Abstract
Warfarin dose requirements have been associated with 2 main haplotypes in VKORC1, but the responsible polymorphisms remain unknown. To search for regulatory polymorphisms, we measured allelic mRNA expression of VKORC1 in human liver, heart, and B lymphocytes. The observed 2-fold allelic mRNA expression imbalance narrowed possible candidate SNPs to −1639G>A and 1173C<T. This genotype effect was observed selectively in the liver but not in heart or lymphocytes. In vitro expression of VKORC1 gene constructs, including coding and promoter regions, failed to reveal any genotype effect on transcription and mRNA processing. Chromatin immunoprecipitation with antibodies against acetyl-histone3 and K4-trimethyl-histone3 revealed preferential association of the promoter −1639 G allele with active chromatin, consistent with enhanced mRNA expression. The minor −1639 A allele generates a suppressor E-box binding site, apparently regulating gene expression by a mechanism undetectable with reporter gene assays. A clinical association study demonstrated that promoter SNP −1639G>A, and the tightly linked intron1 SNP 1173C>T, predict warfarin dose more accurately than intron 2 SNP 1542G>C in blacks. Increased warfarin dose requirement in blacks was accounted for by lower frequency of the −1639 A allele. Therefore, −1639G>A is a suitable biomarker for warfarin dosing across ethnic populations.
Introduction
Vitamin K epoxide reductase complex subunit 1 (VKORC1), a key enzyme in recycling reduced vitamin K, plays an essential role in γ-carboxylation of vitamin K-dependent coagulation factors, and other proteins, such as vitamin K-dependent matrix protein Gla.1,2 VKORC1 is the target of coumarin-based anticoagulants, eg, warfarin, a widely prescribed therapy for prevention of thromboembolic diseases.3
Since the cloning of VKORC1 in 2004,4,5 frequent VKORC1 single nucleotide polymorphisms (SNPs) and haplotypes have been consistently associated with warfarin dose required to therapeutic anticoagulation.6-13 Specifically, 5 SNPs define 2 major haplotypes in European Americans rs719616114 (381T>C or −4931T>C); rs9923231 (3673G>A or −1639G>A); rs9934438 (6484C>T or 1173C>T); rs8050894 (6853G>C or 1542G>C) and rs2359612 C>T (7566C>T or 2255C>T).11 Haplotype A carrying the minor alleles was associated with lower mRNA expression and lower warfarin maintenance dose, compared with the major allele haplotype B,11 but the functional SNP(s) and molecular genetic mechanism(s) underlying the association are unclear.
Recently, the US Food and Drug Administration included with warfarin product labeling consideration of CYP2C9 and VKORC1 genotypes for optimizing warfarin dosing schedules. Use of genetic biomarkers could reduce the time needed to attain anticoagulation target levels (eg, International Normalized Ratio values between 2 and 3 for most indications),15,16 thereby reducing the incidence of adverse effects, but the optimal SNP for genotyping remain uncertain. Because of high linkage disequilibrium (LD) between the 5 main VKORC1 SNPs in white and Asian populations, several studies have relied on any one single SNP, or on several SNPs representing the main haplotypes, as a predictor(s) for warfarin dose.7,17-20 However, failure to select the functionally relevant SNP could reduce the predictive value of VKORC1 genotyping in ethnic populations with lower LD of the 5 haplotype-defining SNPs.21-23
The 5-SNP haplotype A is common in Chinese (87%) and Malays (65%),23 whereas haplotype B prevails in European Americans (64%)11 and Indians (82%).23 Although all 5 SNPs are generally in strong LD in white and Asian populations, haplotype structures are more diverse in African populations (data from HapMap24 ). This raises the question of how to select a single SNP or haplotype for optimal prediction of warfarin maintenance dose in populations with mixed genetic background, and avoid errors in individual subjects where the chosen biomarker SNP is not concordant with the functional polymorphism(s).
To optimize VKORC1 as a genetic biomarker, it is therefore important to identify the relevant polymorphism(s) and to understand the molecular mechanisms underlying functional activity. Previous work had suggested that haplotype A was correlated with a lower mRNA level,11 consistent with promoter SNP −1639 A showing lower activity than −1639 G in a luciferase assay.7 However, a subsequent study failed to detect different promoter activity,9 leaving the question open as to whether −1639G>A is functional. Intronic SNPs also have the potential to affect transcription and mRNA processing. The effects of intronic SNPs on mRNA levels had been investigated thus far in only one study showing that SNP 1173C>T in intron 1 did not affect mRNA splicing in an in vitro transfection assay.10
To search for functional polymorphisms that determine VKORC1 mRNA expression, we have measured allelic mRNA expression and splicing in human liver, heart, and Epstein-Barr virus-transformed B lymphocytes (Coriell Institute Cell Repositories, Camden, NJ), and correlated the allelic expression with genotypes. Because recent studies have suggested that in vitro reporter gene assays may not accurately represent gene regulation and expression in vivo,25 we have also used chromatin immunoprecipitation (ChIP) in human liver samples followed by allelic DNA quantification to assess the effect of promoter SNPs on gene transcription. Moreover, we have also explored the role of DNA methylation and mRNA splicing in VKORC1 mRNA expression in human livers. Having identified a probably regulatory polymorphism in VKORC1, we then performed a genotype association study to determine whether this regulatory polymorphism was predictive of warfarin maintenance dose in whites and blacks.
Methods
Tissue samples
Sixty-five human liver autopsy samples and 65 left ventricle heart tissues were obtained from the Cooperative Human Network Midwestern and Western Division. EB-transformed B lymphocytes were obtained from the Coriell Institute Cell Repositories. The Ohio State University Institutional Review Board (IRB) approved the tissue studies. The IRB of the University of Florida and the University of Illinois at Chicago approved the clinical association studies. Informed consent was obtained in accordance with the Declaration of Helsinki.
