Abstract
In 1951 William Dameshek classified polycythemia vera (PV), essential thombocytosis (ET), and primary myelofibrosis (PMF) as pathogenetically related myeloproliferative disorders (MPD). Subsequent studies demonstrated that PV, ET, and PMF are clonal disorders of multipotent hematopoietic progenitors. In 2005, a somatic activating mutation in the JAK2 nonreceptor tyrosine kinase (JAK2V617F) was identified in most patients with PV and in a significant proportion of patients with ET and PMF. Subsequent studies identified additional mutations in the JAK-STAT pathway in some patients with JAK2V617F− MPD, suggesting that constitutive activation of this signaling pathway is a unifying feature of these disorders. Although the discovery of mutations in the JAK-STAT pathway is important from a pathogenetic and diagnostic perspective, important questions remain regarding the role of this single disease allele in 3 related but clinically distinct disorders, and the role of additional genetic events in MPD disease pathogenesis. In addition, these observations provide a foundation for development of small molecule inhibitors of JAK2 that are currently being tested in clinical trials. This review will discuss our understanding of the pathogenesis of PV, ET, and PMF, the potential role of JAK2-targeted therapy, and the important unanswered questions that need to be addressed to improve clinical outcome.
Introduction
It is fitting that the myeloproliferative disorders (MPD) be discussed on the occasion of the 50th Anniversary of the American Society of Hematology. Polycythemia vera (PV), essential thrombocytosis (ET), and primary myelofibrosis (PMF) serve as a superb paradigm for progress in our understanding and treatment of hematologic disorders over the past 50 years. These entities had been described well over 50 years ago, but clinically prescient insights from William Dameshek in 1951 suggested their interrelatedness. Subsequently, concepts and methods for determining clonal origin of cells based on X-inactivation in females were developed, and demonstrated that these MPD were indeed clonally derived malignancies. These seminal observations in turn fueled an intensive search for the acquired somatic mutations causally implicated in disease pathogenesis. These efforts were facilitated in part by technologic advances in genome and biologic sciences, including the use of small interfering RNA and high-throughput DNA sequencing. In 2005, PV, ET, and PMF were shown to be caused, in most cases, by mutations that constitutively activate the tyrosine kinase JAK2. In-depth study of these disease alleles has begun to shed light on the molecular mechanisms of disease, as well as the complex interplay between genotype and phenotype, with a single mutation contributing to 3 clinically distinct phenotypes. Finally, and perhaps of most importance, JAK2 is a promising candidate for molecularly targeted therapies that are currently in phase 1/2 trials, demonstrating the remarkable pace at which drug development can proceed after target identification and validation. Thus, in 50 years we have come full circle from clinical description of disease, to allele discovery and characterization, to targeted therapies in humans with MPD.
This story begins in 1892, when Louis Henri Vaquez first described polycythemia vera in a patient with marked erythrocytosis and hepatosplenomegaly that he postulated was the result of hematopoietic cell proliferation.1 Subsequently, William Osler described a set of patients with erythrocytosis and splenomegaly, which he referred to as “Vaquez's disease.”2 Myelofibrosis was initially described and characterized by Gustav Hueck, a German physician who noted the presence of bone marrow fibrosis and extramedullary hematopoiesis in patients with PMF.3 Of the different MPD, ET was the last to be described, when in 1934 Emil Epstein and Alfred Goedel recognized that patients with thrombocytosis without marked erythrocytosis constituted a distinct clinical syndrome.4
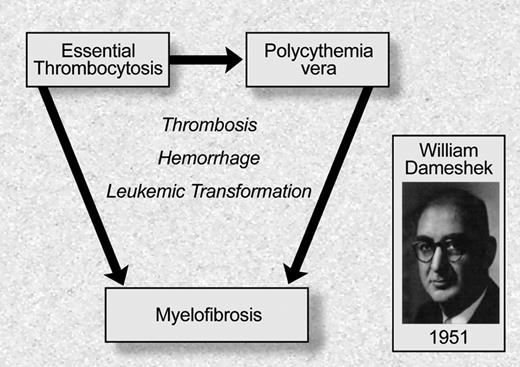
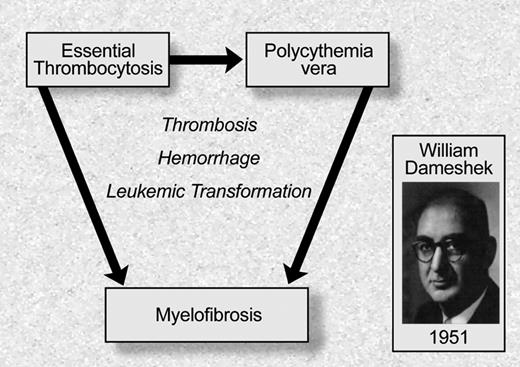
Although the different MPD had been recognized as distinct clinical entities over the previous half century, William Dameshek, the founding editor of Blood, was the first to recognize that these entities should be classified as a set of phenotypically related “myeloproliferative disorders”5 (Figure 1). He noted that although erythrocytosis is characteristic of PV, many PV patients have “pancytosis” with proliferation of the erythroid, megakarycyte, and granulocyte lineages. Moreover, he also noted that patients with PV often develop bone marrow fibrosis, leuko-erythroblastic changes in their peripheral blood, and increasing splenomegaly consistent with progression to “terminal myelofibrosis.” Dameshek argued in this editorial5 that, given the difficulties in distinguishing between PV, PMF, and other MPD, it might be easiest to consider these disorders as “closely interrelated” disorders characterized by bone marrow proliferation, “perhaps due to a hitherto undiscovered stimulus.” This insight has recently proven to be remarkably prescient, and an example of the importance of clinical observation as it relates to the genetic underpinnings of disease. Although the nomenclature and definitions of the different MPD have changed in the ensuring decades, the seminal observation that these are clinically and biologically related proliferative bone marrow syndromes has been substantiated by the recent elucidation of the genetic basis of MPD.
