Abstract
Vertebrate blood formation occurs in 2 spatially and temporally distinct waves, so-called primitive and definitive hematopoiesis. Although definitive hematopoiesis has been extensively studied, the development of primitive myeloid blood has received far less attention. In Xenopus, primitive myeloid cells originate in the anterior ventral blood islands, the equivalent of the mammalian yolk sac, and migrate out to colonize the embryo. Using fluorescence time-lapse video microscopy, we recorded the migratory behavior of primitive myeloid cells from their birth. We show that these cells are the first blood cells to differentiate in the embryo and that they are efficiently recruited to embryonic wounds, well before the establishment of a functional vasculature. Furthermore, we isolated spib, an ETS transcription factor, specifically expressed in primitive myeloid precursors. Using spib antisense morpholino knockdown experiments, we show that spib is required for myeloid specification, and, in its absence, primitive myeloid cells retain hemangioblast-like characteristics and fail to migrate. Thus, we conclude that spib sits at the top of the known genetic hierarchy that leads to the specification of primitive myeloid cells in amphibians.
Introduction
The existence of embryonic macrophage-like cells has been reported in all animal models and its conservation suggests important roles during development. Studies in zebrafish and Xenopus have provided important insights into the mechanisms responsible for primitive hematopoiesis.1-3 Until recently, however, most of these studies have focused on the specification and differentiation of primitive erythrocytes. Comparatively, little is known about the mechanisms and gene networks responsible for the development of primitive macrophages and neutrophils. In Xenopus, primitive myeloid cells arise from the anterior ventral blood island (aVBI), the embryonic region equivalent to the zebrafish anterior lateral plate mesoderm (ALPM) and the mammalian yolk sac. In zebrafish, the ALPM, which contains the primitive myeloid-forming compartment, is physically separated from the primitive erythroid-forming compartment, the posterior lateral plate mesoderm (PLPM). However in Xenopus, the primitive myeloid-forming aVBI, is juxtaposed next to the erythroid-forming compartment, the posterior ventral blood island (pVBI). In both fish and frogs, these tissues arise from distinct locations in the embryo. Whereas the pVBI and PLPM arise from ventral gastrula mesoderm, the aVBI and ALPM arise from dorsal gastrula mesoderm.4-6 Experiments using a variety of model organisms have pointed to a conserved gene regulatory network leading to hematopoietic specification, with the transcription factor scl representing one of the central genes in the hematopoietic specification cascade.7-10 In frog embryos, scl expression appears first in the aVBI and only at a later time point in the pVBI with its characteristic V shape. At stage 20, as scl expression is down-regulated and restricted to bilateral stripes in the aVBI, scl is concomitantly up-regulated in the pVBI forming the tissue that will give rise to the first erythroid cells. The analysis of gene expression profiles and distinct origins of the aVBI and pVBI reveals 2 distinct temporal hematopoietic events that occur in juxtaposed mesodermal tissues. The aVBI tissue at the neurula stage also expresses other transcription factors typical of both hematopoietic and endothelial lineages such as fli1, lmo2, runx1, or hhex and the coexpression of these markers represents the hemangioblast stem cell–like state.11-15 Very little is know about the transcriptional regulation that controls primitive myelopoiesis in Xenopus, however it has been shown in zebrafish that the balance between primitive myelopoiesis and primitive erythropoiesis is dictated by the antagonistic interplay between the activity of 2 transcription factors spi1 (also known as pu.1), which promotes myelopoiesis, and gata1, which promotes erythropoiesis.16 To date, neither spi1 nor any other specific regulator of primitive myeloid development has been characterized in Xenopus. In this study, we identified spib, a gene related to spi1, and show that spib marks primitive myeloid cell progenitors and that it is indispensable for primitive myeloid development. Our data indicate that spib acts upstream of spi1 in the molecular hierarchy of primitive myeloid development and suggest that spib is necessary for the transition from a hemangioblast-like cell state to differentiation of myeloid lineages
Methods
Bioinformatics of spib and spi1
spib and spi1 genomic sequences were identified in scaffold 106 and 907, respectively. Information collated from Joint Genome Institute (http://genome.jgi-psf.org/Xentr4),17 ) Ensembl (http://www.ensembl.org),18 and Metazome (http://www.metazome.net)19 was used to determine transcript structure and synteny between known vertebrate spib and spi1 genes. Protein and ETS domain sequence alignments were performed using ClustalW20 and TreeView X,21 and PEST sequences were determined with PESTfind algorithm.22 Gene names and symbols abide by the Xenopus nomenclature guidelines,23,24 which suggest that gene names and symbols should correspond to the gene names and symbols of the human orthologs. In addition, we have attempted to use primarily the official names and symbols of the genes for each of the mentioned model organisms.
Whole mount in situ hybridization
Whole mount in situ hybridization (WMISH) was performed using standard protocols26 using a BioLane HTI robot (Hölle & Hüttner AG, Tübingen, Germany, http://www.h-net.com). Antisense probes were synthesized using MegaScript kit (Ambion, Austin, TX) and Digoxigenin (Roche, Indianapolis, IN) and purified on Bio-Spin columns (BioRad, Hercules, CA). BM Purple (Roche) was used for the substrate and the embryo pigment was bleached under strong light in 0.5 × SSC, 5% formamide, 2% H2O2. The following orthologs of hematopoietic relevant genes were used for antisense probe synthesis: X laevis spiba: IMAGE 5537169, SalI, T7; spibb: IMAGE 6957053, SalI, T7; xpox2: SalI, T727 ; scl: XhoI, SP611 ; runx1, SalI, T728 ; X tropicalis spib: IMAGE 7023083, EcoRV (or SacI to eliminate the ETS domain), T7; mpo (xpox2): IMAGE 5336501, SalI, T7; cebpa: TEgg058d02, EcoRI, T7; spi1: IMAGE 7695805, SalI, T7; mmp7: IMAGE 7005633, EcoRI, T7; lcp1(l-plastin): IMAGE 7022626, EcoRV, T7; scl: TTbA055c20, EcoRI, T7; lmo2: IMAGE 7002909, EcoRV, T7; globin: TTpA004i19, EcoRI, T7; fli1: IMAGE 7621709, SmaI, T7; flk1: IMAGE 7620420, SalI, T7; and gata1: IMAGE 8953859, SmaI, T7. Image acquisition was done using a Leica MZAPO stereoscope and Northern Eclipse software (Empix Imaging, Mississauga, ON).