Clinical association study population
The study samples consisted of 321 white and 125 black patients taking stable doses of warfarin that placed them in their target anticoagulation range and who were enrolled in pharmacogenetic studies at the University of Florida and the University of Illinois at Chicago. Study design and methods for patient enrollment, baseline demographics of the population, and genotyping have been described.13,26 Because of the similarity in study design, data from the 2 centers were combined for the analysis presented herein. The IRB of the University of Florida and the University of Illinois at Chicago approved these studies.
DNA and RNA preparation
Preparation of genomic DNA, RNA, cDNA, and plasmid DNA were performed as described.27,28 After VKORC1 plasmid transfection, cDNA was prepared using a primer that matches pcDNA3 or PGL3 vector sequence 3′ downstream of VKORC1 to avoid synthesis of endogenous VKORC1. The sequence of all primers used in this study will be provided on request.
Quantitative analysis of mRNA expression
Total mRNA levels for VKORC1 were measured by real-time polymerase chain reaction (PCR) analysis as described28 using gene specific primers that span at least one intron to avoid amplification of genomic DNA. β-actin was used as an internal control.
Quantitative analysis of VKORC1 splice variants
VKORC1 splice variants were measured using PCR with a fluorescently labeled primer as described.29
Genotyping
Six SNPs (−4931T>C, −1639G>A, 1173C>T, 1542G>C, 2255C>T, and 3730G>A, translational start site taken as position 1) in VKORC1 were genotyped in liver genomic DNA using a multiplexed SNaPshot assay as described.28 For clinical association studies, genomic DNA was isolated from buccal cells, and SNPs −1639G>A, 1173C>T, and 1542G>C were genotyped by PCR and restriction fragment length polymorphism or pyrosequencing as described.13,26
Quantitative analysis of allelic ratio in genomic DNA and mRNA using SNaPshot
The detailed method has been published.27 We used a frequent SNP as the marker, 3730G>A (rs7294), located in the 3′-UTR. Therefore, only those samples heterozygous for this SNP can be assayed for allelic mRNA expression. Allelic gDNA ratios, normalized to 1, served as internal control. Deviations of allelic mRNA ratios from 1 (after normalization to DNA ratios), ie, allelic expression imbalance (AEI), indicate the presence of cis-acting polymorphisms in VKORC1 that affect mRNA expression or turnover.
Quantitative analysis of DNA methylation
DNA methylation of the VKORC1 promoter region was determined by single nucleotide extension after bisulfite treatment of genomic DNA as described.30 Sodium bisulfite modification of genomic DNA was performed as described.31 After bisulfite treatment, 2 CpG islands in promoter regions (CpG island 1, 4787-5040; CpG island 2, 3811-4077) were amplified by PCR using methylation specific primers, and the PCR products were subjected to primer extension assay (SNaPshot) as described previously.
ChIP in liver samples heterozygous for promoter SNP −1639G>A followed by allelic DNA analysis
ChIP in liver was performed as described by Huang et al,32 using micrococcal nuclease digestion of chromatin fibers into mononucleosomes. Then chromatin was immunoprecipitated with antibodies against K4 trimethyl-H3 (K4Me3-H3) or acetyl-H3 (Ace-H3). DNA was purified from the immunoprecipitates, PCR amplified, and subjected to SNaPshot using −1639G>A SNP as marker. The ratio of G/A from CHIP DNA was normalized to that of input DNA, which in turn had been normalized to 1.
Cell culture and transfection
Cells were cultured at 37°C in a humidified incubator at 5% CO2 in Ham F-12 (CHO), DMEM/F12 (HEK), or DMEM (BAEC and HepG2) supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin. The day before transfection, cells were plated into 24-well plates. Transfection was performed using lipofectamine or Fugene (HepG2) according to the manufacturer's protocol. Cells were harvested 48 hours after transfection for total RNA preparation or luciferase assay.
VKORC1 reporter gene assays
Promoter fragments with different length (Figure 4B) were amplified from genomic DNA using PCR and cloned into PGL3 basic vector (Promega, Madison, WI) upstream of luciferase gene using XhoI and HindIII cloning sites. The constructs were transfected into HEK293, CHO, BAEC, and HepG2 cells. As control, Renilla luciferase constructs were cotransfected with PGL3 fused constructs at a 1:20 ratio. Cells were harvested after 48 hours and transferred to 96-well plates, and luciferase activity was detected with Dual-Glo luciferase assays (Promega) on a fluorescence plate reader (PerkinElmer Life and Analytical Sciences, Waltham, MA). DNA sequence was confirmed to be free of random mutation by DNA sequencing. For each construct, we selected 2 clones for transfection. A total of 4 independent transfections and triplicate luciferase assays for each transfection were performed for each construct and cell line. Because there were no differences between different clones of the same constructs, the results from the 2 clones were combined.
VKORC1 gene constructs
VKORC1 gene fragments from E1-E3 (sequence from 5294 to 9102, gene access number AY587020) were PCR amplified from genomic DNA samples with different haplotypes: haplotype A containing the minor alleles of 3 intronic SNPs (1173 T, 1542 C, and 2255 T) and haplotype B with the major alleles (1173 G, 1542 G, and 2255 C). These were cloned into pcDNA3 vector using KpnI and EcoRI cloning sites. Multiple clones were selected, and random mutations created by PCR amplification were fixed by restriction enzymes digestion and fragment exchange between different clones. Corrected haplotypes were confirmed by DNA sequencing. To replace luciferase sequence with the VKORC1 fragment, pGL3 was digested with XbaI restriction enzymes followed by DNA polymerase I large Klenow fragment treatment to create blunt ends. Then the vector was treated with HindIII to release the luciferase gene. The VKORC1 fragment in pcDNA3 was excised with HindIII and EcoRV and cloned into the modified pGL3 vector. Then VKORC1 promoter fragments harboring the SNP −1639G>A alleles were cloned into this modified VKORC1-pGL3 vector separately, using XhoI and HindIII cloning sites. The resulting constructs were tested for expression of VKORC1 mRNA (haplotype A or B) driven by the VKORC1 promoter (−1639 G allele or −1639 A allele; Figure 4C). Two clones for each construct were selected and tested.
