Abstract
The mechanism by which the glucocorticoid (GC) dexamethasone induces apoptosis in multiple myeloma (MM) cells is unknown, although previous work suggests that either transactivation through the glucocorticoid response element (GRE), transrepression of NF-κB, phosphorylation of RAFTK (Pyk2), or induction of Bim is important. We studied this question by ectopically expressing mutant glucocorticoid receptors (GRs) in the dexamethasone-resistant MM1R cell line, which has lost its GR. Lentiviral-mediated reexpression of wild-type GR restored GRE transactivation, NF-κB transrepression, RAFTK phosphorylation, Bim induction, and dexamethasone-induced apoptosis. We then reexpressed 4 GR mutants, each possessing various molecular effects, into MM1R cells. A perfect correlation was present between induction of GRE transactivation and induction of apoptosis. In contrast, NF-κB transrepression and RAFTK phosphorylation were not required for apoptosis. Although not required for dexamethasone-mediated apoptosis, NF-κB inhibition achieved by gene transfer suggested that NF-κB transrepression could contribute to apoptosis in dexamethasone-treated cells. Dexamethasone treatment of MM1R cells expressing a mutant incapable of inducing apoptosis successfully resulted in RAFTK (Pyk2) phosphorylation and Bim induction indicating the latter GR-mediated events were not sufficient to induce apoptosis. MM1R cells expressing mutant GRs will be helpful in defining the molecular mechanisms of dexamethasone-induced apoptosis of myeloma cells.
Introduction
Dexamethasone is one of the most effective therapeutic agents in multiple myeloma (MM). The ability of dexamethasone to induce apoptosis in MM cells after in vitro culture suggests that the efficacy of this drug in vivo in patients is also related to its apoptosis-inducing effects. Although it is widely accepted that the ability of glucocorticoids (GCs) like dexamethasone to achieve MM cell apoptosis is mediated via initial binding to their cognate receptor, the GC receptor (GR), the critical downstream pathways are still unknown. The GR is a ligand-activated transcription factor of the nuclear receptor family. After ligand binding, the GR translocates to the nucleus where it binds to GC response element (GRE) sequences in genes resulting in transactivation. Indeed, in several models of GC-induced apoptosis of malignant and nonmalignant lymphoid cells, GR-induced transactivation has been indicted mechanistically, presumably by induction of proapoptotic genes.1-4 However, ligand binding to the GR also results in down-regulation of the NF-κB transcription factor. This so-called transrepression has also been theorized to explain GC-induced MM cell apoptosis, presumably because of an inhibition of antiapoptotic NF-κB–targeted genes.5-9 A third possible molecular mechanism of MM apoptosis was presented by Chauhan et al,10,11 implicating GR-induced phosphorylation and activation of the RAFTK kinase (also known as Pyk2). Overexpression of a kinase inactive RAFTK blocked dexamethasone-induced apoptosis of MM cells. A fourth contributing event is induction of the proapoptotic protein, Bim, in GC-treated cells.4,12,13
A series of MM cell lines, developed by the group at Northwestern University,14 has been used as a model for testing many potential anti-MM agents and investigating the mechanism of dexamethasone-induced MM cell apoptosis. These cell lines from a single patient mirror the progression of disease from GC sensitivity to resistance. The MM1S line is extremely sensitive to dexamethasone-induced apoptosis and the MM1R line is completely resistant, as it has lost normal expression of the GR. We exploited the resistant, GR-null MM1R MM line through stable transfection of wild-type versus mutant GRs with subsequent testing of dexamethasone-induced apoptosis. The mutant GRs were variably deficient in their ability to induce GR-mediated transactivation, NF-κB transrepression, phosphorylation of RAFTK, or induction of Bim. The results indicate a perfect correlation between the ability of GRs to transactivate through the GRE and to induce apoptosis in myeloma cells. Furthermore, although inhibition of NF-κB by gene transfer was sufficient to induce myeloma cell apoptosis, neither NF-κB transrepression nor RAFTK phosphorylation was required for dexamethasone-induced apoptosis. Finally, neither RAFTK phosphorylation nor BIM gene induction was sufficient for apoptosis.
Methods
Cell lines and viral vectors
MM1S and MM1R cell lines were kind gifts from Dr Steven Rosen and Dr Nancy Krett at Northwestern University, Chicago, IL. Human MM1S (dexamethasone-sensitive) and MM1R (dexamethasone-resistant) MM cells were grown in RPMI 1640 media supplemented with 10% heat inactivated fetal-bovine serum,100 units/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine. Opm-2, ANBL-6, RPMI 8226, U266, AF-10, and ARH77 were obtained from ATCC (Manassas, VA). Wild-type ectopic human GR expression in the MM1R cell line was achieved by infecting the cell line with a lentiviral vector. This vector was constructed by cloning in the human GR-α (obtained as a Gateway clone from Invitrogen, Carlsbad, CA) into a replication defective lentiviral construct. 293FT cells were transiently transfected and supernatant with viral particles was collected. Titration of viral particles was performed by infecting SCC12, a head and neck cell line and counting the blasticidin resistant colonies. MM1R cells were infected with the viral particles at an multiplicity of infection (MOI) of 10 and blasticidin selection was started 72 hours later.