Philadelphia chromosome negative myeloproliferative disorders: classification based on William Dameshek. Illustration by Debra Tyler.
Philadelphia chromosome negative myeloproliferative disorders: classification based on William Dameshek. Illustration by Debra Tyler.
Early investigation
Although the discovery of the Philadelphia chromosome6 and the 9:22 translocation7 provided initial insights into the molecular pathogenesis of CML (discussed in detail elsewhere in this series), our understanding of the molecular pathogenesis of Philadelphia-chromosome negative MPD began with a series of elegant studies by Fialkow, Adamson, Axelrad, and other investigators in the 1970s. In 1967, Fialkow and colleagues studied female CML patients with polymorphisms at the G6PD locus. They observed that a single G6PD isoform was present in all red blood cells and granulocytes, consistent with clonally derived myelopoiesis.8 In 1976, Adamson, Fialkow, and colleagues used the same strategy to demonstrate a single G6PD isoform in red blood cells, granulocytes, and platelets from patients with PV, consistent with panmyeloid clonal expansion.9 Subsequent studies using polymerase chain reaction (PCR)–based assays confirmed this initial observation,10 and extended these findings to ET and MF.11,12 In addition, analysis of recurrent cytogenetic alterations, in particular deletions on the long arm of chromosome 20, demonstrate that clonal alterations can occur in MPD cells with multilineage potential.13 Taken together, these data indicated that PV, ET, and PMF originates in a multipotent hematopoietic progenitor, regardless of the predominant lineage(s) involved in the clinical phenotype.
A second seminal observation was made by Jaroslav Prchal and Arthur Axelrad, when they noted that in vitro culture of bone marrow cells from PV patients, but not from normal volunteers, gave rise to erythroid colonies in the absence of exogenous cytokines.14 This phenomenon of endogenous erythroid colony (EEC) formation is characteristic of PV, and is observed in nearly all patients with PV who have not been exposed to cytoreductive therapy. EEC formation is also observed in a subset of patients with ET and PMF,14,15 consistent with the clinical and pathogenetic overlap of these 3 disorders. Although subsequent studies have suggested that the “endogenous” growth of EECs derived from MPD cells might reflect increased responsiveness to limiting concentrations of erythropoietin (EPO),16 this study served as the initial demonstration that the bone marrow proliferation observed in PV, ET, and PMF is a cell-autonomous characteristic of the MPD clone, as had been suggested by Dameshek.
Clinical trials: Polycythemia Vera Study Group and beyond
Clinical investigation in these disorders entered the modern era with the creation of the Polycythemia Vera Study Group (PVSG) in 1967 by Louis Wasserman. Under his leadership, the PVSG performed a series of randomized trials in PV, first demonstrating that phlebotomy was superior to phlebotomy plus chlorambucil or P32, due to an increased incidence of leukemic transformation in patients treated with chlorambucil or P32.17 The PVSG subsequently reported that hydroxyurea was associated with a reduced risk of thrombosis compared with a historical series of patients managed with phlebotomy,18 and that high-dose antiplatelet therapy was associated with an increased risk of bleeding in PV.19 Although the PVSG no longer actively conducts clinical trials in MPD, the accomplishments of the PVSG represent an important milestone for MPD clinical research, as the PVSG was the first clinical trial group dedicated to systematic investigation of clinical therapies for the treatment of MPD. Subsequent to the PVSG, there have been several landmark MPD clinical trials that have served to define the current standard of care for the treatment of PV and ET. Landolfi and colleagues performed a randomized trial in PV demonstrating that low-dose aspirin therapy in PV is associated with a reduced risk of thrombotic complications without associated bleeding risks.20 This observation has led to the use of low-dose aspirin for prophylaxis of thrombosis in PV. In addition, randomized trials have demonstrated that hydroxyurea, in combination with antiplatelet therapy, reduces thrombotic complications in ET in comparison to placebo or to anagrelide.21,22 Although these studies have provided important insight into the management of PV and ET, only one novel agent, anagrelide, has been approved for the treatment of these disorders in the past 25 years, and to date there are no randomized trials available to define the optimal management of PMF. There is therefore a need for new therapies for patients with PV, ET, and PMF, which would ideally be based on genetic insight into disease pathogenesis.