Data analysis
Data are expressed as means plus or minus SD. Statistical analysis of gene expression, promoter activity, and allelic DNA or RNA ratios was performed using Prism (GraphPad Software, San Diego, CA). Association between AEI and genotypes, and 2 loci interaction were analyzed using Helix Tree software (Golden Helix, Bozeman, MI). For association of VKORC1 genotypes and warfarin maintenance dose, we used generalized linear model analysis to assess the individual effects of each polymorphism on warfarin dose requirements as described.13
Results
Tissue-specific AEI of VKORC1
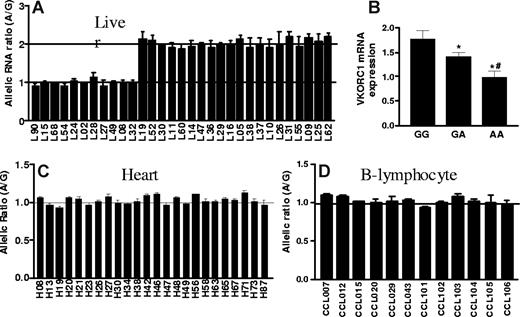
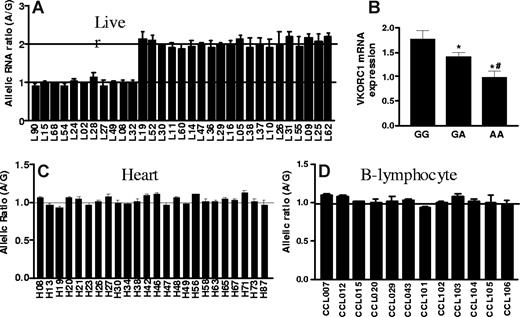
To test whether cis-acting polymorphisms affect mRNA levels, we measured allelic mRNA expression ratios with SNapShot19,27,28,33 in human liver autopsies, using marker SNP 3730A>G in the 3′-UTR. Among 31 liver samples heterozygous for the marker SNP, 20 showed AEI, with a 2-fold difference between alleles (Figure 1A). To avoid interference from pseudogenes highly homologous to VKORC1,5 we PCR-amplified the entire VKORC1 cDNA using specific primers. Reverse dideoxynucleotide extension primers (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) yielded similar AEI results (Figure 1A shows combined results). The marked division between subjects with no AEI (allelic ratio = 1) and those showing AEI with approximately 2-fold allelic ratios suggest the presence of only one main polymorphism/haplotype accounting the observed allelic variability in expression.
Allelic expression imbalance and mRNA levels of VKORC1 in human liver, left ventricular heart and B-lymphocytes. (A) Allelic expression imbalance (AEI) in liver. Genomic DNA and cDNA were amplified by PCR, and allelic mRNA and DNA ratios were measured in samples heterozygous for marker SNP (3730A>G, rs7294) located at 3′-UTR of VKORC1. The A/G ratio for DNA was set to 1, and ratios for cDNA were normalized to that of DNA. Three different conditions were used to measure AEI and the results were combined: E1, cDNA was amplified using primers within exon3; E2 and E3, complete cDNA was amplified. For AEI measurement in E1 and E2, the forward extension primer was used, and for E3, the reverse extension primer. (B) VKORC1 mRNA expression in 65 human liver samples grouped by −1639G>A (or 1173C>T) genotypes. (C) and (D) AEI in left ventricular heart and B-lymphocytes. Data are means (± SD), *P < .05 versus GG, #P < .05 versus GA, ANOVA with Dunnett post test.
Allelic expression imbalance and mRNA levels of VKORC1 in human liver, left ventricular heart and B-lymphocytes. (A) Allelic expression imbalance (AEI) in liver. Genomic DNA and cDNA were amplified by PCR, and allelic mRNA and DNA ratios were measured in samples heterozygous for marker SNP (3730A>G, rs7294) located at 3′-UTR of VKORC1. The A/G ratio for DNA was set to 1, and ratios for cDNA were normalized to that of DNA. Three different conditions were used to measure AEI and the results were combined: E1, cDNA was amplified using primers within exon3; E2 and E3, complete cDNA was amplified. For AEI measurement in E1 and E2, the forward extension primer was used, and for E3, the reverse extension primer. (B) VKORC1 mRNA expression in 65 human liver samples grouped by −1639G>A (or 1173C>T) genotypes. (C) and (D) AEI in left ventricular heart and B-lymphocytes. Data are means (± SD), *P < .05 versus GG, #P < .05 versus GA, ANOVA with Dunnett post test.
Because regulatory polymorphisms can have tissue-specific effects, we also measured allelic VKORC1 mRNA expression in left ventricular heart tissues and in transformed B lymphocytes (Coriell Institute Cell Repositories; Figure 1C,D). No significant AEI was detectable, demonstrating that the genotype effect on mRNA expression is tissue-specific.