GR mutants
Mutations in the GR DNA binding domain (S425G, L436V, and N454D/A458T) have been described earlier.6 We generated the mutants by in vitro site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) of our human GR lentiviral expression construct. The oligonucleotide sequences for site-directed mutagenesis were the same as described earlier.6 The mutant C476W/R479Q was described in a separate report.15 It was also generated by site-directed mutagenesis using the after oligonucleotide sequences: 5′-AAACTGCCCAGCATGGCGCTATCAAAAATGTCTTCAG-3′ and 5′-CTGAAGACATTTTTGATAGCGCCATGCTGGGCAGTTT-3′. All the mutants were sequenced, and MM1R cells were infected with different GR mutant lentiviral vectors with subsequent selection in blasticidin to obtain cells with mutant GR expression.
Apoptosis assay
Activated caspase 3 apoptosis kit from BD Biosciences PharMingen (San Diego, CA) was used to perform the apoptosis assays. Cells were treated with various concentration of dexamethasone (Sigma-Aldrich, St Louis, MO) for 48 to 60 hours and then analyzed by flow cytometric analysis for expression of activated caspase 3.
Reporter assays
GRE reporter assay was performed by transiently transfecting cells with 1 μg TK-Renilla luciferase (thymidine kinase promoter, transfection control) plasmid and 4 μg GRE-firefly luciferase reporter plasmid. Transfection was performed with cells in Opti-MEM media (Invitrogen) in 6-well dishes with 5 μL Lipofectamine 2000 (Invitrogen) as per manufacturer protocol; 10−6 M dexamethasone was added 6 hours after transfection for 12 hours and then cells harvested for reporter assay. Dual luciferase assay was performed with the kit from Promega. TK-Renilla luciferase expression is used to control for transfection efficiency. GRE reporter expression is reported as fold increase in cells relative to cells not treated with dexamethasone (arbitrarily = 1). NF-κB transrepression assays were performed by transiently transfecting cells with 1 μg TK-Renilla luciferase (transfection control), 4μg NF-κB firefly luciferase reporter plasmid, and 10 ng rel A expression plasmid, PLG033-P65-SP10 (gift of Dr Matthew Rettig, University of California–Los Angeles). Six hours after transfection, cells were washed, dexamethasone 10−6 M was added, and cells analyzed 12 hours later. TK-Renilla luciferase expression is once again used to control for transfection efficiency. NF-κB transrepression is calculated by the relative change in rel A–induced firefly luciferase expression achieved by dexamethasone treatment.
Immunoblot analysis
Cells were washed with phosphate-buffered saline and lysed in lysis buffer from Cell Signaling Technology (Danvers, MA); a 20 μg sample was loaded per lane, and proteins were separated by electrophoresis, transferred to polyvinylidene difluoride membrane, and analyzed by immunoblot analysis. The antibodies used were phospho-RAFTK antibody (Tyr-402) from Cell Signaling Technology, RAFTK and BIM antibody from Santa Cruz Biotechnology (Santa Cruz, CA). The antigen-antibody complexes were visualized by chemiluminescence (GE Healthcare, Little Chalfont, United Kingdom).
Statistics
Significant differences between groups were determined by the t test.