Discovery of JAK2V617F
Although several studies had noted dysregulated expression of specific genes in MPD, including the thrombopoietin receptor (MPL),23 BCL-XL,24 and PRV-1,25 the genetic basis for these disorders remained elusive for more than a half century after Dameshek's seminal editorial in Blood. In 2005, the observations of Dameshek, Fialkow, Adamson, and Prchal converged with the identification of the JAK2V617F allele in the majority of patients with PV, ET, and PMF.26-29 A variety of genetic, functional and genomic approaches allowed the different groups to identify the identical mutation in JAK2 in these disorders. The group led by William Vainchenker observed that small molecule or siRNA-mediated inhibition of JAK2 in PV hematopoietic progenitors abrogated EEC formation,30 which led them to examine JAK2 for mutations in PV.26 Anthony Green and colleagues used candidate gene resequencing followed by allele-specific PCR to identify the JAK2V617F allele in PV, ET, and PMF.27 Building on the key observation of Josef Prchal and colleagues that acquired uniparental disomy (UPD) of chromosome 9p24 is common in PV,31 Robert Kralovics, Radek Skoda, and their colleagues sequenced the genes in the minimal region of UPD to identify the JAK2V617F allele.28 Levine and colleagues used an approach based on the previous identification of activating mutations in tyrosine kinases in other MPDs, and performed a systematic survey of the tyrosine kinome in PV using high throughput DNA resequencing that led to the identification of the recurrent mutation in JAK2 in MPD.29
The mutation in JAK2 is a guanine to thymidine substitution that results in a valine to phenylalanine substitution at codon 617 of JAK2. The mutation is not present in the germ line, consistent with the notion that JAK2V617F is acquired as a somatic disease allele in the hematopoietic compartment. To date no alternate mutations have been described at this codon in MPD or in other human malignancies, which is surprising, especially in light of a recent report demonstrating that substitution of any of 4 alternative residues (tryptophan, methionine, isoleucine, and leucine) for valine at codon 617 results in constitutive JAK2 activation and in hematopoietic transformation in vitro.32 In addition, alternate activating mutations that activate JAK2, including the T875N kinase domain mutation,33 the IREED deletion,34 and the less common JAK2 exon 12 mutations35 have been observed in patient samples and in cell lines although each of these alleles is exceedingly rare in frequency compared with JAK2V617F. Future studies may elucidate the functional basis for a single recurrent mutation in JAK2 that is observed in such a high proportion of patients with PV, ET, and PMF. Although JAK2 signaling is constitutively activated by a variety of genetic and epigenetic mechanisms in human malignancies, including lymphoma and myeloma,36,37 the JAK2V617F allele is exclusive to myeloid malignancies,38,39 suggesting there are distinct mechanisms that activate JAK2 in different neoplasms. This is particularly notable in light of the recent discovery of novel activating mutations in the pseudokinase domain of JAK2, centered around codon 683, in Down syndrome–associated acute lymphoblastic leukemia.40
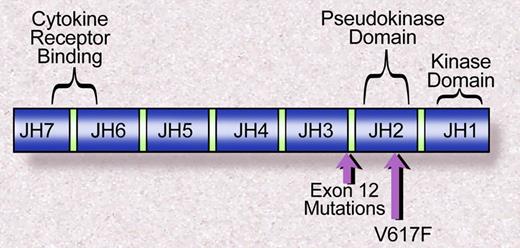
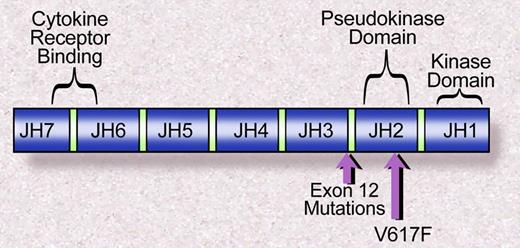
The Janus family of nonreceptor tyrosine kinases includes JAK1, JAK2, JAK3, and TYK2. Each of the JAK kinases has 7 JAK homology domains (Figure 2). The JHI domain, at the C terminus, contains the catalytic domain, whereas the N-terminal JH7 domain has been shown to be essential for association with cytokine receptors.41 Valine 617 lies within the JH2, or pseudokinase domain, which has significant homology to the JH1 domain but lacks catalytic activity. The crystal structure of the JH1 domains of JAK2 and JAK3 have been solved,42,43 and that has permitted modeling of the structure of JH2 based on a high degree of sequence homology between JH1 and JH2. However, the structure of the full-length JAK2, or the JAK2 JH1 and JH2 domain together, has not been solved. Thus the precise mechanism of kinase activation mediated by JAK2V617F is not fully understood. However, based on the observation that deletion of the JH2 domain leads to increased JAK2 kinase activity,44 it has been suggested that the JH2 domain negatively regulates JH1 kinase activity, similar to the autoinibitory role of juxtamembrane domains in receptor tyrosine kinases, such as FLT3.45 Indeed, a computational model of the structure of JAK2 suggests that valine 617 closely associates with the JH1 activation loop.46 However, direct structural insight is needed to elaborate the role of the JAK2 JH2 domain, and of valine 617 specifically, in the regulation of JAK2 kinase activity.