Search for functional polymorphisms responsible for AEI of VKORC1
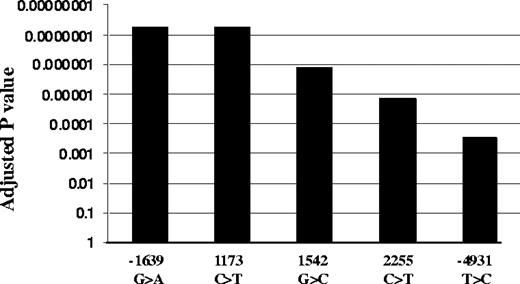
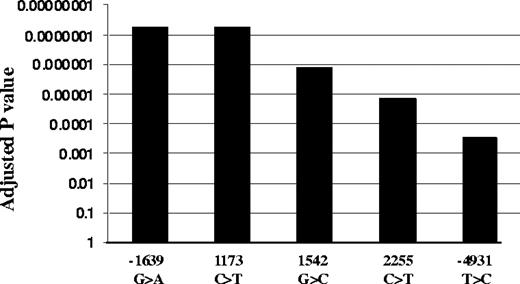
We genotyped all 5 highly linked SNPs, −4931T>C (∼5kb upstream), −1639G>A (promoter, position −1639), 1173C>T (intron1), 1542G>C (intron2), and 2255C>T (intron2; translational start site taken as position 1),11 in the same 31 liver samples. Functional SNPs must be heterozygous in samples showing AEI and homozygous in samples lacking AEI. All tissues with AEI were heterozygous for SNPs −1639G>A and 1173C>T, whereas samples without AEI were all homozygous for these 2 SNPs (Table S1). In contrast, 3 samples showing AEI were homozygous for SNP −4931T>C (L26, L37, and L52), whereas 3 AEI-negative samples were heterozygous for SNPs −4931T>C (L49), 1542G>C (L49), and 2255C>T (L68, L90), essentially ruling these SNPs out as functional. SNPs scanning found AEI to be linked to SNPs −1639G>A and 1173C>T with the lowest P value, whereas SNP −4931T>C has the highest P value (Figure 2). Two-loci analysis did not reveal any interaction between the 5 SNPs. These results indicate that −1639G>A or 1173 C>T appears to be functional, whereas the other 3 SNPs do not contribute to AEI. Consistent with this result, VKORC1 mRNA expression is higher in livers of −1639 GG/1173 CC than −1639 GA/1173 CT or −1639 AA/1173 TT carriers (Figure 1B). Sequencing of the VKORC1 locus (sequence from 41 to 9285 in AY587020) in 2 liver samples with AEI and 2 samples without AEI did not reveal any other polymorphisms that correlated with AEI. In contrast to the results with liver samples, there was no observed AEI in B lymphocytes or in autopsy heart tissues, regardless of genotype (Table S2).
Association of VKORC1 SNPs with AEI. Adjusted P values were used to correct for multiple comparisons using Helix Tree software.
Association of VKORC1 SNPs with AEI. Adjusted P values were used to correct for multiple comparisons using Helix Tree software.
Splice variants of VKORC1 and intronic SNPs
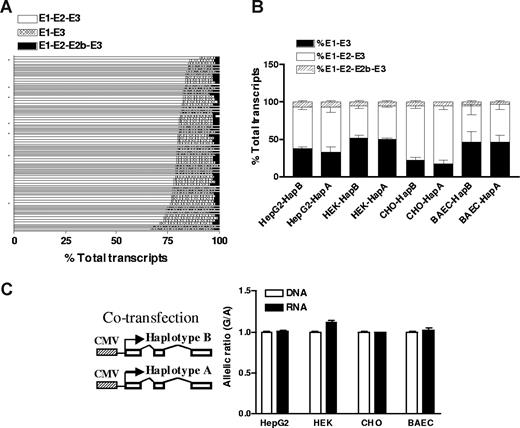
To test whether intronic SNPs affect VKORC1 before RNA splicing, we measured splice variants of VKORC1 mRNA in liver samples. Two alternative splice variants had been reported for VKORC1: one missing exon 2 and the other having an extra exon 2b. Using fluorescently labeled primers as described previously for the L-type calcium channel CACNA1C,29 we measured all 3 splice variants. Shown in Figure 3A, the major isoform, E1-E2-E3, accounts for more than 70% (77% ± 5%; range, 63%-90%) of total transcripts, and E1-E3 for approximately 20% (17% ± 4%; range, 8%-32%) of total transcripts, whereas the splice variant with exon 2b was less than 5%. The formation of different splice variants does not correlate with any SNPs tested in this study, indicating that they do not affect VKORC1 splicing. To test whether VKORC1 RNA splicing is tissue specific, we also measured VKORC1 splice variants in heart, lung, kidney, small bowel, brain, B lymphocytes, HEK293 cells, HepG2 cells, and peripheral blood cells. All these tissues/cells express detectable amounts of VKORC1 mRNA (reverse transcription–PCR Ct < 25). The splicing patterns in these tissues/cells are similar to that of liver, except that brain and HEK293 cells display higher levels of the E1-E3 splice variant (∼50% of total transcripts), indicating that the splicing of VKORC1 varies to some degree between tissues.
The effects of VKORC1 SNPs on RNA splicing and mRNA levels. (A) Relative amounts of 3 splice variants of VKORC1 in 65 liver samples. The splice variants were measured by PCR using fluorescently labeled primers. (B) Splice variants of VKORC1 in 4 cell lines after transfection with the entire transcribed region of VKORC1. Haplotype A (1173 T, 1542 C, 2255 T) or haplotype B (1173 C, 1542 G, 2255 C) in pcDNA vector was transfected separately into different cell lines, and VKORC1 splice variants measured. (C) Allelic RNA expression of VKORC1 in 4 cell lines after cotransfection with VKORC1 haplotype A and B, using 3730A>G as marker. Haplotype B contains the 3730 G allele, whereas haplotype A contains the 3730 A allele. Data are means plus or minus SD.
The effects of VKORC1 SNPs on RNA splicing and mRNA levels. (A) Relative amounts of 3 splice variants of VKORC1 in 65 liver samples. The splice variants were measured by PCR using fluorescently labeled primers. (B) Splice variants of VKORC1 in 4 cell lines after transfection with the entire transcribed region of VKORC1. Haplotype A (1173 T, 1542 C, 2255 T) or haplotype B (1173 C, 1542 G, 2255 C) in pcDNA vector was transfected separately into different cell lines, and VKORC1 splice variants measured. (C) Allelic RNA expression of VKORC1 in 4 cell lines after cotransfection with VKORC1 haplotype A and B, using 3730A>G as marker. Haplotype B contains the 3730 G allele, whereas haplotype A contains the 3730 A allele. Data are means plus or minus SD.