Results
Ectopic expression of wild-type GR in MM1R cells reestablishes an apoptotic response to dexamethasone
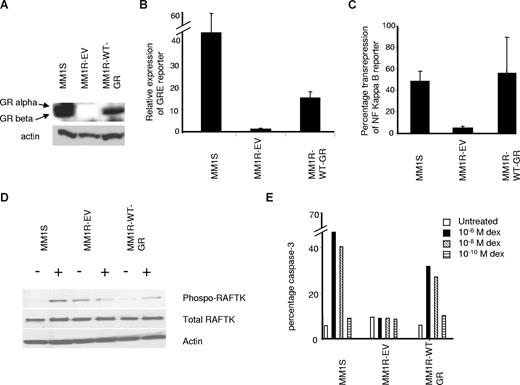
The alpha isoform of the GR is the predominant isoform present in dexamethasone-sensitive cells, whereas the β isoform is unable to bind ligand and is not involved in any transcriptional activity.16,17 Thus, we stably expressed wild-type GR-α in MM1R cells (Figure 1, MM1R-WT-GR) through lentiviral infection and compared them with empty vector lentivirus-infected MM1R cells (MM1R-EV, empty vector) and dexamethasone-sensitive MM1S cells. As previously published18 and confirmed in Figure 1A, the resistant MM1R cell line (MM1R-EV in Figure 1A) has lost the expression of both GR-α and -β isoforms as the anti-GR antibody recognizes the 2 isoforms in MM1S cells. In addition, when exposed to dexamethasone, this cell line is deficient in GR-induced transactivation (Figure 1B, 1.2-fold increase when exposed to dexamethasone compared with a 40-fold increase in MM1S cells), NF-κB transrepression (Figure 1C), RAFTK phosphorylation (Figure 1D), and an apoptotic response (Figure 1E). MM1R cells with ectopic expression of wild-type GR-α (MM1R-WT-GR) resulted in GR-α expression, which was comparable with that of the sensitive MM1S cells (Figure 1A). When these MM1R-WT-GR cells were exposed to dexamethasone, significant GRE reporter expression was also demonstrated (Figure 1B), although slightly less than what is induced in MM1S cells (15-fold increase when exposed to dexamethasone). Dexamethasone also was capable of achieving NF-κB transrepression in MM1R-WT-GR cells (Figure 1C). The ectopically expressed wild-type GR receptor also allowed dexamethasone-induced RAFTK phosphorylation (Figure 1D). These results confirmed that the expressed GR was functional. Finally, when exposed to dexamethasone, MM1R-WT-GR cells were also now capable of undergoing apoptosis, assayed by flow cytometric analysis of activated caspase 3 (Figure 1E). The apoptosis results indicate that the absolute dexamethasone-resistant phenotype was the result of loss of the GR in MM1R cells and that, once the GR was reexpressed, significant dexamethasone-induced apoptosis occurred. More importantly, they indicated that this model could be used to test mutant GRs for their ability to induce apoptosis in MM cells.
Characterization of MM1R cells expressing wild-type GR. (A) Western blot analysis of MM1S, MM1R-EV, and MM1R with wild-type GR-α (MM1R-WT-GR). Two bands in MM1S cells correspond to GR-α and beta isoforms. (B) GRE reporter expression by dexamethasone treatment (10−6 M for 12 hours) comparing MM1S cells to MM1R cells with empty vector (MM1R-EV) and MM1R-WT-GR. Data shown as fold induction calculated as described in “Reporter assays” and reported as mean plus or minus SD; n = 3. MM1R-WT-GR cells demonstrate significantly increased dexamethasone-induced reporter expression compared with MM1R-EV cells (P < .05). (C) NF-κB transrepression assay, reported as percentage repression of reporter expression on dexamethasone treatment compared with the untreated control (mean ± SD). MM1R-WT-GR cells demonstrate significantly greater NF-κB repression compared with MM1R-EV cells (P < .05). (D) Western blot analysis of MM1R cells for total RAFTK, phospho-RAFTK, and actin with and without dexamethasone treatment (10−6 M) for 14 hours. (E) Percentage of cells undergoing apoptosis. Caspase 3 assay with flow cytometric analysis of cells treated with 3 concentrations of dexamethasone and analyzed at 60-hour time point. Experiment was done twice with similar results.
Characterization of MM1R cells expressing wild-type GR. (A) Western blot analysis of MM1S, MM1R-EV, and MM1R with wild-type GR-α (MM1R-WT-GR). Two bands in MM1S cells correspond to GR-α and beta isoforms. (B) GRE reporter expression by dexamethasone treatment (10−6 M for 12 hours) comparing MM1S cells to MM1R cells with empty vector (MM1R-EV) and MM1R-WT-GR. Data shown as fold induction calculated as described in “Reporter assays” and reported as mean plus or minus SD; n = 3. MM1R-WT-GR cells demonstrate significantly increased dexamethasone-induced reporter expression compared with MM1R-EV cells (P < .05). (C) NF-κB transrepression assay, reported as percentage repression of reporter expression on dexamethasone treatment compared with the untreated control (mean ± SD). MM1R-WT-GR cells demonstrate significantly greater NF-κB repression compared with MM1R-EV cells (P < .05). (D) Western blot analysis of MM1R cells for total RAFTK, phospho-RAFTK, and actin with and without dexamethasone treatment (10−6 M) for 14 hours. (E) Percentage of cells undergoing apoptosis. Caspase 3 assay with flow cytometric analysis of cells treated with 3 concentrations of dexamethasone and analyzed at 60-hour time point. Experiment was done twice with similar results.