Embryo manipulations
Xenopus laevis embryos were used in homotypic transplants and kept at 16°C unless stated otherwise. Microruby, a fluorescent cell tracer dextran was injected into one-cell embryos (10 nL, 2%; Invitrogen, Eugene, OR; D7162). aVBIs were transplanted between stages 14 and 16; donor and host embryos were placed side by side, ventral side up, in adjacent chambers made with a fire-polished glass Pasteur pipette into plasticine coated with 0.1% BSA (nontoxic, nonsticky; Eberhard Faber, Neumarkt, Germany). aVBIs were cleanly cut using a Gastromaster (Xenotek Engineering, Belleville, IL), maintaining consistent sizes, exchanged between donor and host embryos and left to heal undisturbed in 0.4 × MMR, 1% Ficoll for 2 to 3 hours. Imaging was performed in anesthetized embryos (< 10 minutes, 0.02% MS222) with an Olympus IX70 inverted fluorescent microscope (Watford, United Kingdom), using Northern Eclipse software for time-lapse image acquisition, and movies were assembled using ImageJ software (National Institutes of Health, Bethesda, MD). Embryonic wounds were made in sterile 0.1 × MMR with a Gastromaster tip. Infection assays were performed by injection of a suspension of E coli expressing GFP (a gift from Jordan Raff).
Morpholino oligonucleotides
Loss-of-function experiments were performed in the diploid frog Xenopus tropicalis kept at 23°C. The following antisense morpholino oligonucleotides (MO) were obtained against Xenopus tropicalis spib (GeneTools, Philomath, OR): ATG*** (3-mismatch): ACGGTA CTGACT CCAACC TGACCAT; ATG: ACCGTA GTGAGT CCAAGC TGAGCAT; and e1i1 (exon1–intron1): CTAAAG CCCATC ACTTAC CGTAGTG. They were dissolved and stored according to the manufacturer's instructions and injected at the 1-cell stage; embryos were kept in 0.01 × MMR.
RNA analysis
mRNA was extracted using Trizol from a pool of 5 Xenopus tropicalis embryos. SuperScript II (Invitrogen) was used to do reverse cDNA synthesis and the primers used were as follows: spib forward: TGCCCA CAAAAT GCTCAG CTTG, spib reverse: TGTCGT ATGTCT CGTAAC CAGG; spi1 forward: AGAAAC ACACAG AAATCC CTTTATG, spi1 reverse: CGCTTG ATGTAA GAAACC CAATAAG; cebpa forward: GGGATG TGCCCT TTATTG TCAG, cebpa reverse: GCAAGG ATGTCC AACATG AAGC; mpo forward: AACAGA CCCTGG ACAACC AACG, mpo reverse: ACCTGG CTGGCA TCAACA TAAG; mmp7 forward: CCAAGT GGTAGA CACCGC AATAC, mmp7 reverse: ACATCA AGGCTC GTGGGT CAGTAG; lcp1 forward: GAGACC GAGAAA CTCAAC AACGC, lcp1 reverse: CCCATA AGGCAA GCAAAG ACTGTC; and globin forward: ACAAAC AGGGCA TTGATT AATGCT, globin reverse: AATGTC TCCGCT GTCTCT GGA. For semiquantitative reverse-transcription–polymerase chain reaction (RT-PCR) analysis, the number of cycles required for each primer pair was determined empirically and controls without reverse transcriptase were included as well as cDNA dilutions to ensure that the PCR reaction was in the log-linear range. Real-time quantitative RT-PCR (qRT-PCR) analysis was performed in individual Xenopus tropicalis embryos using an Opticon II (MJ Research, Watertown, MA) adding to the PCR mix a 1:2000 dilution of SYBR (Sigma-Aldrich, St Louis, MO). For each primer pair, the PCR product was examined by gel electrophoresis and its melting curve to ensure a single fragment of the predicted molecular weight. The data for each sample were normalized to the expression level of ornithine decarboxylase (ODC) and calculated by the 2−ΔΔCt method, normalized to expression levels at stage 10.
Sudan Black
Neutrophils can be visualized using Sudan Black histochemical stain in whole mount embryos (Sigma-Aldrich). Briefly, after 1 hour of fixation embryos were stained, washed in 100% EtOH, and then hydrated to PBS, washed, and bleached under strong light (0.5 × SSC, 5% formamide, 2% H2O2).
Results
Isolation of spib
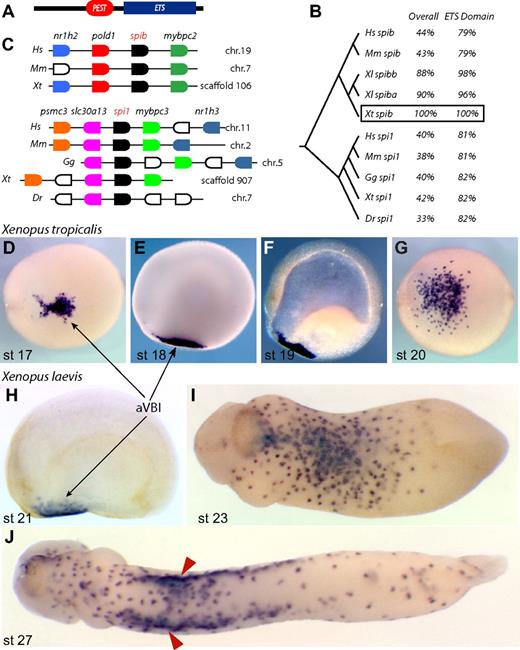
In an unrelated screen,29 we identified a novel transcript expressed in the anterior ventral blood islands at neurula stage (Figure 1). In tailbud embryos, expression of this gene was observed in a punctate pattern reminiscent of primitive myeloid markers such as XLURP-1, XPOX2, and MMP7.27,30,31 Sequence analysis showed that this gene belonged to the spi subfamily of ETS transcription factors, with a C-terminal ETS domain and a PEST domain (Figure 1A,B). This architecture is conserved in all members of the spi subfamily, of which spi1 (also known as pu.1 and Sfpi1) is the most well-characterized member. To determine to which member of the spi subfamily our gene belonged, we used genome and synteny information to conclude that our gene is the Xenopus tropicalis ortholog of spib, given that it is located between the pold1 and mybpc2 genes (Figure 1C). We also isolated the Xenopus tropicalis ortholog of spi1, which shows remarkable synteny with the human, mouse, and chick genomes (Figure 1C). Interestingly, however, we noted that spib sequences appear to be absent from the chicken and fish genomes.
