Abstract
RUNX3/AML2 is a Runt domain transcription factor like RUNX1/AML1 and RUNX2/AML3. Regulated by 2 promoters P1 and P2, RUNX3 is frequently inactivated by P2 methylation in solid tumors. Growing evidence has suggested a role of this transcription factor in hematopoiesis. However, genetic alterations have not been reported in blood cancers. In this study on 73 acute myeloid leukemia (AML) patients (44 children and 29 adults), we first showed that high RUNX3 expression among childhood AML was associated with a shortened event-free survival, and RUNX3 was significantly underexpressed in the prognostically favorable subgroup of AML with the t(8;21) and inv(16) translocations. We further demonstrated that this RUNX3 repression was mediated not by P2 methylation, but RUNX1-ETO and CBFβ-MYH11, the fusion products of t(8;21) and inv(16), via a novel transcriptional mechanism that acts directly or indirectly in collaboration with RUNX1, on 2 conserved RUNX binding sites in the P1 promoter. In in vitro studies, ectopically expressed RUNX1-ETO and CBFβ-MYH11 also inhibited endogenous RUNX3 expression. Taken together, RUNX3 was the first transcriptional target found to be commonly repressed by the t(8;21) and inv(16) fusion proteins and might have an important role in core-binding factor AML.
Introduction
The Runt domain transcription factor family includes RUNX1/AML1, RUNX2/AML3, and RUNX3/AML2. These transcription factors share a conserved Runt domain for binding to a 6-base pair (bp) DNA sequence (TGT/cGGT) and heterodimerization with core-binding factor β (CBFβ). CBFβ does not bind DNA directly but increases the ability of RUNX proteins to bind DNA and regulate transcription.1 RUNX1 and RUNX2 are essential for hematopoiesis and osteogenesis, respectively.2,3 Moreover, RUNX1 regulates neuron and muscle function.4,5 On the other hand, RUNX3 is involved in neurogenesis and thymopoiesis and acts as a tumor suppressor in gastric cancer.6-8 In addition, RUNX3 is inactivated frequently by promoter methylation and less frequently by gene deletion, point mutations, and protein mislocalization in various solid tumors.9 RUNX3 and RUNX1 show prominent expression in hematopoietic cells and different subsets of neurons,4,10 while RUNX2 is expressed mainly in bone and also other tissues including hematopoietic stem cells.11 The expression of RUNX3 and RUNX1 can be induced by retinoic acid, suggesting that these transcription factors may jointly regulate retinoic acid–mediated hematopoietic differentiation.10 The functional overlap was supported by the observations that hematopoietic defects due to RUNX1 deficiency could be rescued by RUNX3.12,13 Further evidence suggesting a role of RUNX3 in hematopoiesis was reduction of mature blood cell formation in zebrafish by RUNX3 depletion.14 The role of RUNX3 in tumorigenesis and its potential involvement in hematopoiesis suggest a role of this transcription factor in hematological malignancies. However, genetic alterations of RUNX3 have not been reported in acute myeloid leukemia (AML).15
t(8;21)(q22;q22) and inv(16)(p13;q22) are the 2 most common translocations, representing about 25% of AML cases, and are associated with the French-American-British (FAB) M2 and M4Eo subtypes.16 t(8;21) fuses the N-terminal portion of RUNX1 including its Runt domain to most of the eight-twenty-one (ETO)/MTG8 protein.17 ETO has homology to the Drosophila Nervy protein and can recruit histone deacetylase (HDAC) complex to repress transcription.18 RUNX1-ETO has been postulated as a dominant RUNX1 inhibitor because it can repress genes that are normally activated by RUNX1.19-21 In addition, RUNX1-ETO may indirectly repress transcription through interaction with other transcription factors like E proteins22 and PU.1.23 On the other hand, inv(16) fuses the first 165 amino acids of CBFβ to the coiled-coil region of the smooth muscle myosin heavy chain (MYH11).24 Like RUNX1-ETO, CBFβ-MYH11 has a dominant negative role on RUNX1-mediated transactivation by sequestrating RUNX1 (and potentially other RUNX proteins) from its targets in the DNA.25 In addition, CBFβ-MYH11 can recruit corepressors via its C-terminal domain and cooperate with RUNX1 to repress transcription.26,27 At the molecular level, both t(8;21) and inv(16) disrupt the RUNX1/CBFβ complex that regulates hematopoiesis. Thus, it has long been speculated that CBF-AML may involve common downstream targets that participate in leukemogenesis.
The RUNX3 gene is regulated by 2 promoters P1 and P2.28 The P2 promoter contains a large CpG island that is the target of frequent methylation in solid tumors.9 On the other hand, the P1 promoter contains 2 consensus RUNX binding sites that are conserved in similar positions in the RUNX1 and RUNX2 promoters, suggesting possible cross-regulation among these transcription factors.28 Accordingly, RUNX3 has been shown to repress the RUNX1 promoter in B cells via the conserved RUNX sites.29 Likewise, the conserved RUNX sites in the RUNX2 promoter may auto-regulate its activity.30 In this study, we provided evidence that these RUNX sites mediated transcriptional repression of RUNX3 by the t(8;21) and inv(16) fusion proteins in AML. By contrast to findings in solid tumors, RUNX3 P2 methylation was absent in AML, further emphasizing the distinct mode of RUNX3 repression by these fusion proteins. Our findings thus illustrate a novel transcriptional mechanism of RUNX3 inactivation in human cancers and suggest RUNX3 as a common critical target in CBF-AML.