Characteristics of mutant GRs
MM1R cells were next transfected with mutant GRs via lentiviral infection. We used 4 mutants: S425G, L436V, N454D/A458T, and C476W/R479Q. The mutant S425G has a single amino acid change in the DNA binding domain of the GR (serine to glycine at position 425) and is reported to be deficient in its ability to transrepress NF-κB,6 although it effectively transactivates through the GRE.15 L436V also has a single amino acid change (leucine to valine at position 436). It is capable of inducing GRE transactivation as well as NF-κB transrepression.6,15 N454D/A458T (asparagine to aspartic acid at position 454 and alanine to threonine at position 458) is a mutant with 2 amino acid changes, one of which, A458T, is in the D loop of the GR, a region involved in the dimerization of the receptor.6,15 This mutant is reported to be capable of NF-κB transrepression but not inducing GRE transactivation.6,15 The mutant C476W/R479Q, with 2 amino acid change (cysteine to tryptophan at position 476 and arginine to glutamine at position 479), converts one of the coordinating cysteine residues to tryptophan, thus disrupting the second zinc finger of the GR.15 In previous reports, this particular mutant did not transactivate GRE.15
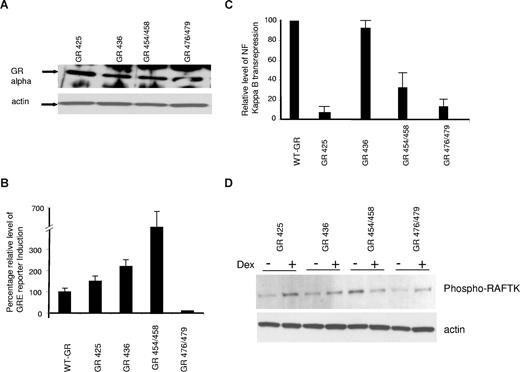
As shown in Figure 2A, comparable expression of an imunodetected GR-α was present in all the transfected mutant MM1R cells. These mutant receptors were variably efficient in achieving GR transactivation, NF-κB transrepression, and RAFTK phosphorylation (Figures 2B-D). In Figure 2B and C, the ability of the mutant receptors to transactivate through the GRE or transrepress NF-κB is shown as percentage of control (100%) where the control is the activity of the wild-type GR receptor, MM1R-WT-GR, studied concurrently with the mutant receptors. The S425G cells could transactivate in a GRE reporter assay similar to the MM1R-WT-GR cells (Figure 2B) but could not transrepress the NF-κB reporter (Figure 2C). L436V cells exhibited more GRE transactivation (220% of control, Figure 2B) and similar transrepression (100% of control, Figure 2C) as MM1R-WT-GR cells. N454D/A458T cells clearly demonstrated much higher levels of GRE transactivation compared with the MM1R-WT-GR cells but with significantly lower transrepression of the NF-κB activity (30% of WT GR cells). C476W/R479Q mutant cells did not transactivate GRE or transrepress NF-κB in the reporter assays. Immunoblot assay demonstrated that dexamethasone was capable of inducing RAFTK phosphorylation in all mutant GR clones except in N454/A458T (Figure 2D).
Analysis of MM1R cells expressing GR mutants. (A) Western blot analysis of MM1R cells expressing 4 different GR mutants for GR expression with actin control. (B) Fold induction of GRE reporter expression with dexamethasone treatment of MM1R cells expressing wild-type GR or 4 GR mutants. The relative level of GRE reporter expression on dexamethasone treatment of MM1R-WT-GR cells was arbitrarily made 100%. Data from 3 experiments (mean ± SD). (C) Relative NF-κB transrepression with dexamethasone treatment. The data shown as NF-κB reporter inhibition relative to the repression observed in MM1R-WT-GR cells (arbitrarily 100%). Data are mean plus or minus SD of 3 separate experiments. (D) Western blot analysis of GR mutants, in the presence or absence of dexamethasone 10−6 M for 14 hours. (Top panel) Phospho-RAFTK. (Bottom panel) Actin.
Analysis of MM1R cells expressing GR mutants. (A) Western blot analysis of MM1R cells expressing 4 different GR mutants for GR expression with actin control. (B) Fold induction of GRE reporter expression with dexamethasone treatment of MM1R cells expressing wild-type GR or 4 GR mutants. The relative level of GRE reporter expression on dexamethasone treatment of MM1R-WT-GR cells was arbitrarily made 100%. Data from 3 experiments (mean ± SD). (C) Relative NF-κB transrepression with dexamethasone treatment. The data shown as NF-κB reporter inhibition relative to the repression observed in MM1R-WT-GR cells (arbitrarily 100%). Data are mean plus or minus SD of 3 separate experiments. (D) Western blot analysis of GR mutants, in the presence or absence of dexamethasone 10−6 M for 14 hours. (Top panel) Phospho-RAFTK. (Bottom panel) Actin.