Janus homology domains of the JAK family of kinases. The V617F mutation is within the JH2, or pseudokinase domain, whereas the exon 12 mutations in JAK2 are proximal to the JH2 domain. Illustration by Debra Tyler.
Janus homology domains of the JAK family of kinases. The V617F mutation is within the JH2, or pseudokinase domain, whereas the exon 12 mutations in JAK2 are proximal to the JH2 domain. Illustration by Debra Tyler.
The JAK kinases, through their association with cytokine receptors and receptor tyrosine kinases, are central to cytokine signaling and to signal transduction in all mammalian cells. Genetic and biochemical data demonstrate that JAK kinase activation is required for cytokine receptor signaling, and that the disparate physiologic effects of different cytokine receptors is in part due to the ability of these cytokine receptors to engage different JAK kinases after cytokine stimulation. Most cytokine receptors can associate with more than one JAK kinase, but it has been shown that Jak2-deficient myeloid progenitors fail to respond to EPO, thrombopoietin, or granulocyte-monocyte colony stimulating factor, and that Jak2 deficiency results in an absence of definitive erythropoiesis.47 These data suggest that JAK2 is the predominant JAK kinase involved in myeloid cell proliferation and differentiation. It is perhaps not surprising, therefore, that a gain-of-function mutation in JAK2 is observed in a spectrum of myeloid malignancies,38,48,49 and that JAK2V617F+ cells are hypersensitive to cytokine stimulation.29,50
One mutation, 3 diseases
The identification of JAK2V617F provided important insight into the pathogenesis of PV, ET, and PMF, but many important questions remain regarding the pathogenesis of these neoplasms. First and foremost among these is the contribution of a single disease allele to the development of 3 related, but clinicopathologically distinct, myeloid disorders. It was noted in the initial studies describing JAK2V617F that subsets of patients were homozygous for the JAK2V617F allele, consistent with loss of heterozygosity (LOH) at the JAK2 locus.27-29,50 Unlike classical LOH observed with tumor suppressor genes, in which one allele is inactivated by mutation and the second by deletion, LOH and resultant JAK2V617F homozygosity is copy neutral—the result of acquired UPD at chromosome 9p24.31 The genetic data are consistent with the notion that JAK2V617F gene dosage influences MPD phenotype, as homozygous JAK2V617F mutant erythroid colonies are observed in most patients with PV, even in patients with a low JAK2V617F allele burden, but are only rarely observed in ET.51 It is therefore likely that chromosome 9p24 UPD and resultant JAK2V617F homozygosity is a relatively early event in PV pathogenesis, and that the increase in allele burden over time in PV represents clonal selection of the preexisting JAK2V617F homozygous mutant clone. Data from murine models of Jak2V617F-mediated MPD are also consistent with the notion that gene dosage contributes to MPD phenotype. Initial in vivo studies using a retroviral bone marrow transplant (BMT) assay demonstrated that overexpression of Jak2V617F in the hematopoietic compartment results in a fully penetrant MPD phenotype notable for polycythemia and leukocytosis without associated thrombocytosis.52,53 In these reports, there was strain-specific variation in the degree of leukocytosis and myelofibrosis, suggesting an additional host-modifying influence on disease phenotype. More recently, 2 groups have generated transgenic models of JAK2V617F-mediated disease.54,55 The lower level of JAK2V617F expression in these transgenic models results in an ET-like phenotype with thrombocytosis and mild leukocytosis, but not polycythemia. Taken together, the human and murine data suggest a single copy of the JAK2V617F allele does not saturate activation of JAK2 signaling, and that there are qualitative and/or quantitative differences in constitutive signaling in JAK2V617F-mutated cells depending on gene dosage, which affect MPD phenotype. Future studies using more accurate genetic “knock-in” models in which Jak2V617F is expressed from the endogenous promoter, and of primary patient samples, are needed to elucidate differences in signaling in cells heterozygous and homozygous for JAK2V617F, and to determine the contribution of JAK2V617F gene dosage in MPD pathogenesis.