Xenopus spib identification. (A) Conserved protein motifs in spib and spi1; the PEST degradation domain and ETS DNA-binding domain. (B) ClustalW tree alignment on vertebrate spib and spi1 protein sequences and percentage identity over their entirety and DNA-binding domains. (C) Genome organization around the known vertebrate spib and spi1 genes, showing synteny between various vertebrate genomes. Genes shown in the same color represent putative orthologous genes. Expression pattern of Xenopus tropicalis spib and (D-G) Xenopus laevis spib paralogs (SpiBa and SpiBb, H-J) is indistinguishable and marks a population of primitive myeloid progenitors in the anterior ventral blood islands (aVBIs). spib expression is transient and first detected at stage 17. (F) Cleared embryo shows only mesodermal expression, and (J) spib expression is eventually down-regulated after spib-expressing cells associate with vitelline veins (arrowheads). Anterior is to the left, and dorsal to the top in all lateral views; panels D, G, and I,J represent ventral views. The following database sequences were used: Hs spib Q01892, Mm spib O35906, Xl spiba BC046671, Xl spibb BC130210, Xt spib IMAGE 7023083, Hs spi1 P17947, Mm spi1 P17433, Gg spi1 NP_990354, Xt spi1 BC098077, and Dr spi1 CAD58735 (outgroup NP_001008139 and P14921).
Xenopus spib identification. (A) Conserved protein motifs in spib and spi1; the PEST degradation domain and ETS DNA-binding domain. (B) ClustalW tree alignment on vertebrate spib and spi1 protein sequences and percentage identity over their entirety and DNA-binding domains. (C) Genome organization around the known vertebrate spib and spi1 genes, showing synteny between various vertebrate genomes. Genes shown in the same color represent putative orthologous genes. Expression pattern of Xenopus tropicalis spib and (D-G) Xenopus laevis spib paralogs (SpiBa and SpiBb, H-J) is indistinguishable and marks a population of primitive myeloid progenitors in the anterior ventral blood islands (aVBIs). spib expression is transient and first detected at stage 17. (F) Cleared embryo shows only mesodermal expression, and (J) spib expression is eventually down-regulated after spib-expressing cells associate with vitelline veins (arrowheads). Anterior is to the left, and dorsal to the top in all lateral views; panels D, G, and I,J represent ventral views. The following database sequences were used: Hs spib Q01892, Mm spib O35906, Xl spiba BC046671, Xl spibb BC130210, Xt spib IMAGE 7023083, Hs spi1 P17947, Mm spi1 P17433, Gg spi1 NP_990354, Xt spi1 BC098077, and Dr spi1 CAD58735 (outgroup NP_001008139 and P14921).
Using WMISH, we determined the expression pattern of Xenopus laevis spib paralogs and Xenopus tropicalis spib, from the neurula stages through the tailbud stages (Figure 1D-J). These data revealed that, in both species, spib was expressed in the progenitors of primitive myeloid cells, originating from the aVBI from stage 17. At the tailbud stages, spib-expressing cells dispersed throughout the embryo before the establishment of a functional vasculature (Figure 1). In tadpoles, a subset of spib-expressing cells was observed associated with the nascent vitelline veins that surround the pVBI (Figure 1J red arrowheads). Soon after, the expression of spib was down-regulated as myeloid cells differentiate (data not shown).