Dexamethasone-apoptosis induced in mutant GR expressing MM1R cells
Dexamethasone treatment of cells was performed using different concentrations of dexamethasone and by analyzing cells at 2 time points, 48 and 60 hours. On dexamethasone treatment for 60 hours, S425G, L436V, and N454D/A458T cells undergo apoptosis in a concentration-dependent fashion (optimal with 10−6 M and minimal with 10−10 M) as shown in Figure 3 (bottom panel). In contrast, no cell death was seen in C476W/R479Q cells. When cells were analyzed at an earlier time point of 48 hours (Figure 3 top panel), N454D/A458T cells were clearly less sensitive to dexamethasone treatment compared with S425G and L436V. Only 2% to 5% cells were caspase 3 positive with N454D/A458T positive compared with 25% to 30% S425G and L436V. Figure 3 also summarizes the data for these mutants in regards to GRE transactivation, NF-κB transrepression, and RAFTK phosphorylation in response to dexamethasone treatment (Figure 3 bottom). As can be seen by comparing apoptosis induction to the biochemical properties of the mutant receptors, there is a correlation between apoptosis and the ability of GRs to induce GRE transactivation, although the apoptosis induced in N454D/A458T cells is somewhat delayed in its kinetics. Apoptosis induction (by 60 hours) is achieved with the 3 mutant receptors that also are capable of activating the GRE. In contrast, NF-κB transrepression and RAFTK phosphorylation are not required for apoptosis as they are both absent in the apoptosis-inducing N454D/A458T cells and NF-κB repression is also absent in the apoptosis-inducing S425G cells.
Percentage apoptosis as determined by caspase 3 assay and flow cytometric analysis. Cells were treated with 3 different concentrations of dexamethasone for 48 hours (top panel) and 60 hours (middle panel). The experiment was performed twice with similar results. (Bottom panel) Table describing the properties of transactivation, transrepression, and RAFTK phosphorylation for the cells expressing GR mutants.
Percentage apoptosis as determined by caspase 3 assay and flow cytometric analysis. Cells were treated with 3 different concentrations of dexamethasone for 48 hours (top panel) and 60 hours (middle panel). The experiment was performed twice with similar results. (Bottom panel) Table describing the properties of transactivation, transrepression, and RAFTK phosphorylation for the cells expressing GR mutants.
Genetic down-regulation of NF-κB in MM1R cells is sufficient to induce apoptosis
These results with the S425G and N454/A458 mutants clearly demonstrate that NF-κB transrepression is not required for dexamethasone-induced MM cell apoptosis. However, it is possible that such transrepression could mediate a degree of apoptosis that might be additive with that induced by GRE transactivation. To test this possibility, we genetically down-regulated NF-κB activity by infection of the N454D/A458T and C476W/R479Q cell lines with a replication defective IκB superrepressor (IκBSR)–expressing adenovirus.19 This gene expresses a phosphorylation-resistant IκB, which cannot dissociate from NF-κB, and competitively inhibits the endogenous IκB. For control purposes, a similar adenovirus lacking a transgene was used (“CMV” in Figure 4). In initial experiments, with an adenovirus expressing GFP as a marker gene, we observed an infection efficiency of more than 80% when a MOI of 100 was used. As shown in Figure 4A, expression of the IKBSR markedly inhibited NF-κB activation and inhibition was related to the MOI of the viral vector. An MOI of 100 completely abrogated NF-κB reporter expression in both MM1R mutants, whereas an MOI of 25 had lesser effect. As previously shown (Figure 3), N454D/A458T cells exhibit a delayed apoptotic response to dexamethasone with minimal apoptosis at 48-hour time point compared with the 60-hour time point. NF-κB inhibition in these cells was sufficient to induce apoptosis as shown in Figure 4B, compared with the control adenovirus infection. Of note, use of IκBSR at MOI of 100 has significantly more effect as an apoptosis inducer compared with MOI of 25. Thus, NF-κB inhibition alone is sufficient to achieve apoptosis in these cells. Furthermore, when dexamethasone is added to the NF-κB–inhibited N454D/A458T cells, an increase in 48-hour apoptosis is detected (Figure 4B left panel, dotted line). The increase in apoptosis is significant (P < .05) at MOI of 100. Because dexamethasone is incapable of any 48-hour apoptosis in control infected cells without inhibition of NF-κB activity, these data suggest that NF-κB activity in the N454D/A458T mutant cells causes the delay in apoptosis and, if NF-κB can be inhibited (by IκBSR in this experiment), dexamethasone can induce apoptosis more rapidly at 48 hours, possibly via GRE transactivation. Although NF-κB inhibition is also sufficient to induce apoptosis in the C476W/R479Q mutant cells with correlation to MOI used (Figure 4 right panel), in contrast to the results with N454D/A458T, the addition of dexamethasone has no further enhancing apoptotic effect in these cells, which lack GRE transactivation (right panel dotted line).
Inhibition of NF-κB activity in MM1R cells expressing GR mutants, GR N454D/A458T, and GR C476W/R479Q. (A) Inhibition of NF-κB activity in cells infected with adenoviral vector expressing Iκ superrepressor. Cells expressing mutant GR N454D/A458T and C476W/R479Q were infected with either a control adenovirus (CMV) with no transgene or adenovirus expressing Iκ superrepressor at an MOI of 100 and 25, and 24 hours later transfected with NF-κB luciferase reporter, rel A expression plasmid (PLG033-P65-SP10), and TK-Renilla luciferase reporter (“Reporter assays”). Twelve hours after transfection, cells were analyzed by a dual luciferase assay. Data shown as ratio of firefly to Renilla luciferase, relative to the cells not infected with viral vector. (B) Cells expressing mutant GR N454D/A458T and C476W/R479Q were infected with either a control adenovirus (CMV) with no transgene or adenovirus expressing Iκ superrepressor at MOI 5, 25, and 100 and simultaneously treated with 10−6 M dexamethasone. Caspase assay was performed at 48 hours after infection and initiation of dexamethasone treatment.