In addition to JAK2V617F gene dosage, several lines of evidence suggest that additional inherited alleles contribute to the pathogenesis of PV, ET, and PMF. The most compelling evidence comes from families with a predisposition for the development of MPD. In many of these kindreds the inheritance pattern is consistent with autosomal dominant inheritance with incomplete penetrance, whereas in other families the inheritance pattern is less evident.56,57 Regardless of the mode of inheritance, in all familial JAK2V617F+ MPD cases evaluated to date, JAK2V617F is acquired as a somatic allele, consistent with the existence of heretofore-unidentified MPD predisposition alleles. The presence of the identical somatic mutation in JAK2 in most affected individuals suggests that these inherited MPD alleles may directly or indirectly amplify JAK2 signaling. This would then lead to a greater selective advantage for hematopoietic progenitors that acquire activating mutations in the JAK2 signaling pathway. In support of this hypothesis is the recent description of families where some individuals at risk develop JAK2V617F+ MPD, and others acquire somatic mutations in Exon 12 of JAK2.58 In addition, we have observed that 5% to 10% of patients with PV, ET, and PMF have a first degree relative with an MPD, suggesting that there are low-penetrance inherited alleles that contribute to MPD pathogenesis in some patients with presumed sporadic disease (R.L.L.,M. Wadleigh, and D.G.G., unpublished observations, July 1, 2008).
In addition to predisposition alleles, it is also possible that there are inherited modifiers, which may not predispose to MPD development, but instead contribute to the clinical phenotype of JAK2V617F+ MPD. Pardanani and colleagues analyzed 32 single nucleotide polymorphisms (SNPs) in JAK2, MPL, EPOR, and GCSFR in 179 patients with MPD, and found that specific SNPs in JAK2 were associated with PV or with ET.59 These findings need to be confirmed in independent cohorts, and the mechanism by which these SNPs influence phenotype remains to be delineated. Nonetheless these data provide evidence that host genetic variation contributes to the phenotype of JAK2V617F+ MPD. With the advent of high throughput SNP genotyping platforms, it will be possible to extend these findings to the whole genome, and to identify modifier alleles that influence MPD phenotype, progression, and complications.
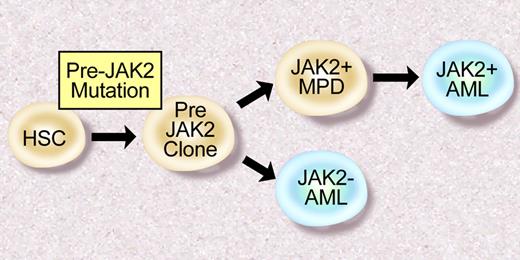
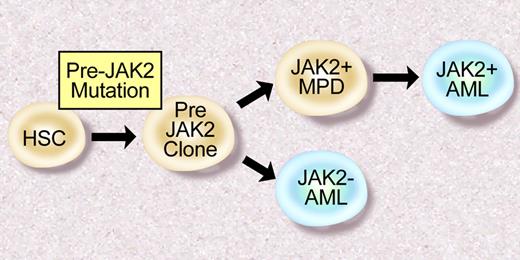
There is also reason to suspect there are additional somatic mutations that contribute to the pathogenesis of JAK2V617F+ PV, ET, and PMF. First, clonogenic cytogenetic abnormalities have been observed in patients with MPD.60 Furthermore, in some cases, such as deletion 20q, these abnormalities are frequently observed in JAK2V617F+ MPD.61 In addition, molecular analysis of 2 patients with JAK2V617F and 20q deletions demonstrated that all hematopoietic cells had 20q loss, whereas a smaller proportion of cells were JAK2V617F+.62 These data, as well as clonality analysis of a large MPD cohort,63 suggest there are “pre-JAK2 alleles” responsible for clonal hematopoiesis in excess of the JAK2V617F mutated subclone. Moreoever, many patients with a JAK2V617F+ MPD transform to a JAK2V617F− acute leukemia,61,64 consistent with the presence of a JAK2V617F− transformed hematopoietic progenitor (Figure 3). To date, no group has identified JAK2V617F− MPD patients who subsequently acquire the JAK2V617F allele during disease progression, suggesting the “pre-JAK2” clone does not manifest as a clinical MPD. The identity of these “pre-JAK2” alleles, and their relationship to JAK2 signaling and MPD pathogenesis, is the subject of current and future investigation by many investigators.
Model of leukemic transformation in MPD. Some patients develop JAK2V617F+ AML after JAK2V617F+ MPD, consistent with a JAK2V617F+ progenitor that undergoes leukemic transformation. However, some patients with JAK2V617F+ MPD transform to a JAK2V617F− AML, consistent with a “pre-JAK2” clone, which is susceptible to leukemic transformation. Illustration by Debra Tyler.