Primitive myeloid cell behavior
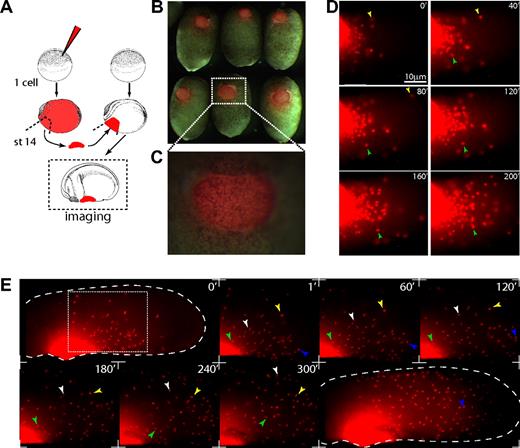
To analyze the initial steps of primitive myeloid cell migration, we performed homotypic transplantations of aVBIs from microruby-injected embryos into stage-14 and -16 host embryos. This allowed us to observe the migration of primitive myeloid cells away from the aVBI, using time-lapse fluorescence video microscopy (Figure 2). By stages 19 and 20, migration of microruby-positive cells was observed as single individual cells leaving the transplant (Figure 2D,E; Videos S1,S2, available on the Blood website; see the Supplemental Materials link at the top of the online article). The overall morphology and cell population behavior throughout the colonization process was very dynamic. It started with a “blebbing” cell behavior with low motility as cells left the aVBI, changing to a behavior with higher protrusive activity and low migratory speeds (Videos S1,S2). As the distance from the aVBI increased the cells changed to a highly motile bipolar behavior, which allowed the cells to colonize the whole embryo within 4 to 8 hours (Figure 2E; Video S3). Once that process was completed, a third phase ensued as isolated primitive myeloid cells began to patrol the embryo, coincident with less protrusive activity in the cells. Throughout these migratory behaviors, primitive myeloid cells were capable of undergoing cell divisions and some were phagocytic (data not shown). In summary, the behavior of these cells is consistent with the previously described Xenopus primitive macrophages or zebrafish primitive macrophages and neutrophils.27,31-34
Primitive myeloid migration time-lapse video microscopy. (A) Experimental setup, stage-14 to -16 anterior ventral blood islands were transplanted from microruby-injected to noninjected embryos. Transplanted cells become migratory and leave the transplant to colonize the embryo in 4 to 8 hours. Throughout this period, primitive myeloid cells show different behaviors (Videos S1Video 2. Lateral migration 4× (MOV, 4.82 MB)–S3). (B,C) Stage-18 brightfield/fluorescent composite of transplanted embryos. (D,E) Stills from supplementary movies. (Panel D and Video S2) Ventral view of primitive myeloid cells leaving the transplanted aVBI with “blebbing” behavior and low migratory speeds. (Panel E and Video S3) Primitive myeloid cells leaving the transplanted aVBI (stage 26, lateral view). At this stage, cells acquire elongated cell morphology and higher motility. Large dashed line shows the embryo contour, and the light dashed square shows the enlarged region shown in still frames. Colored arrowheads point and track the same cell. (D,E) Anterior view is shown to the left; dorsal, to the top. Time is shown in minutes. Images in panels B and C were obtained on a fluorescence stereoscope Leica MZ FLIII (Wetzlar, Germany) attached to a Sony CCD camera DXC-950 image capture system controlled by Northern Eclipse software 7.0 (Empix Imaging, Mississauga, ON). For panels D and E, the same image capture system was attached to an Olympus IX70 inverted fluorescent microscope; 0.1× MMR was used as imaging medium. (D) Total magnification 300× objective (Olympus LCPlan 20×/0.4 NA). (E) Total magnification 40× objective (Olympus UPlanFL 4×/0.13 NA). Photoshop CS2 (Adobe Systems, San Jose, CA) or ImageJ 1.38 (National Institutes of Health, Bethesda, MD) were used for image or time-lapse video processing.
Primitive myeloid migration time-lapse video microscopy. (A) Experimental setup, stage-14 to -16 anterior ventral blood islands were transplanted from microruby-injected to noninjected embryos. Transplanted cells become migratory and leave the transplant to colonize the embryo in 4 to 8 hours. Throughout this period, primitive myeloid cells show different behaviors (Videos S1Video 2. Lateral migration 4× (MOV, 4.82 MB)–S3). (B,C) Stage-18 brightfield/fluorescent composite of transplanted embryos. (D,E) Stills from supplementary movies. (Panel D and Video S2) Ventral view of primitive myeloid cells leaving the transplanted aVBI with “blebbing” behavior and low migratory speeds. (Panel E and Video S3) Primitive myeloid cells leaving the transplanted aVBI (stage 26, lateral view). At this stage, cells acquire elongated cell morphology and higher motility. Large dashed line shows the embryo contour, and the light dashed square shows the enlarged region shown in still frames. Colored arrowheads point and track the same cell. (D,E) Anterior view is shown to the left; dorsal, to the top. Time is shown in minutes. Images in panels B and C were obtained on a fluorescence stereoscope Leica MZ FLIII (Wetzlar, Germany) attached to a Sony CCD camera DXC-950 image capture system controlled by Northern Eclipse software 7.0 (Empix Imaging, Mississauga, ON). For panels D and E, the same image capture system was attached to an Olympus IX70 inverted fluorescent microscope; 0.1× MMR was used as imaging medium. (D) Total magnification 300× objective (Olympus LCPlan 20×/0.4 NA). (E) Total magnification 40× objective (Olympus UPlanFL 4×/0.13 NA). Photoshop CS2 (Adobe Systems, San Jose, CA) or ImageJ 1.38 (National Institutes of Health, Bethesda, MD) were used for image or time-lapse video processing.
Primitive myeloid cell are recruited to embryonic wounds
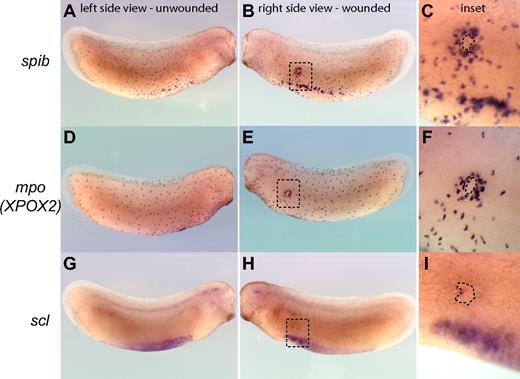
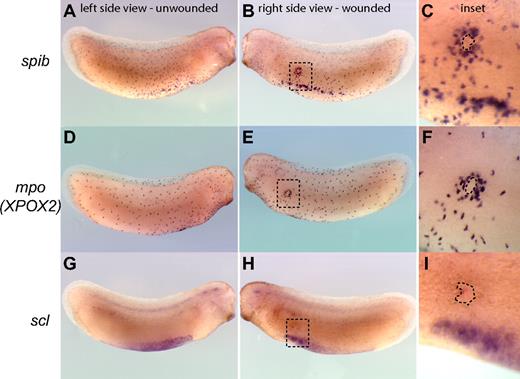
The recruitment of myeloid cells to sites of injury is a hallmark of the inflammatory and wound-healing responses and can be used as a measure of functional myeloid cells in embryos. Within this framework, we sought to test the function of primitive myeloid cells by evaluating whether they are able to respond to wounds or E coli infection in Xenopus tailbud-stage embryos before a functional vasculature is established. We found that primitive myeloid cells, as identified by the expression of spib and mpo but not scl, were efficiently recruited to Xenopus embryonic wounds in tailbud-stage embryos (Figure 3A-F). Indeed, primitive myeloid cells started to migrate toward wound sites within 2 to 3 minutes after injury or infection, with recruitment of myeloid cells to the wound edge completed by 3 hours (Figure 3; Video S4). In summary, primitive myeloid cells are capable of responding to wounding or infection before a functional vasculature is established in the embryo, demonstrating that primitive myeloid cells differentiate and become functional very early in embryos.