In another test for effects of the 3 intronic SNPs on VKORC1 mRNA splicing, we PCR-amplified the entire transcribed VKORC1 gene (sequence from 5294 to 9102, AY587020, including all introns but lacking the promoter region) from samples carrying haplotype A (1173 T, 1542 C, 2255 T, minor alleles) or haplotype B (1173 C, 1542 G, 2255 C, major alleles), and cloned them into pcDNA3 (Figure 3). These constructs were transfected separately into HEK293, CHO, HepG2, and BAEC cells, and VKORC1 splice variants were measured. All these cells express similar amounts of transfected VKORC1 (Ct ∼ 20). Shown in Figure 3B, there were no differences in splice variants after transfection with haplotype A and haplotype B constructs in all 4 cells lines tested, supporting the finding that intronic SNPs 1173C>T, 1542G>C, and 2255C>T do not affect VKORC1 splicing.
To test whether intronic SNPs affect RNA processing other than splicing, thereby causing changes in overall mRNA levels, we cotransfected the same amounts of full-length VKORC1 constructs of haplotype A and B (driven by CMV promoter) into different cells (Figure 3C) and measured the allelic ratio of RNA and plasmid DNA using SNP 3730A>G in the 3′-UTR as marker. To avoid interference from endogenously expressed VKORC1, the primer for cDNA synthesis matched transcribed pcDNA vector sequence 3′ downstream of VKORC1. Allelic mRNA ratios were the same as the transfected plasmid DNA allele ratios in the 4 cell lines tested, again showing that haplotype B and haplotype A behaved similarly (Figure 3C). These results indicate that intronic SNPs of VKORC1 do not affect mRNA levels and therefore probably do not account for the observed AEI of VKORC1 in liver.
Testing −1639G>A promoter SNP on VKORC1 mRNA expression using reporter gene assay
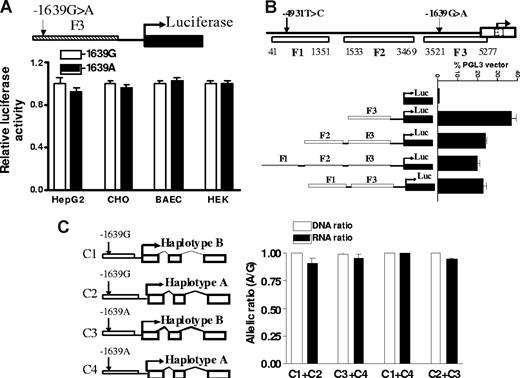
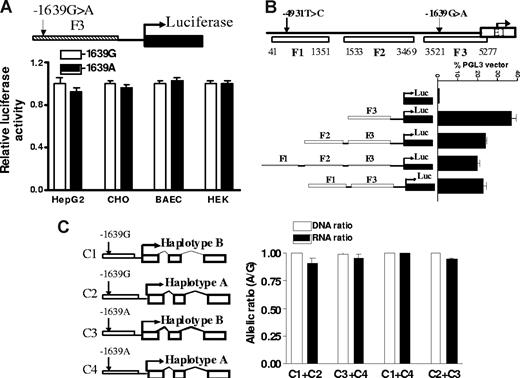
DNA fragments (F1-3) of promoter regions containing −1639G>A as reported7 were PCR-amplified and cloned into pGL3 vector, and transcriptional activity was measured with a luciferase reporter gene assay (Figure 4). In contrast to the result of Yuan et al,7 but consistent with Bodin et al,9 we were unable to detect significant differences between F3 fragments containing −1639 G and −1639 A in 4 cell lines tested, including HepG2 cells, a cancer cell line derived from liver (Figure 4A). To test interactions between the −4931T>C and promoter SNP −1639G>A, we also amplified DNA fragment F1, from position 41 to 1351 containing −4931T>C and ligated it to the 5′ end of F3. Shown in Figure 4B, this upstream fragment did not increase promoter activity of F3, indicating lack of enhancer activity for F1. Moreover, there is no difference between F1 containing the −4931 T or −4931 C allele on the promoter activity of F3 containing either −1639 G or −1639 A alleles (data not shown), indicating that −4931T>C was not functional in this assay.
The effects of promoter SNP on VKORC1 mRNA expression. (A) Transcriptional activity of promoter fragments containing −1639 G or −1639 A, measured by luciferase reporter gene assays in 4 different cell lines. (B) Effects of 5′ upstream fragments F1 and F2 on transcription activity of proximate promoter fragment F3. (C) Allelic mRNA expression of VKORC1 in HepG2 cells after cotransfection with VKORC1 gene constructs. The constructs combine −1639 G or −1639 A alleles with either haplotype A or B of the transcribed region. Data are means plus or minus SD.
The effects of promoter SNP on VKORC1 mRNA expression. (A) Transcriptional activity of promoter fragments containing −1639 G or −1639 A, measured by luciferase reporter gene assays in 4 different cell lines. (B) Effects of 5′ upstream fragments F1 and F2 on transcription activity of proximate promoter fragment F3. (C) Allelic mRNA expression of VKORC1 in HepG2 cells after cotransfection with VKORC1 gene constructs. The constructs combine −1639 G or −1639 A alleles with either haplotype A or B of the transcribed region. Data are means plus or minus SD.