Inhibition of NF-κB activity in MM1R cells expressing GR mutants, GR N454D/A458T, and GR C476W/R479Q. (A) Inhibition of NF-κB activity in cells infected with adenoviral vector expressing Iκ superrepressor. Cells expressing mutant GR N454D/A458T and C476W/R479Q were infected with either a control adenovirus (CMV) with no transgene or adenovirus expressing Iκ superrepressor at an MOI of 100 and 25, and 24 hours later transfected with NF-κB luciferase reporter, rel A expression plasmid (PLG033-P65-SP10), and TK-Renilla luciferase reporter (“Reporter assays”). Twelve hours after transfection, cells were analyzed by a dual luciferase assay. Data shown as ratio of firefly to Renilla luciferase, relative to the cells not infected with viral vector. (B) Cells expressing mutant GR N454D/A458T and C476W/R479Q were infected with either a control adenovirus (CMV) with no transgene or adenovirus expressing Iκ superrepressor at MOI 5, 25, and 100 and simultaneously treated with 10−6 M dexamethasone. Caspase assay was performed at 48 hours after infection and initiation of dexamethasone treatment.
Apoptosis and NF-κB transrepression in additional myeloma cell lines
To test whether our findings on the role of NF-κB in dexamethasone-induced apoptosis of MM1S cells could be generalized, we also studied other myeloma cell lines. Cells were treated with 10−6 M dexamethasone and analyzed for apoptosis by caspase 3 assay after 72 hours of dexamethasone treatment. ANBL-6 cells were grown in the presence of 100 units of IL-6 per mL and IL-6 was withdrawn 24 hours before the addition of dexamethasone. Three cell lines, U266, AF-10, and ARH77, were found to be resistant to dexamethasone-induced apoptosis, whereas Opm-2, 8226, and ANBL-6 cell lines were dexamethasone-sensitive (dark bars in Figure 5 representing percentage of cells positive for caspase 3). When these cell lines were tested for NF-κB transrepression, only AF-10 and ARH77 demonstrated transrepression with dexamethasone treatment (30% and 60% decrease in reporter activity, respectively). Thus, in the 3 cell lines (Opm-2, 8226, and ANBL-6), which are sensitive to dexamethasone-mediated apoptosis, NF-κB transrepression is not required for apoptosis. In addition, in AF-10 and ARH77 cell lines, transrepression is not sufficient for apoptosis. These data further support the experiments with GR mutants in the MM1S model.
Dexamethasone induced apoptosis and NF-κB transrepression in different myeloma cell lines. U266, Opm-2, 8226, ANBL-6, AF-10, and ARH77 cell lines were treated with 10−6 M dexamethasone for 72 hours and percentage of cells undergoing apoptosis identified by caspase 3 assay. Bar diagram shows percentage of cells undergoing apoptosis (■). NF-κB transrepression experiment was performed as described in “Reporter assays.” □ shows the percentage decrease in the NF-κB reporter activity on the addition of dexamethasone. Both the experiments were done twice with similar results.
Dexamethasone induced apoptosis and NF-κB transrepression in different myeloma cell lines. U266, Opm-2, 8226, ANBL-6, AF-10, and ARH77 cell lines were treated with 10−6 M dexamethasone for 72 hours and percentage of cells undergoing apoptosis identified by caspase 3 assay. Bar diagram shows percentage of cells undergoing apoptosis (■). NF-κB transrepression experiment was performed as described in “Reporter assays.” □ shows the percentage decrease in the NF-κB reporter activity on the addition of dexamethasone. Both the experiments were done twice with similar results.
BIM induction
We also studied the role of BIM induction in the process of dexamethasone-mediated apoptosis in these cell lines. BIM is a proapoptotic gene containing a BH3 domain that is transcribed as 3 major splice variants EL, L, and S, which encode distinct proteins. The BimS protein has been reported to the most strongly proapoptotic of the 3 isoforms.20 The 3 major isoforms of BIM are expressed in myeloma cells, and its expression is negatively regulated by IL-6,21 a viability factor that protects against dexamethasone induced-apoptosis. Furthermore, Bim induction has been indicted in dexamethasone-induced apoptosis of lymphoid leukemia cells as well.4,12,13 As expected, immunoblot analysis of our cells confirms that dexamethasone induces Bim expression in MM1S cells, there is no effect in MM1R cells (MM1R-EV cells in Figure 6A), and reexpression of wild-type GR reestablishes dexamethasone-induced Bim induction. This is most evident in the immunodetection of BimL and BimS. In addition, all the mutant clones are successful in enhancing BimL and BimS expression when exposed to dexamethasone (Figure 6B). Because cells expressing the C476W/R479Q mutant GR are resistant to dexamethasone-mediated apoptosis (Figure 3) but nevertheless able to induce Bim expression, it is clear that Bim expression is not sufficient for dexamethasone-induced apoptosis.