Recruitment of primitive myeloid cells to embryonic wound sites in Xenopus laevis. spib (A-C), mpo (XPOX2) (D,E), and scl (G-I) whole mount in situ hybridization in wounded embryos. Three hours is sufficient for the recruitment of myeloid cells expressing spib or mpo to the scrape wound outline (dashed line contour in the inset). scl-expressing cells are not recruited to the wound site, and primitive myeloid cell recruitment occurs before the appearance of a fully functional vascular network.
Recruitment of primitive myeloid cells to embryonic wound sites in Xenopus laevis. spib (A-C), mpo (XPOX2) (D,E), and scl (G-I) whole mount in situ hybridization in wounded embryos. Three hours is sufficient for the recruitment of myeloid cells expressing spib or mpo to the scrape wound outline (dashed line contour in the inset). scl-expressing cells are not recruited to the wound site, and primitive myeloid cell recruitment occurs before the appearance of a fully functional vascular network.
Characterization of primitive myeloid molecular markers in Xenopus tropicalis
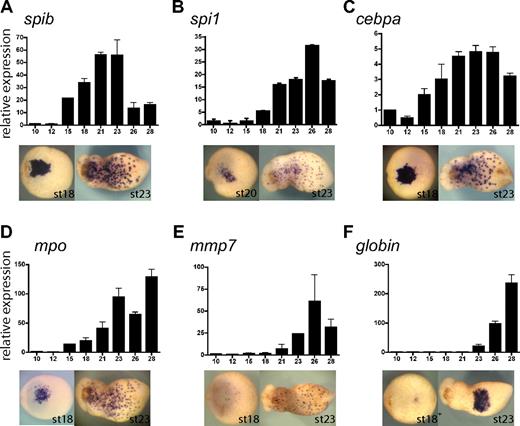
To study the function of spib in primitive myeloid differentiation, we isolated and determined the temporal and spatial pattern of expression of the following primitive myeloid molecular markers in Xenopus tropicalis: cebpa, spi1, mpo, and mmp7 (Figure 4A-E). Expression of all these markers was restricted to the aVBI at neurula stages, whereas at tadpole stages expression was observed as a characteristic myeloid punctate pattern. Noticeably, spib was expressed earlier than all the markers, with the exception of cebpa (CCAAT/enhancer binding protein alpha) (Figure 4A-E), suggesting that spib is one of the earliest markers of primitive myeloid cells.
Xenopus tropicalis primitive myeloid marker expression analysis by RT-PCR and WMISH. (A-F) spib, spi1, cebpa, mpo, mmp7, and globin single embryo qRT-PCR and ventral views of WMISH at stages 18 and 23. Among the transcription factors, (A) spib is the first to be specifically expressed in primitive myeloid cells, (B) followed by spi1. (C) Earlier expression of cebpa is due to its broad mesodermal expression before its restriction to the aVBI. (D) mpo is a neutrophil marker; (E) mmp7, a macrophage marker; and (F) globin, a erythroid marker. Error bars represent the SD of at least 3 independent experiments.
Xenopus tropicalis primitive myeloid marker expression analysis by RT-PCR and WMISH. (A-F) spib, spi1, cebpa, mpo, mmp7, and globin single embryo qRT-PCR and ventral views of WMISH at stages 18 and 23. Among the transcription factors, (A) spib is the first to be specifically expressed in primitive myeloid cells, (B) followed by spi1. (C) Earlier expression of cebpa is due to its broad mesodermal expression before its restriction to the aVBI. (D) mpo is a neutrophil marker; (E) mmp7, a macrophage marker; and (F) globin, a erythroid marker. Error bars represent the SD of at least 3 independent experiments.
In zebrafish, spi1 (pu.1) marks primitive myeloid cells and acts as a key regulator of primitive myeloid development.16,35-37 We found that the Xenopus tropicalis ortholog of spi1 was also expressed in primitive myeloid cells (Figure 4B). However, qRT-PCR data showed that the onset of spi1 expression begins several stages after the onset of spib expression (ie, stage 18 for spi1 versus stage 15 for spib; Figure 4A,B). Furthermore, spib transcripts could be robustly detected by WMISH at stage 17, whereas spi1 could be detected only 3 hours later at stage 20. Xenopus tropicalis mpo is the ortholog of X laevis XPOX2 and zebrafish mpx. It encodes a myeloperoxidase, which is expressed in neutrophil granules during embryogenesis and in adult mammalian granulocytes.27,38 Using double WMISH, we found that mpo was expressed in the aVBI in an overlapping pattern to that of spib, suggesting that both genes are expressed in the same cells (Figure S1). mpo was also the earliest specific myeloid marker after spib (Figure 4D). Therefore, we have used mpo as an early differentiation marker of primitive myeloid cells, possibly of the granulocytic lineage. As a primitive macrophage marker we have used mmp7, a secreted metalloproteinase involved in extracellular matrix remodeling. Homologues of this gene have been previously used as macrophage differentiation markers.30,33,39,40 Relative to other myeloid markers, mmp7 is expressed considerably later and with a similar timing to the erythroid lineage marker, globin (Figure 4E,F). In general, our findings on the timing of expression and the functional assays we have performed suggest that primitive myeloid cells differentiate and are functional before primitive erythroid cells.
