our separation to the differential expression of a single surface antigen (TER-119), we avoid the use of peptidases and additional manipulations required when multiple antibodies are used during magnetic bead-column separation.40,41 Finally, GFLC can be applied to any system in which a cell surface marker is expressed differentially (eg, differentiation, oncogenesis, or changes in cell state such as activation or apoptosis).
Possible pathways for the reduction of H3 K79 dimethylation
There are 2 potential mechanisms for the reduction of H3 K79 methylation in the β-globin locus with high-level expression. Active demethylation by residue-specific histone demethylases has been described, though not for H3 K79 (for a review, see Shi42 ). Our results raise the possibility that a demethylase for K79 of H3 may exist and may be involved in removing methylation from H3 upon high-level transcription. Another possible model that also explains the observed inverse correlation of expression and enrichment of H3 K79 dimethylation involves transcription-dependent histone H3 deposition and turnover.27-32 Unlike several other histone modifying enzymes, Dot1 (or mDot1L), which is responsible for H3 K79 methylation, can only modify histones incorporated into chromatin.1,43,44 Thus, in contrast to H3 K4 dimethylation, there may not be a pool of free premodified H3 K79 dimethylated histones. If histones must be incorporated to be modified, and passage of polymerase complexes displaces histones, one would predict decreased H3 K79 dimethylation in a transcription-level–dependent manner. Our genome-wide analysis in Drosophila cells suggested3 that levels of expression of most active genes are correlated with levels of H3 K79 methylation. However, many of these fly genes may not be driven by strong enhancers such as the LCR in mammalian cells. We suggest that with particularly high rates of enhancer-driven transcription in mammalian cells, as seen with the LCR, histone displacement could become predominant over histone modification, resulting in loss of the modification. Consistent with this model, H3 K4 dimethylation, which occurs efficiently on the free form of core histones in vitro, as well as on incorporated histones,43,45,46 is maintained on highly transcribed WT templates (Figure 1). Thus, the histone modification status of active gene loci reflects several competing and dynamic processes, in addition to simple modification or demodification reactions.
Targets of H3 K79 dimethylation
The inverse correlation between higher-level expression and H3 K79 dimethylation raises the question of how this modification is involved in the activation process. Analyses of this histone mark during the cell cycle may provide some insight into this question. Recent reports suggest that both H3 K79 dimethylation in mammals and H3 K76 dimethylation, the corresponding mark in Typanosoma brucei, occur during M phase.43,47 Levels of H3 K79 dimethylation in several gene loci are up-regulated during the G2/M phase, and reduced during the G1 phase, when pol II recruitment, H3 acetylation, and H3 K4 methylation occur.9 This suggests that H3 K79 dimethylation is involved in propagating the active transcriptional state from G2 to the following G1 phase (ie, short-term “memory”).9
Previously, we proposed that ΔLCR alleles represent an intermediate (or poised) phase of the transactivation process of the β-globin locus. Therefore, an alternative role of H3 K79 dimethylation may be to transiently mark an intermediate phase of transcriptional activation. The transcription activation cycle is composed of numerous steps, including preinitiation complex formation, transcription initiation, pol II pausing, capping, elongation, termination, reinitiation, etc. If H3 K79 dimethylation only transiently marks some of these events, then levels of this modification may be dependent on how long a gene remains at this step, and may not be correlated with actual expression levels. As reported previously,14 in the globin locus, pol II is released from the promoter proximal region of the ΔLCR allele much more slowly than from the WT allele. Thus, an attractive hypothesis is that high-level H3 K79 dimethylation marks this poised state.
Taken together, we propose that this mark is involved in gene activation, but does not have to be maintained in a template when actual transcription is engaged in a template. Multiple competing and dynamic processes, in addition to simple modification or demodification reactions, may determine the modification states.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Mr Tim Waters and Dr Rebecca McHugh (Miltenyi Biotec) for technical advice, and Rachel Byron, Rebecca Carroll (Groudine lab, FHCRC), Weldon E. Debusk, Kristen K. White, and Andrew J. Berger (flow cytometry lab, FHCRC) for technical assistance.
This work was supported by fellowships from American Society of Hematology and the Leukemia & Lymphoma Society (to T.S.), and National Institutes of Health grants DK071868-01 (M.A.B.), DK50107 (E.H.B), DK 44746, and HL57620 (M.G.). H.I. is a Predoctoral Fellow of the American Heart Association.
National Institutes of Health
Authorship
Contribution: T.S. designed and performed the research, analyzed data, and wrote the paper; J.H., H.I., and T.R. performed research; E.H.B. supervised the research; M.A.B. designed the research, analyzed data, and wrote the paper; and M.G. supervised the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mark Groudine, Division of Basic Sciences, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, Mailstop A3-025, Seattle, WA 98109-1024; e-mail: markg@fhcrc.org.

![Figure 5. DNA-RNA immunoFISH analyses of GFLC fraction 2 from ΔLCR/WT heterozygous fetal liver cells. 2 Z-sections, which contain allelic signals derived from the same cell, are shown (panels A and B, and panels C and D). Cells were stained by DAPI (A,C), anti–H3 K79 dimethylation (blue in panels B and D). The cells are also hybridized with a β-globin BAC (green in panels B and D) and the second intron of the adult β-globin (red in panels B and D) probes for detecting the loci and β-globin primary transcripts, respectively. Merged images of the alleles (green), primary transcripts (red), and H3 K79 dimethylation foci (blue) are shown. A part of each color channel from merged images in panels B and D is magnified and shown in panels E and F, respectively. Both color (top row) and gray (bottom) images are shown. The WT (green; panels B and E) and ΔLCR (green; panels D and F) alleles in heterozygous cells were identified based on primary transcript signals. Frequencies of colocalization of the WT and ΔLCR alleles are plotted (G). Each bar represents the average of 2 separate data sets (N = 20 + 26 [WT]; N = 28 + 29 [ΔLCR]). Error bars represent SD. Scale bars equal 10 μm. Image acquisition information can be found in Document S1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/112/2/10.1182_blood-2007-12-128983/4/m_zh80140820880005.jpeg?Expires=1768020593&Signature=wUUloy6lGwmhTDepl-7P162S2RXBPVF45Oe65Yqbmjd1mkXC~~b-6b~TXP~mXpdXrC1X4GJ2sO~Wwb7Tl2e6FYx4RsAoRg17wb-MJEpifc2c-CHqnJIIvvHx48KuebYjHUlraDsU-KyVbEpS--Pyp5PwWM8HphA-RPwbQKXqDPgEx1uwZVhCHJLA68Htnp3WSBxRr13ASkwrNJRS18i-nC12mac8cHcKq6787dpv2TiAG52nJgySK6uIn~UJTeOfhRcB6EMRYQ~keRy2oqd8M3AubzCuXetNdbSwLqJnVCcNd0hJ2LulgvkGBtfNl-T3gaxDdmxlPc5Y68qlaWTVHw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)