Because AEI of VKORC1 is tissue-specific (Figure 1), failure to observe −1639G>A effects could have resulted from the inappropriate cell models used. To test whether the transformed hepatic HepG2 cells are similar to liver in regulating VKORC1 expression, we measured allelic mRNA expression ratios of endogenous VKORC1, which is heterozygous for the marker SNPs and all 5 candidate SNPs tested. We observed significant AEI (RNA ratio (A/G) of 1.5 ± 0.06) for endogenously expressed VKORC1 in HepG2 cells, suggesting that a regulatory polymorphism is active in this cell context. Lack of genotype effects in the reporter gene assay indicates that either −1639G>A alone is insufficient and requires interaction with intronic SNPs, especially intron 1 SNP 1173C>T, or the reporter gene assay is inappropriate for detecting −1639G>A effects.
To test whether there are any enhancer elements within the intronic region that can regulate promoter activity in addition to −1639G>A, we replaced the luciferase gene in pGL3 vector with the entire transcribed VKORC1 region (sequence from 5294 to 9102, AY587020) containing reference sequence (haplotype B) or SNP variants (haplotype A), driven by the VKORC1 promoter containing either −1639 G or −1639 A, respectively (Figure 4C). We cotransfected the same amount of haplotype B and haplotype A plasmids driven by either −1639 G or −1639 A promoter into HepG2 cells and measured allele-specific mRNA expression derived from these 2 constructs using marker SNP 3730A>G. We specifically amplified transfected but not endogenous VKORC1 mRNA using a primer for cDNA synthesis matched to a transcribed sequence of the PGL3 vector. Shown in Figure 4C, after normalization to plasmid DNA ratios, the allelic ratio of RNA is close to 1 for all 4 different combinations, indicating that none of the intronic SNPs affected mRNA expression through interaction with promoter SNP −1639G>A in this extended VKORC1 construct.
Interaction between posttranscriptional histone modifications and regulatory polymorphisms in VOKRC1
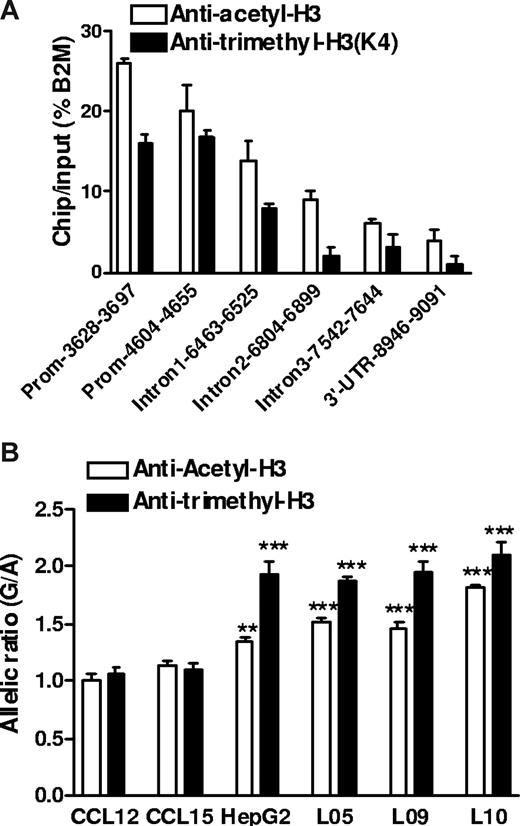
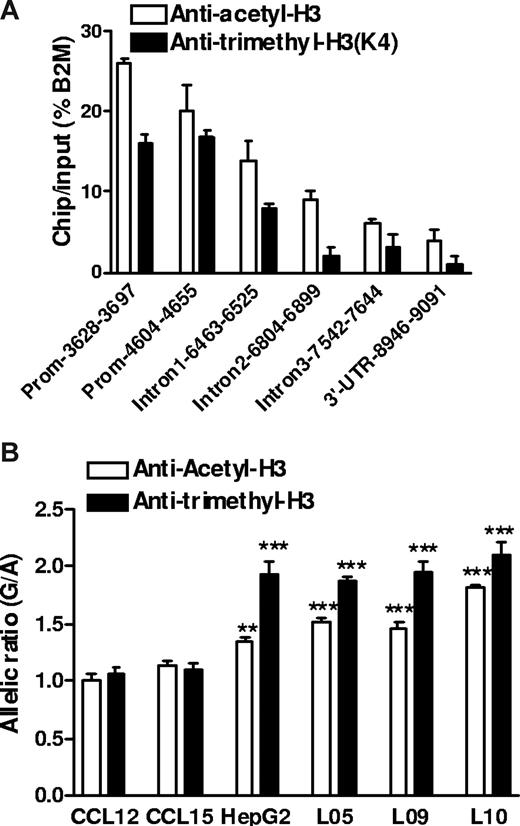
Histone H3 acetylation and H3 K4 trimethylation are associated with active gene expression,34 and moreover, could interact with promoter polymorphisms. −1639G>A creates an E-box site for transcription factors that tend to suppress gene expression and recruit histone modifying factors including histone deacetylases.35 Because circular plasmid DNA does not support histone-chromatin formation as genomic DNA, this effect could escape detection by reporter gene assays. We performed ChIP assays in human liver samples heterozygous for −1639G>A using antibodies against acetyl-histone H3 (Ace-H3) or K4-trimethyl histone H3 (K4Me3H3, also associated with active chromatin). First, we determined which regions of VKORC1 were recovered from the precipitated fractions, using quantitative PCR (Figure 5A). Whereas DNA sequence along the entire VKORC1 gene was associated with Ace-H3 or K4Me3H3 immunoprecipitates, the promoter region had the highest binding activity with ChIP/input of 15% to 25% of the housekeeping gene β2-microglobulin. Second, we determined the allelic ratio of genomic DNA coimmunoprecipitated with Ace-H3 or K4Me3H3, using −1639G>A as marker. Normalized to the genomic DNA ratio without ChIP, the allelic −1639 G/-1639 A ratio was approximately 2 in K4Me3H3 precipitates and approximately 1.5 in Ace-H3 precipitates (Figure 5B). This result was consistent in 3 liver samples heterozygous for −1639G>A and was also observed in HepG2 cells, but not in 2 transformed B lymphocytes that did not display AEI of VKORC1 mRNA as shown in Figure 1D. These results demonstrate that allele −1639 G is more likely to associate with transcriptionally active chromatin Ace-H3/ K4Me3H3 in liver and HepG2 cells, thereby producing more mRNA than the −1639 A allele and accounting for the observed AEI of VKORC mRNA in human liver. However, because of the high LD between −1639G>A and 1173C>T in intron 1, also detected in the immunoprecipitates (Figure 5A), we cannot completely rule out an effect of 1173C>T on the regulation of chromatin structure promoted by −1639G>A.