BIM induction in MM1S cells and MM1R cells expressing different GR mutants with dexamethasone treatment (10−6 M for 14 hours). (A) Western blot analysis of BIM induction in MM1S, MM1R-EV, and MM1R with wild-type GR (MM1R-WT-GR). With actin control. (B) MM1R cells with 4 GR mutants and actin control.
BIM induction in MM1S cells and MM1R cells expressing different GR mutants with dexamethasone treatment (10−6 M for 14 hours). (A) Western blot analysis of BIM induction in MM1S, MM1R-EV, and MM1R with wild-type GR (MM1R-WT-GR). With actin control. (B) MM1R cells with 4 GR mutants and actin control.
Discussion
Glucocorticoids are important apoptosis inducers in MM and lymphoid leukemias. We have used a strategy exploiting the dexamethasone-resistant GR-null MM1R MM cell line with reexpression of mutant GRs. This MM1R line has been compared with its dexamethasone-sensitive parent MM1S line in several studies10,18 specifically addressing the mechanism of dexamethasone-induced apoptosis in myeloma. After creating mutations in the DNA-binding domain of the wild-type GR and reexpressing mutant GRs, we could test their variable ability to induce apoptosis. We selected these mutation sites because of their known effects on downstream induction of GRE transactivation or NF-κB transrepression6,15 in other model systems. We also tested their ability to induce phosphorylation of the RAFTK kinase as previous work10,11 had implicated that pathway in dexamethasone-induced MM cell apoptosis. Although the ligand-activated GR can also transrepress the AP-1 transcription factor,7,15 we did not investigate this effect, as it is unlikely to be important in MM cell apoptosis because many studies in MM cells22,23 document a proapoptotic effect of activating AP-1 and an antiapoptotic effect of inhibiting AP-1. Our results indicate a perfect correlation between GRE transactivation and apoptosis induction. Furthermore, NF-κB transrepression and RAFTK phosphorylation were not required for GC-apoptosis.
The zinc finger DNA binding domain of the GR is responsible for transactivation and transrepression.6,15 Mutants S425G and L436V are located in the N-terminal Zn finger structure, and Heck et al15 reported increased transactivation potential of these 2 mutant GRs compared with the wild-type GR. We also observe (Figure 2) that mutant GRs have more transactivation potential than wild-type GR (150% and 220% of wild-type GR). Mutant N454D/A458T has one of its mutations in the dimerization domain of the GR (D-loop mutant),6,15 which is required for cooperative binding of the GR to the GRE. We were surprised to note a significant GRE transactivation by this mutant, as this was not observed in a previous study15 with reporter constructs with single or multiple GRE elements. However, Dahlman-Wright et al24 demonstrated that DNA binding and transactivation were noted even in D-loop mutants. It is possible that this inconsistency is target cell specific where the coactivators and or corepressors recruited to DNA by D-loop mutants vary from cell type to cell type and this recruitment regulates gene transactivation. In any regard, our data are clear that the N454D/A458T mutant in MM cells is successful in inducing GRE transactivation subsequent to GC ligand binding. The mutant GR C476W/R479Q changes a cysteine residue that is necessary to tetrahedrally bind Zn in the second Zn finger. This disruption results in lack of transactivation (Figure 2), and these results suggest that the overall conformation of the second Zn finger structure in the GR is more important for GRE transactivation in MM cells than the dimerization loop. Our results indicate that GRE transactivation by the mutant GRs correlates well with the induction of MM cell apoptosis. However, apoptosis induction is more marked in S425G and L436V mutants compared with the N454D/A458T mutant, and the latter is more successful at GRE transactivation. Indeed, when assayed at an early time point (48 hours), the N454D/A458T mutant is unsuccessful at initiating apoptosis. One possible explanation for these results is an interaction between the ability of GR to induce GRE activation and either transrepress NF-κB (as in L436V mutant) or activate RAFTK (as in both S425G and L436V). Thus, the lesser degree of apoptosis in the double-mutant N454D/A458T may be explained by its inability to achieve some other effects that could contribute to apoptosis. Further support for the additive role of NF-κB transrepression to GC apoptosis is described in the next paragraph.