spib is required for primitive myeloid cell differentiation
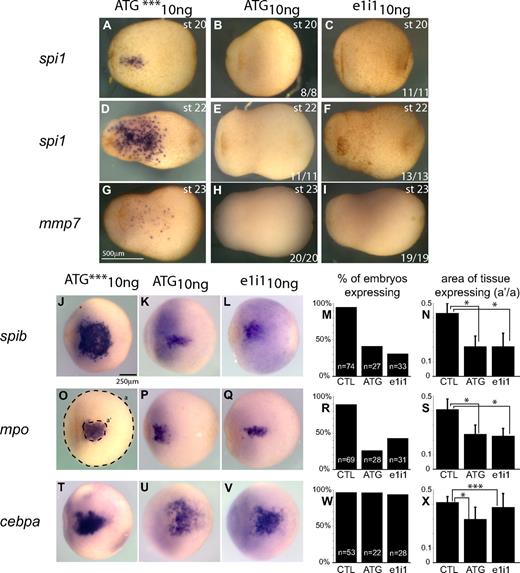
The timing and pattern of spib expression prompted us to characterize its role during primitive myelopoiesis in Xenopus tropicalis. We used an antisense morpholino oligonucleotide loss-of-function approach designed to block either the translation or splicing of spib transcripts (spibATG10ng and spibe1i110ng, respectively; Figure S2A). Embryos injected with morpholinos targeting translation or splicing of spib appeared morphologically normal at tailbud stages (Figure 5). However, injection of both types of MOs resulted in an elimination or severe reduction in the expression of all primitive myeloid markers tested, with the exception of cebpa (Figure 5). spi1, mpo, and mmp7 and other myeloid markers such as lcp1 (X tropicalis l-plastin) were all absent or markedly reduced from late neurula to tailbud stages, when examined by either WMISH or semiquantitative RT-PCR (Figure 5 and data not shown). Given that both morpholino oligonucleotides gave rise to very similar repression of the myeloid markers, whereas the mismatch negative control oligonucleotide (ATG***) had no effect, we conclude that the MO injection resulted in a specific inhibition of spib function. As expected, the ATG morpholino had little effect on the spib RNA levels as judged by RT-PCR, whereas the splicing morpholino significantly decreased the expression of mature spib transcripts (compare Figure 5P and 5Q). However, based on in situ hybridization, we observed considerably fewer spib-positive cells outside of the aVBI region in spibATG10ng morphants, suggesting that once myeloid development was blocked the cells did not migrate properly.
spib is necessary for primitive myeloid differentiation. spib-depleted embryos show absence or reduced expression of mpo, spi1, and mmp7 but not cebpa. (A-O) WMISH and (P,Q) RT-PCR analysis on spib ATG and e1i1 morphants. ATG*** is a 3-mismatch ATG morpholino. (P) spib ATG morpholino reduces but does not eliminate all spib transcripts, (Q) whereas spib e1i1 morpholino eliminates functional spib mRNA at 10 ng. (A-F) Severe reduction on the number of spib- and mpo-expressing cells and (J-O) marked absence of spi1 and mmp7 expression on spib morphants. (G-I) Under the same experimental conditions, cebpa-expressing cells are not affected. All embryos are shown in lateral views with anterior to the left. The number of representative embryos is shown at the bottom right corner of each panel.
spib is necessary for primitive myeloid differentiation. spib-depleted embryos show absence or reduced expression of mpo, spi1, and mmp7 but not cebpa. (A-O) WMISH and (P,Q) RT-PCR analysis on spib ATG and e1i1 morphants. ATG*** is a 3-mismatch ATG morpholino. (P) spib ATG morpholino reduces but does not eliminate all spib transcripts, (Q) whereas spib e1i1 morpholino eliminates functional spib mRNA at 10 ng. (A-F) Severe reduction on the number of spib- and mpo-expressing cells and (J-O) marked absence of spi1 and mmp7 expression on spib morphants. (G-I) Under the same experimental conditions, cebpa-expressing cells are not affected. All embryos are shown in lateral views with anterior to the left. The number of representative embryos is shown at the bottom right corner of each panel.
Of all the markers we analyzed, only cebpa expression was relatively unaffected by spib loss of function, although cebpa-expressing cells were found to cluster around the vitelline veins (Figure 5M-O red arrowheads). Together with the fact that cebpa is expressed prior to spib, this result suggests that cebpa may act upstream of spib in the hierarchy of myeloid development. However, the markers expressed well after spib, such as spi1 and mmp7, are particularly sensitive to spib depletion (Figure 5G-L,P-Q; Figure 6D-I). This demonstrates that spib is necessary for spi1 and mmp7 expression, suggesting that spib functions upstream of spi1 and mmp7. Furthermore, loss of primitive myeloid cell differentiation was sustained until tailbud stages as observed by the reduction in the number of neutrophils at stage 42 (Figure S2B).
Effect of spib knockdown in primitive myeloid progenitors. (A-I) Primitive macrophages defined by the expression of spi1 and mmp7 are absent from spib-depleted embryos. (J-L,O-Q,T-V) WMISH analysis of aVBI in spib morphants; at the earliest point we can identify a pool of primitive myeloid progenitors by the coexpression of spib, mpo, and cebpa. (M,R,W) Quantification of the number of embryos that express spib, mpo, and cebpa and (N,S,X) size of primitive myeloid progenitor pool, by the area of tissue-expressing progenitor markers. Error bars represent the SD of the number of embryos analyzed (n). (O) Amount of tissue was calculated using area ratios of a and a′. Similar results were obtained in either ATG or e1i1 morphants and are in contrast with the uninjected and ATG mismatch control group (CTL). Student t test P values lower than .001 are marked as * and considered statistically significant; P values higher than .05 are not statistically significant and marked as ***.