Model of leukemic transformation in MPD. Some patients develop JAK2V617F+ AML after JAK2V617F+ MPD, consistent with a JAK2V617F+ progenitor that undergoes leukemic transformation. However, some patients with JAK2V617F+ MPD transform to a JAK2V617F− AML, consistent with a “pre-JAK2” clone, which is susceptible to leukemic transformation. Illustration by Debra Tyler.
JAK2V617F− MPD
Although the majority of patients with PV and a significant proportion of patients with ET and PMF are JAK2V617F+, there are a subset of patients with these disorders who are JAK2V617F−, even when sensitive, allele specific assays are used.27,63 Analysis of large patient cohorts has allowed investigators to delineate differences in laboratory and clinical parameters between JAK2V617F+ and JAK2V617F− MPD patients,65 but these differences are too small to separate MPD patients into clinically distinct subsets based on JAK2V617F mutational status. This led investigators to hypothesize that JAK2V617F− MPD results from somatic mutations that activate JAK2 signaling in an analogous fashion to the JAK2V617F kinase.
Scott et al used a candidate gene approach to search for alleles that activate JAK signaling in the small proportion of patients with JAK2V617F− PV.35 DNA sequence analysis of all exons of JAK2 resulted in the identification of somatic mutations in exon 12 of JAK2 in 10 patients with JAK2V617F− PV. Unlike JAK2V617F in which a single allele has been identified, at least 8 distinct mutations in exon 12 of JAK2 have been reported, including missense mutations, deletions, and insertions involving residues 538 to 543.58 As has been observed for JAK2V617F, expression of JAK2 exon 12 mutations transforms hematopoietic cells to cytokine independent growth, and activates downstream signaling pathways including STAT5, AKT, and MAP kinase signaling. In addition, expression of JAK2 exon 12 mutations in vivo in the murine BMT assay results in a MPD phenotype similar to that observed with JAK2V617F, including marked polycythemia and extramedullary hematopoiesis. These data demonstrate that JAK2 exon 12 mutations are bona fide activating alleles in JAK2V617F− PV. Although JAK2V617F is observed in PV, ET, and PMF, exon 12 mutations to date have only been identified in patients with JAK2V617F− erythrocytosis, and in most cases are observed in patients with idiopathic erythrocytosis (IE) without associated thrombocytosis or leukocytosis. This has important clinical and biologic implications, as testing for JAK2 exon 12 mutations may be of specific value in patients who present with IE,66 as opposed to classical PV with pan-myeloid expansion. Moreover, these data suggest that different activating mutations in JAK2 are associated with different clinical phenotypes.
Alternate alleles in JAK2 or other JAK kinases have not been identified in JAK2V617F− ET and MF, suggesting that mutations in other genes important in signal transduction are present in this subset of patients. The role of JAK2 in EPO, TPO, and GCSF cytokine signaling, and the presence of activating mutations in cytokine receptors in patients with inherited erythrocytosis or thrombocytosis,67,68 suggests that mutations in myeloid specific cytokine receptors might contribute to the pathogenesis of JAK2V617F− ET and MF. Indeed, a point mutation at codon 515 of MPL has been identified in 5% to 10% of JAK2V617F− ET and PMF.69,70 The mutations at codon 515 substitute either leucine or lysine for trytophan. Although in vitro studies demonstrate that expression of MPLW515L constitutively activates JAK2 and downstream signaling pathways, as observed for JAK2V617F, expression of MPLW515L in vivo results in a distinct phenotype notable for marked thrombocytosis and marked myelofibrosis. It has been suggested that the RWQFP motif, at the transmembrane-juxtamembrane junction, which includes W515, maintains MPL in an inactive conformation in the absence of ligand stimulation, which presumably is altered by mutations at codon 515.71 However, as is the case for JAK2V617F and JAK2 exon 12 mutations, structural insight is needed to understand the mechanism of activation of MPLW515 mutations. In addition, recent clinical studies observed that MPLW515+ ET is associated with higher platelet counts than JAK2V617F+ or JAK2/MPL wild-type ET,72,73 and that endogenous megakaryocyte colonies, but not endogenous erythroid colonies, can be grown from MPLW515+ patient cells. In PMF, MPLW515+ disease is associated with increased anemia and with an increased need for transfusions,74 consistent with reduced erythropoiesis compared with JAK2V617F+ PMF. These data suggest that MPLW515+ hematopoietic progenitors have a bias toward the megakaryocyte lineage, and that there are differences in signaling between JAK2V617F and MPLW515L/K that influence clinical phenotype.