The role of NF-κB transrepression in GC-mediated MM cell apoptosis has been assumed from a host of other studies that demonstrate MM cell apoptosis when NF-κB is inhibited.25-27 However, there are additional studies that demonstrate a lack of apoptosis in NF-κB inhibited MM cells.28 It is possible that MM cells containing genetic lesions that result in constitutive heightened NF-κB activity are somewhat addicted to that transcription factor for survival and rapidly die when activity is inhibited. Such genetic lesions have recently been described,29,30 and the MM1S model contains such lesions. Our results are clear that NF-κB transrepression is not required for dexamethasone-induced apoptosis. However, it may contribute to apoptosis and, to test this, we genetically inhibited NF-κB in N454D/A458T and C476W/R479Q cells. Inhibition of NF-κB with IκB superrepressor, IκBSR, is in itself able to trigger apoptosis (Figure 4), a finding consistent with other studies and scores the importance of this pathway in myeloma cells. The addition of dexamethasone to N454D/A458T cells with a repressed NF-κB pathway significantly increases apoptosis when assayed at 48 hours (Figure 4B left panel). In contrast, 48 hours of exposure to dexamethasone in control infected cells does not induce apoptosis. These results suggest that NF-κB activity is a survival factor in MM1R cells and can modulate apoptosis mediated by other effects of dexamethasone, probably mediated by GRE transactivation. In contrast, in the C476W/R479Q mutant with lack of GRE transactivation, addition of dexamethasone does not increase apoptosis in NF-κB–inhibited cells, supporting this notion. Thus, the data suggest that, in the N454D/A458T cells, both pathways, GRE transactivation and NF-κB inhibition, can achieve apoptosis in an additive and interactive way. Furthermore, the inability of this mutant GR to transrepress NF-κB results in a delayed and somewhat decreased apoptotic response compared with the L436V mutant. Apoptosis and NF-κB transrepression studies were also extended to other myeloma cell lines (Figure 5). The data provide further evidence that dexamethasone-mediated apoptosis in Opm-2, 8226, and ANBL-6 cell lines is not the result of a concurrent inhibition of NF-κB activity.
Previous studies also support a role for the RAFTK in dexamethasone-induced MM cell apoptosis.10,11 Dexamethasone induces a posttranslational modification of phosphorylation, and blocking this event with an RAFTK mutant prevented apoptosis.10,11 Our results demonstrate that RAFTK phosphorylation was neither sufficient by itself nor required for GC-induced apoptosis. It is possible, however, that RAFTK phosphorylation participates in apoptosis when an additional GR-mediated event occurs, such as GRE transactivation. Such an additive apoptotic effect could explain the higher degree of apoptosis in the S425G mutant compared with the C476W/R479Q mutant.
Previous literature also suggests that induction of the proapoptotic protein Bim may be important for GC-induced apoptosis. Dexamethasone treatment induces Bim expression, and Bim knock-out mice demonstrate a delayed apoptotic response to dexamethasone in leucocytes and thymocytes. Furthermore, in ALL cell lines, Bim silencing results in dexamethasone resistance.4 In MM cells, Mcl1, an antiapoptotic protein, binds and neutralizes Bim and thus prevents apoptosis.21 During apoptosis, the disappearance of Mcl1 allows Bim to function as a proapoptotic protein. In our study, Bim induction is seen in MM1S cells, is absent in MM1R-EV cells, and is induced in MM1R-WT-GR cells (Figure 6A). Induction of all the isoforms is seen with a more pronounced effect in BimL and BimS isoforms. This induction is also seen in all the mutant GR-expressing cells when exposed to dexamethasone (Figure 6B). It is clear that Bim is induced by a GR mutant that is deficient in GRE transactivation (C476W/R479Q). This is consistent with the finding that the BIM gene does not contain a GRE.31 Apparently, the ability of dexamethasone to induce expression of Bim is the result of the ability of GRs, including all our GR mutant GRs, to interact positively with other transcriptional factors. As mentioned, BimS is the most proapoptotic of the 3 isoforms,20 and its induction is observed in response to dexamethasone treatment. However, a lack of correlation between apoptosis and Bim induction is seen as mutant C476W/R479Q also induces Bim but is resistant to apoptosis, indicating that its up-regulation alone is not sufficient for apoptosis.
In conclusion, we have studied the mechanism of dexamethasone-induced apoptosis in a myeloma model by analyzing cells expressing different GR mutants. By comparing the transactivation and repression characteristics of the GR mutants, we find that GRE transactivation correlates with the induction of apoptosis. NF-κB inhibition by itself is also able to induce apoptosis but is not essential for apoptosis in dexamethasone-treated cells. This model of isogenic cell lines expressing mutant GRs could be further used to examine the key determinants of dexamethasone-induced apoptosis. Gene expression profiling of these mutant GR expressing cells treated with or without dexamethasone may help pinpoint the genes critical for the apoptotic response.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by research funds from the Veterans Health Administration and grant RO1 CA111448 awarded by the National Institutes of Health.
National Institutes of Health
Authorship
Contribution: S.S. designed and performed experiments and wrote the manuscript; and A.L. analyzed data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sanjai Sharma, West Los Angeles VA Medical Center, Department of Hematology Oncology, 11301 Wilshire Blvd, Bldg 304, E1-115, Los Angeles, CA 90073; e-mail: sasharma@mednet.ucla.edu.