Histone modification and regulatory polymorphism in VKORC1. (A) Relative amount of DNA fragments from different regions of the VKORC1 locus precipitated by antiacetyl-H3 and antitrimethyl-H3 antibodies in human liver samples. ChIP assays in frozen liver tissues were performed after micrococcal nuclease digestion, and precipitated DNA was recovered and quantified by real-time PCR. β2-microglobulin (B2M) serves as internal control. (B) Allele-specific DNA analysis after ChIP with antiacetyl-H3 and antitrimethyl-H3 in human livers, HepG2 cells, and B lymphocytes, using promoter SNP −1639G>A as a marker. The allelic DNA ratio from control DNA (no ChIP) was set as 1, and allelic DNA ratios from precipitates were normalized to the ratio of input. Data are means plus or minus SD (**P < .01, ***P < .001, compared with input ratio, ANOVA with Dunnett posttest).
Histone modification and regulatory polymorphism in VKORC1. (A) Relative amount of DNA fragments from different regions of the VKORC1 locus precipitated by antiacetyl-H3 and antitrimethyl-H3 antibodies in human liver samples. ChIP assays in frozen liver tissues were performed after micrococcal nuclease digestion, and precipitated DNA was recovered and quantified by real-time PCR. β2-microglobulin (B2M) serves as internal control. (B) Allele-specific DNA analysis after ChIP with antiacetyl-H3 and antitrimethyl-H3 in human livers, HepG2 cells, and B lymphocytes, using promoter SNP −1639G>A as a marker. The allelic DNA ratio from control DNA (no ChIP) was set as 1, and allelic DNA ratios from precipitates were normalized to the ratio of input. Data are means plus or minus SD (**P < .01, ***P < .001, compared with input ratio, ANOVA with Dunnett posttest).
Lack of effect of genomic DNA-CpG methylation on VKORC1 expression
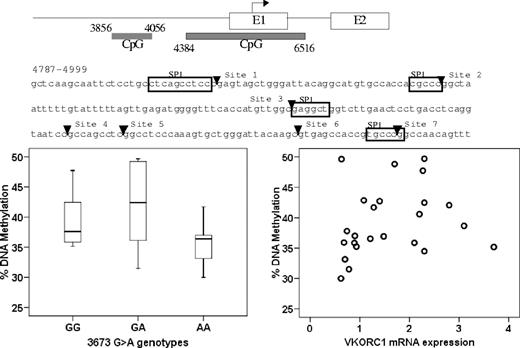
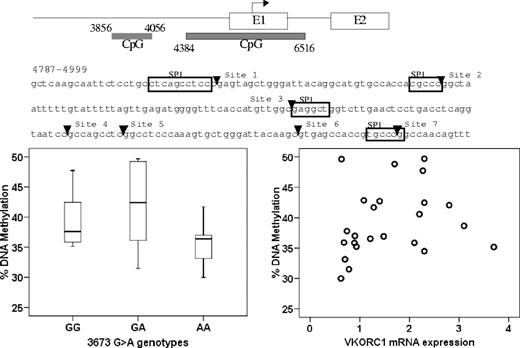
DNA methylation at CpG island plays a role in regulating gene expression.36-38 Therefore, we measured the extent of CpG methylation of 2 CpG islands in the promoter region, CpG island 1 (3856-4056) and CpG island 2 (4384-6516). Because methylation within CpG island 1 was low (data not shown), we focused on CpG island 2, quantitatively measuring the methylation of 7 CpG sites close to a putative SP-1 binding sites within the 4781-4999 region. Average DNA methylation in B lymphocytes was slightly higher than in liver (48% ± 3% vs 39% ± 6%). However, the level of DNA methylation did not correlate with genotype of −1639G>A or other intronic SNPs, or with VKORC1 mRNA expression (Figure 6). This result indicates that DNA methylation has little, if any, effect on VKORC1 expression, and the studied polymorphisms do not change the DNA methylation pattern in VKORC1.
No correlation between CpG island methylation in the VKORC1 promoter region and −1639G>A genotype or VKORC1 mRNA expression. The horizontal line represents the median, the box the 25th percentile, and the whiskers the 75th percentile.
No correlation between CpG island methylation in the VKORC1 promoter region and −1639G>A genotype or VKORC1 mRNA expression. The horizontal line represents the median, the box the 25th percentile, and the whiskers the 75th percentile.
Association between SNPs −1639G>A, 1173C>T, and 1542G>C with warfarin dose in whites and blacks
We compared the association of −1639G>A, 1173C>T (intron1), and 1542G>C (intron 2) with warfarin maintenance dose in clinical studies. Shown in Table 1 for white patients, all 3 SNP genotypes were significantly associated with warfarin dose, with similar R2 value because of the high LD between these 3 SNPs in the white population. However, in black patients, only promoter SNP −1639G>A and intron 1 SNP 1173C>T were highly linked and significantly associated with warfarin dose, whereas intron 2 SNP 1542G>C was not. These data further support SNP −1639G>A as the functional SNP but do not exclude 1173C>T. Moreover, the higher mean warfarin dosage required for blacks (mean = 44.6 mg/wk) compared with whites (mean = 36.7 mg/wk) appears to be accounted for by the lower frequency of the −1639 A and 1173 T alleles that require reduced dosing. No difference in steady-state dosing was observed between the ethnic populations when grouped by VKORC1 genotype.