These studies collectively suggest that constitutive activation of JAK2 signaling is a common pathogenetic event in PV, ET, and PMF, and that additional alleles in the JAK-STAT signaling pathway may be identified in JAK2/MPL− MPD. However, direct evidence of JAK2 activation in JAK2/MPL− MPD has not been reported, and a recent study noted reduced STAT3 phosphorylation and lower expression of the JAK-STAT target genes Pim-1 and SOCS-2 in JAK2V617F− ET.75 The development of specific JAK2 inhibitors (see below) will facilitate investigation relating to the role of JAK2 activation in MPD patients without known mutations by determining whether these patients are sensitive to JAK2 inhibition in preclinical and clinical studies.
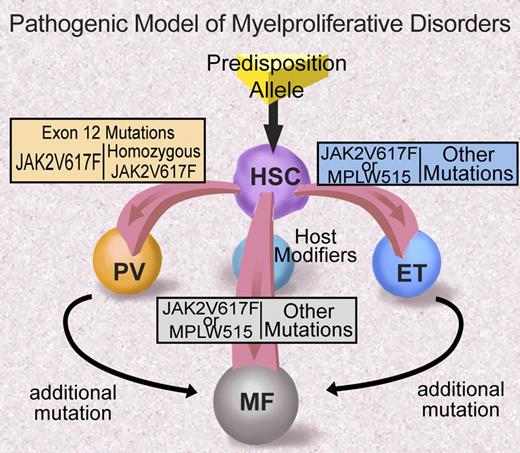
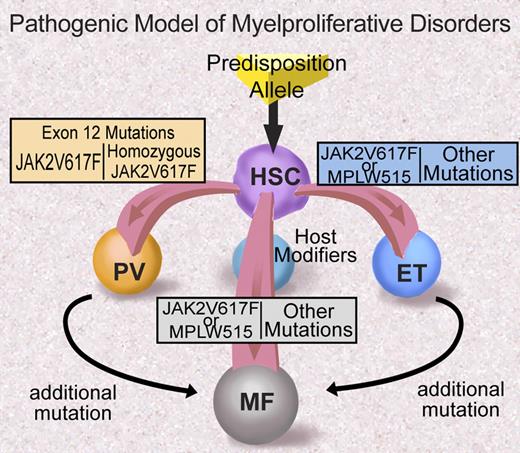
Many questions remain regarding the role of JAK2 exon 12 mutations and MPLW515 mutations in MPD pathogenesis (Figure 4). Most importantly, the mechanism through which these particular alleles are associated with specific clinical phenotypes—in contrast with JAK2V617F that is associated with a more promiscuous clinical phenotype—is not known and remains to be delineated. Possible explanations for the differences in clinical phenotype include qualitative and/or quantitative differences in signaling by different JAK2/MPL alleles, differences in negative feedback, and/or differences in JAK2 and MPL expression in different lineages. Future investigation will likely elucidate the basis for differences in clinical phenotype observed for different alleles in the JAK2 signaling pathway.
Proposed model of MPD pathogenesis (adapted from ASH Education Program 2006). Illustration by Debra Tyler.
Proposed model of MPD pathogenesis (adapted from ASH Education Program 2006). Illustration by Debra Tyler.
JAK2, signal transduction, and MPD pathogenesis
JAK2 was first identified in 1993 as the tyrosine kinase involved in growth hormone receptor and erythropoietin receptor signaling,76,77 and the signaling pathways downstream of JAK2 have been extensively investigated. However, the role and requirement of different signaling pathways in JAK2V617F-mediated hematopoietic transformation has not been fully elucidated, and is the subject of current investigation by many different laboratories. Among the signaling pathways activated by JAK2V617F are the Stat family of transcription factors, including STAT5A and STAT5B.78 It has been surmised that STAT5 activation is required for JAK2V617F-mediated transformation based on in vitro and in vivo studies. Expression of constitutively active STAT5 or the STAT5 target gene BCL-XL in human hematopoietic progenitors is sufficient to induce EEC formation.79 In addition, studies in Stat5ab-deficient mice demonstrated that STAT5A and STAT5B are necessary for TEL-JAK2–mediated myeloproliferation.80 Similar experiments will undoubtedly determine whether STAT5 signaling is required for hematopoietic transformation by JAK2V617F, and additional genetic and pharmacologic studies are needed to ascertain the relevance of MAP kinase, PI3 kinase, and other signaling pathways activated by JAK2V617F to MPD pathogenesis.
In addition, the role of JAK2V617F and other disease alleles in clinically relevant MPD complications, including thrombosis, myelofibrosis, and leukemic transformation, remains unknown. Recent studies suggest that leukocytosis is an important risk factor for thrombosis in MPDs,81,82 and a recent study observed increased expression of genes indicative of neutrophil activation in JAK2V617F+ ET neutrophils, suggesting that there is a relationship between neutrophil activation, thrombosis, and JAK2 signaling in MPDs.83 In addition, the contribution of JAK2 signaling to the development of myelofibrosis is not known. Previous studies have observed abnormalities in GATA1 and CXCR4/CXCL12 in murine models of myelofibrosis and in human PMF,84-86 however, the precise role of these abnormalities in the fibrotic process, and whether these abnormalities are direct or indirect effects of activated JAK2 signaling in hematopoietic progenitors, requires further evaluation. Moreover, given that a significant proportion of patients with a JAK2V617F+ MPD subsequently transform to JAK2V617F− AML,61,64 it may be that JAK2 signaling is dispensable for leukemic transformation, although this needs to be tested using in vitro and in vivo systems.