Effect of spib knockdown in primitive myeloid progenitors. (A-I) Primitive macrophages defined by the expression of spi1 and mmp7 are absent from spib-depleted embryos. (J-L,O-Q,T-V) WMISH analysis of aVBI in spib morphants; at the earliest point we can identify a pool of primitive myeloid progenitors by the coexpression of spib, mpo, and cebpa. (M,R,W) Quantification of the number of embryos that express spib, mpo, and cebpa and (N,S,X) size of primitive myeloid progenitor pool, by the area of tissue-expressing progenitor markers. Error bars represent the SD of the number of embryos analyzed (n). (O) Amount of tissue was calculated using area ratios of a and a′. Similar results were obtained in either ATG or e1i1 morphants and are in contrast with the uninjected and ATG mismatch control group (CTL). Student t test P values lower than .001 are marked as * and considered statistically significant; P values higher than .05 are not statistically significant and marked as ***.
We next asked when could we first detect the effect of knocking down spib function during primitive myeloid differentiation? For this, we focused on the earliest time point at which these cells can be molecularly identified in Xenopus tropicalis: that is, stage 18, using spib, mpo, and cebpa as markers; stage 20, using spi1 as a marker; and stage 23, using mmp7 as a marker. As before, the most significantly affected markers were spi1 and mmp7, which are also the markers that are expressed the latest, and both were absent in all the experimental embryos (Figure 6A-I). This finding is interesting in light of previous reports suggesting that spi1 acts as a master regulator of not only adult myelopoiesis but also of primitive myelopoiesis.16,41 In contrast, the effects on earlier markers were more variable. We quantified both the percentage of embryos expressing spib, mpo, or cebpa, as well as the area of aVBI tissue expressing those markers in control versus spib-depleted embryos (Figure 6J-X). The majority of spib-depleted embryos fail to express either spib or mpo (Figure 6M,R). However, in the few embryos that still express spib and mpo (20% to 40% depending on the morphant), the area of expression is significantly reduced (Figure 6K-N,P-S; Student t test, P < .001). This suggests that although spib function may not be absolutely required for its own initial expression, it may play an important role in maintaining its own transcription and that of mpo. Conversely, cebpa expression was neither affected nor reduced in spib morphants (Student t test, P > .05), demonstrating that spib was not required for maintenance or initiation of cebpa expression (Figure 6T-X). In light of these results, we conclude that spib is required very early in the differentiation of primitive myeloid progenitors in Xenopus.
spib morphants do not have expanded posterior blood islands
In Xenopus, primitive myelopoiesis and erythropoiesis occur in juxtaposed locations with distinct developmental timing—the aVBI earlier and pVBI later. Previous experiments in zebrafish have suggested a cross-regulation in the development of these lineages between myelopoietic and erythropoietic compartments, including the negative cross-regulation of spi1 and gata1.16 Due to the similarity between spib and spi1 and the elimination of primitive myeloid development in spibe1i110ng and spibATG10ng morphants, we investigated whether the posterior-erythroid compartment was affected in our spib morphant embryos. We assayed the state of primitive erythroid differentiation, using globin, scl, and lmo2 expression, at stage 26, when erythroid differentiation is under way. Interestingly, no shift in the expression pattern of these genes into the anterior primitive myeloid compartment was detected, nor was a premature expression of hematopoietic genes in the posterior compartment (Figure S3). Therefore, when the differentiation of primitive myeloid cells was impaired, this did not result in an anterior shift or premature erythroid differentiation. Thus, we conclude that blocking myeloid differentiation, by eliminating spib function, did not interfere either with primitive erythroid lineage differentiation or the balance between primitive erythroid and myeloid-forming compartments as assayed by WMISH.
Retention of hemangioblast-like characteristics in spib-depleted embryos
One possible model to explain the phenotype we observed in spib-depleted embryos is that, as primitive myeloid progenitor differentiation is blocked, cell migration away from the aVBI is impaired and progenitor cells remain in an earlier state of hematopoietic differentiation. We first evaluated the migration of primitive myeloid cells in spib morphants by taking advantage of the range of phenotypes obtained with spibe1i110ng and spibATG10ng morphants. In stage 22 control embryos, migration away from the aVBI is well under way; however, in spib-depleted embryos, it is consistently absent (Figure 7A-I). This result is particularly relevant if we compare the phenotype of Xenopus spib morphants with that of the zebrafish spi1 morphants. In zebrafish when myeloid development is blocked by depletion of spi1, the defective primitive myeloid progenitor cells still migrate away from the ALPM.16 In contrast, our data indicate that spib-defective myeloid progenitor cells never migrate out of the aVBI. To analyze whether primitive progenitors were “stuck” in an earlier cellular stage of hematopoietic differentiation, we analyzed the expression of (1) postulated hemangioblast markers such as scl and fli1, (2) hematopoietic stem cell markers such as scl, lmo2, and runx1, (3) endothelial differentiation markers such as fli1 and kdr, or (4) the erythroid marker gata1. We expected that modifications in the patterns of expression of these genes could provide insight in cell fate changes that occur when primitive myeloid development is blocked in vivo. In situ hybridization analysis of spibe1i110ng and spibATG10ng morphants indicated that a triangle-shaped expression domain of scl and fli1 was retained at the border between aVBI and pVBI at stage 22, whereas all other markers analyzed to date were unaffected (Figure 7J-O black and white arrowheads). In this small area, expression of scl and fli1 has normally been down-regulated by stages 22 and 23 (compare Figure 7J-K and M-N). Retention of scl and fli1 expression is suggestive of retention of hemangioblast-like characteristics. However, this is a delay and not a permanent block in hematopoietic differentiation. At stage 26, no differences in the expression of scl and fli1 can be observed between spib-depleted and control embryos (Figure S3 and data not shown). We also did not observe differences in the pattern of expression of the hematopoietic stem cell markers runx1 and lmo2. Overall, these results suggest that in the absence of spib function primitive myeloid progenitors are blocked in their normal differentiation and migration, and they retain for a period of 2 to 3 hours hemangioblast-like characteristics in a small subset of aVBI cells, which would have otherwise differentiated into primitive myeloid cells.