Discussion
VKORC1 variants are now recognized as biomarkers for optimizing the dosage regimen, but which polymorphism is active and whether more than one polymorphism contributes to activity remained uncertain. The results of the present study indicate that promoter SNP −1639G>A is sufficient to account for interindividual variability in VKORC1 activity; however, we cannot entirely exclude a role for 1173C>T in intron1 because of high LD of the 2 SNPs. The −1639 A allele is associated with a 2-fold lower level of VKORC1 mRNA in human liver, consistent with a reduced warfarin maintenance dose in subjects carrying the haplotype marked by −1639 A, as reported in previous studies.11
For optimal use as a biomarker in clinical practice, several algorithms have been developed for personalized warfarin dosing using various VKORC1 polymorphisms of haplotypes.6,19,20 For example, the Marshfield Clinic Research Foundation (Marshfield, WI) had selected SNP 1542G>C (6853G>C, rs8050894) as a marker for VKORC1,20 whereas a group in Singapore has used SNP −4931C>T (381C>T, rs7196161).19 Neither SNP is completely linked with −1639G>A in both white and black populations, having LD r2 values with 1542G>C and −4931C>T of 0.96 and 0.69 in whites and 0.70 and 0.35 in blacks, respectively (calculated from 65 liver tissues). Our clinical association data also showed that −1639G>A is a better predictor for warfarin dose than intron2 SNP 1542G>C in blacks (Table 1) who have more diverse haplotype structures compared with whites and Asians. Because of the very high LD between −1639G>A and 1173C>T in intron1 (LD r2 = 0.946 in white and r2 = 0.902 in black in our study cohort), these 2 SNPs yield the same statistical results. Similarly, previous studies demonstrate strong LD between the −1639G>A and 1173C>T SNPs in Asian populations, with the −1639AA or 1173TT genotype consistently associated with lower warfarin dose requirements.7,23,39 Our allelic expression and in vitro results strongly favor −1639G>A over 1173C T as the active polymorphism. For optimal use of VKORC1 as a clinical biomarker for warfarin dose predictions, we propose that −1639G>A alone is sufficient. Even though other VKORC1 SNPs are in high linkage disequilibrium in some ethnic populations, use of marker polymorphisms, rather than the functionally relevant polymorphism, to predict warfarin dose response could adversely affect warfarin therapy of patients where linkage disequilibrium is disrupted, as is frequently the case in subjects of African descent. Given the large number of patients on warfarin therapy, such adverse effect could be of clinical concern.
Overall mean warfarin doses are higher in blacks compared with whites,40 but the reason for this discrepancy was unclear. We show here that, when grouped by −1639G>A and 1173C>T genotype (Table 1), the mean warfarin doses are identical in the 2 ethnic groups, and the greater overall mean in blacks is caused by the lower frequency of the minor −1639G>A and 1173C>T alleles, which therefore appears to be sufficient to guide warfarin dosing in both populations.
This study also demonstrates that the effect of −1639G>A is tissue-dependent, observable in the liver but not in B lymphocytes and heart tissue. This result cautions one in the interpretation of significant associations reported between the main VKORC1 haplotypes and arterial cardiovascular disease,41,42 as this was thought to require local effects on gene expression and modification of matrix proteins in target tissues. Consistent with this, one association study failed to observe association between VKORC1 genotypes and arterial cardiovascular diseases.43
Our studies further indicate that the regulation of VKORC1 expression by promoter SNP −1639G>A is mediated by a mechanism involving chromatin modification. Using ChIP analysis combined with allelic DNA ratio analysis to detect differences in the association of the −1639G>A alleles with active open chromatin structures characterized by Ace-H3 and K4Me3-H3,34 we found more −1639 G allele than −1639 A allele binding to active histone, indicating allele specific chromatin modification. The −1639G>A substitution creates an E-box binding site (from CGGGTG to CAGGTG), which could attract repressive E-box binding proteins.35 Corepressors often form complexes that contain histone deacetylases,44,45 which render the chromatin transcriptionally inactive through deacetylation of acetylated histones.46 Because histone acetylation is a mark for histone K4 methylation, hence, reducing histone acetylation is often accompanied by decreased histone K4 methylation.47 Hypo-acetylated H3 or K4 hypo-methylated H3 may recruit other histone remodeling proteins and silence gene transcription. No link was observed between histone modification and CpG DNA methylation as an additional epigenetic regulatory pathway.
Previous promoter gene assays have yielded inconsistent results on the role of −1639G>A.7,9 We propose that these inconsistencies arise from a molecular mechanism underlying inhibition of −1639 A promoter activity that involves chromatin modification in the target tissue, a process insufficiently mimicked by transfection of circular plasmid DNA. Others have also reported inconsistent results between ChIP and luciferase reporter gene assays,45,48 so that reporter gene assays may not accurately represent gene regulation and expression in vivo25 and must be viewed with caution.
In conclusion, we have shown that promoter SNP −1639G>A affects mRNA expression of VKORC1 in liver and is associated with different warfarin dose requirement in 2 ethnic groups. To make personalized medicine applicable to all populations with varied genetic background, it is imperative to identify the causative factors as predictive biomarkers.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health (grant HL 74 730) and a new investigator award (L.H.C.) from the American Association of Colleges of Pharmacy and the American Foundation for Pharmaceutical Education.
National Institutes of Health
Authorship
Contribution: D.W. designed and performed research, analyzed data, and wrote the manuscript; H.C. performed research; K.M.M., L.H.C., and J.A.J. designed, performed, and analyzed data related to clinical association study; and W.S. designed research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Danxin Wang, Program in Pharmacogenetics, Department of Pharmacology, College of Medicine, The Ohio State University, 333 West 10th Avenue, Graves Hall, Columbus, OH 43210; e-mail: wang.808@osu.edu.