Targeted therapy for MPDs
The identification of activating mutations in the JAK2 signaling pathway in PV, ET, and PMF provides an opportunity to develop molecularly targeted therapies for patients with these disorders. There is reason to believe that inhibition of JAK2 kinase activity will be of value for patients with JAK2V617F+ MPD, particularly in light of the efficacy of imatinib and second generation ABL kinase inhibitors for the treatment of CML.87-90 This has led multiple groups to develop specific inhibitors of JAK2 kinase activity,91 and based on activity in preclinical models of MPD,92 Phase 1 trials with JAK2 inhibitors have been initiated in PMF and post-PV/ET myelofibrosis.93,94 Although there is considerable optimism that JAK2 inhibitors will offer significant benefit to patients with MPD, there also may be significant toxicities associated with JAK kinase inhibition. Potential side effects of candidate JAK2 inhibitors must be evaluated carefully, particularly in light of the importance of JAK2 kinase signaling to a myriad of cellular functions including normal erythropoiesis and thrombopoiesis. In addition, inhibition of JAK1, JAK3, and TYK2 should be minimized, particularly given the immune deficiencies associated with inherited loss-of-function mutations in JAK3 and TYK2.95-97 Although JAK2 inhibitors are currently being tested in patients with PMF and post-PV myelofibrosis, the indolent nature of PV and ET and the efficacy and safety of existing therapies (hydroxyurea, anagrelide, phlebotomy) for these disorders will necessitate that JAK2 inhibitors have favorable side effect profiles before being tested in PV and ET. In addition, the data from MPD and AML patients consistent with the existence of a “pre-JAK2” malignant clone,61,64 suggests the possibility that JAK2 inhibitor therapy might select for the emergence of JAK2V617F− clones with a propensity for leukemic transformation.
As new disease alleles are identified—either alone or in conjunction with JAK2 mutations—additional therapies may evolve that target these alleles. There is also still room for development of drugs based on empiric response to therapy. For example, there are recent reports of activity of inhibitors of chromatin remodeling proteins such as histone deacetylase inhibitors and HSP90 inhibitors in MPD,98,99 If corroborated, these clinical observations may further inform molecular mechanisms of disease. Moreover, molecular responses have been observed in PV patients treated with pegylated interferon-α-2a,100 suggesting existing therapies might have mutant-selective effects in PV, ET, and PMF.
Future directions
The genetic, biochemical, and functional studies described in this review have provided important insight into the pathogenesis of PV, ET, and PMF. There remain, however, fundamental questions regarding the molecular basis of these disorders. The role of a single disease allele in 3 related but clinically distinct disorders is not well understood, and future studies will undoubtedly identify additional mutations that contribute to the pathogenesis of these disorders. The etiology of JAK2/MPL− MPD requires additional investigation, and our understanding of the mechanism of transformation by JAK2V617F is incomplete. From a clinical perspective, the development of JAK2 inhibitors provides us an opportunity to ascertain whether targeted therapies, based on our understanding of the genetic basis of these disorders, will offer significant benefit to patients with PV, ET, and PMF.
Acknowledgments
We thank the patients, physicians, and investigators who have contributed to our understanding of these disorders, and apologize to our colleagues whose work was not highlighted due to space constraints. The Gilliland Laboratory is supported by the U.S. National Institutes of Health (Bethesda, MD), the Howard Hughes Medical Institute (Boston, MA), the Leukemia & Lymphoma Society (White Plains, NY), the Doris Duke Charitable Foundation (New York, NY), and the Myeloproliferative Disorders Foundation (Chicago, IL). D.G.G. is an Investigator of the Howard Hughes Medical Institute, and a Doris Duke Charitable Foundation Distinguished Clinical Scientist Award recipient. R.L.L. is an American Society of Hematology (Washington, DC) Basic Research Fellow Award recipient, a Doris Duke Charitable Foundation Clinical Scientist Development Award recipient, a Howard Hughes Medical Institute Early Career Award recipient, and is the Geoffrey Beene Junior Chair at Memorial Sloan-Kettering Cancer Center.
National Institutes of Health
Authorship
Contribution: R.L.L. wrote the manuscript and D.G.G. reviewed and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ross. L. Levine, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, Box 20, New York, NY 10065; e-mail: leviner@mskcc.org or ggilliland@rics.bwh.harvard.edu.