Analysis of migratory potential and hemangioblast-like characteristics of primitive myeloid progenitors in spib-depleted embryos. Migration away from the aVBI is the first morphologic sign of primitive myeloid differentiation and is absent in spib-depleted embryos. (A-I) In spib-depleted embryos, we never observed isolated cells far from the aVBI expressing spib, mpo, and cebpa at a time when migration should have already set in. Usual down-regulation of scl and fli (▿, J,M) is eliminated in spib morphants (▼, K,L and N,O). The scl+fli+ hemangioblast-like characteristics are maintained for longer in a small subset of spib-depleted aVBI tissue.
Analysis of migratory potential and hemangioblast-like characteristics of primitive myeloid progenitors in spib-depleted embryos. Migration away from the aVBI is the first morphologic sign of primitive myeloid differentiation and is absent in spib-depleted embryos. (A-I) In spib-depleted embryos, we never observed isolated cells far from the aVBI expressing spib, mpo, and cebpa at a time when migration should have already set in. Usual down-regulation of scl and fli (▿, J,M) is eliminated in spib morphants (▼, K,L and N,O). The scl+fli+ hemangioblast-like characteristics are maintained for longer in a small subset of spib-depleted aVBI tissue.
Discussion
Primitive myeloid cells are the first functional blood lineages in the embryo
Blood cell development starts soon after the end of gastrulation, possibly under similar regulatory and signaling mechanisms that direct adult hematopoietic differentiation. However, there are clear differences between embryonic and adult hematopoiesis. For instance, differentiation of erythroid cells, evaluated by the expression of globin does not start until the mid–tailbud stages (ie, stages 24-25), which is about 12 hours after gastrulation. In addition, the vascular system is not functional until the early tadpole stages (ie, stage 35), which is yet another day later.42 Here we show that primitive myeloid cells become migratory at the end of the neurula stages (ie, stage 20). Furthermore, we found that these early differentiating myeloid lineages are functional, as they can respond to stimuli, such as embryonic wounds or bacterial infections, by quickly and efficiently migrating toward these sites. Therefore, our findings show that primitive myeloid cells are indeed the first blood lineages to become functional in the embryo, and they do so more than a day prior to the establishment of a functional vasculature. How heterogeneous this early population of myeloid cells is and what their varied functions are during development, repair, and embryonic immunity are of great interest, but these are questions that remain to be addressed.
spib is required to exit myeloid progenitor cell state
In both zebrafish and Xenopus, a cell population of dorsal mesodermal origin transiently goes through a hemangioblast-like state, which at present is best characterized by an scl and fli1 double-positive population of cells. These cells are believed to give rise to both vascular endothelium and blood lineages.12-14 In this work, we found a novel role for spib in the transition from a postulated scl+fli1+ hemangioblast-like cell state into a migratory cell population leading to differentiation of embryonic macrophages and neutrophils. In particular, we found that blocking primitive myeloid differentiation and consequently their migration, through the loss of spib function, results in the maintenance of scl and fli1 expression in the aVBI, suggesting that spib may be required for an exit of the hemangioblast-like state toward the primitive myeloid differentiation. This finding is distinct from the proposed role for spi1 during primitive myelopoiesis in zebrafish, where spi1 loss of function promotes erythroid specification at the expense of myeloid specification.16 In Xenopus, inhibition of spib does not lead to erythroid specification, instead, it leads to a delay in the exit of the progenitor state toward a pathway of myeloid differentiation.
spib is a key player in primitive myeloid development
Here we show that spib is one of the earliest genes expressed in the progenitors of primitive myeloid cells in Xenopus, and we also show that spib is required for the specification and differentiation of primitive myeloid cells. A role for Spib in the hematopoietic system has been described in mice, in particular during the differentiation of B and dendritic cells of mice.43,44 However, a role for spib during primitive myeloid differentiation is novel. spib is related to spi1, a well-known and essential gene for both primitive and definitive myelopoiesis.16,45,46 In mice, Spi1 (official name, Sfpi1, but most commonly known as Pu.1) is required for the differentiation of all myeloid cells, even those originating from the yolk sac. However, in-depth evaluation of Spi1-null mice showed that Spi1 is primarily required for terminal primitive myeloid differentiation in the yolk sac, as early markers of primitive myeloid cells, such as GM-CSFR, G-CSFR, and MPO are still present.41,47 Consistent with this finding, we have found that inhibiting the function of spi1 in Xenopus tropicalis does not affect the expression of early myeloid markers, such as spib and mpo, but does decrease the level of later myeloid markers, such as mmp7 (R.M.B.C., X.S., and E.A., unpublished observation, May 2008). Thus, although very important for primitive myeloid differentiation, spi1 is unlikely to be the sole player regulating primitive myeloid development. To this end, our findings show that spib functions upstream of spi1 during primitive myeloid development based on its earlier expression pattern and the fact that spi1 expression is dependent on spib. Furthermore, we found that early and late markers of primitive myeloid cells, with the exception of cebpa, were reduced or inhibited when spib function was knocked down. Whether Spib plays a similarly central role during primitive myeloid specification in mammals is an important question that will require further investigation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Kimberley Mace and Steve Brown for critical reading of the paper, Aldo Ciau-Uitz for the scl and runx1 X laevis plasmids, and Michael Redd for suggesting using Sudan Black.
This work was supported by the Wellcome Trust (082450/Z/07/Z; London, United Kingdom) and The Healing Foundation (London, United Kingdom).
Wellcome Trust
Authorship
Contribution: R.M.B.C. performed and analyzed most of the experiments, prepared the figures, and cowrote the paper; X.S. and Y.C. performed and analyzed the RT-PCR experiments and some of the WMISH experiments; A.M.Z. guided the screen that led to the initial identification of spib; and E.A. guided the project and cowrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Enrique Amaya or Ricardo Costa, The Healing Foundation Centre, Michael Smith Building, Faculty of Life Sciences, University of Manchester, Oxford Road, Manchester, M13 9PT, United Kingdom; e-mail: enrique.amaya@manchester.ac.uk or ricardo.costa@manchester.ac.uk.