Methods
Patient samples and cell culture
Diagnostic bone marrow (BM) from 44 childhood AML patients (23 males and 21 females) collected in Prince of Wales Hospital since December 1996 were recruited in this study, with international review board approval of the Joint Chinese University of Hong Kong-New Territories East Cluster Clinical Research Ethics Committee. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki. The median age of the patients was 8.5 years old (range, 0.7-17 years). The patient cohort consisted of 7 M1, 11 M2, 6 M3, 5 M4, 6 M5, 1 M6, and 8 M7. The median percentage of BM blasts was 80% (range, 15%-99%). G-banded cytogenetic studies were performed as described previously.31 The presence of t(8;21) and inv(16) translocations was confirmed by reverse-transcription–polymerase chain reaction (RT-PCR) detection of RUNX1-ETO and CBFβ-MYH11 fusion transcripts.32 Patients were treated with the modified U.K. MRC AML 12 protocol, according to cytogenetics and their response to induction chemotherapy.33,34 Essentially patients were scheduled to receive 2 courses (course 1: 10 + 3 + 5 and course 2: 8 + 3 + 5) of ADE (cytarabine, daunorubicin, etoposide) induction therapy. After achievement of complete remission (CR), a third course of MACE (amsacrine, cytarabine, etoposide) consolidation therapy was given. Good-risk patients [with t(8;21), t(15;17), and inv(16)] received one further course of MIDAC (mitozantrone, cytarabine). Non–good-risk patients with remission achieved after course 1 chemotherapy received the same 4 courses of chemotherapy, or allogeneic transplant if a matched donor was available. However, non–good-risk patients who achieved remission after course 2 chemotherapy received MACE and then an additional course of chemotherapy CLASP (cytarabine, asparaginase) followed by MIDAC as the fifth course, or an allogeneic transplant if a matched donor was available. For acute promyelocytic leukemia patients, all-trans retinoic acid was given in addition to the chemotherapy described. Of the 44 childhood patients, 35 received the same 4 courses of chemotherapy, 7 went on to transplantation, and 2 had poor response and received 5 chemotherapy courses.
Diagnostic BM from 29 adult AML patients (16 males and 13 females) were also included for RUNX3 expression and DNA methylation studies. The median age of the patients was 47 years old (range, 21-83 years). The patient cohort consisted of 4 M1, 9 M2, 4 M3, 6 M4, 5 M5, and 1 M6. The presence of RUNX1-ETO and CBFβ-MYH11 rearrangements was confirmed by cytogenetic analysis and RT-PCR as described in the first paragraph of this section.
BM mononuclear cells were prepared by centrifugation on Ficoll-Hypaque (GE Healthcare, Little Chalfont, United Kingdom) and stored at −80°C before nucleic acid extraction.
All suspension cell lines (Kasumi-1, THP-1, MV4–11, CMK, NB4, ME-1, HL-60, K562, KCL22, 697, Reh, CEM, MOLT-3, and U937) were obtained from DSMZ (Braunschweig, Germany) and cultured under the recommended conditions. HeLa and A549 cell lines were kindly provided by Dr K.W. Lo (Department of Anatomical and Cellular Pathology, The Chinese University of Hong Kong) and cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA).
RNA extraction and RT-PCR
Total RNA was extracted using TRIZOL (Invitrogen) and treated with DNaseI (USB, Cleveland, OH). Real-time RT-PCR analysis of RUNX3, RUNX1-ETO, CBFβ-MYH11, and GAPDH (internal control) expression was performed using Power SYBR Green PCR Master Mix (ABI, Foster City, CA). Samples were amplified for 40 cycles of 30 seconds at 95°C and 30 seconds at 62°C (RUNX3 and GAPDH) or 15 seconds at 95°C and 1 minute at 60°C (RUNX1-ETO and CBFβ-MYH11). The forward and reverse RUNX3 primers were 5′CAGAAGCTGGAGGACCAGAC3′ and 5′GTCGGAGAATGGGTTCAGTT3′ (180 bp), whereas the GAPDH primers were 5′ATGTTCGTCATGGGTGTGAA3′ and 5′GTCTTCTGGGTGGCAGTGAT3′ (173 bp). Primers used for quantitative RUNX1-ETO and CBFβ-MYH11 (type A transcript) analysis were the same as previously described.35 For each sample, the relative quantity of the mRNAs was measured in triplicate using serial dilutions of cDNA from a pool of normal peripheral blood (PB; n = 10) (RUNX3 and GAPDH), Kasumi-1 (RUNX1-ETO), and ME-1 (CBFβ-MYH11) as calibration curves.
For semi-quantitative RT-PCR, the forward RUNX3 primer was the same as that used for real-time RT-PCR and the reverse primer was 5′CTGGTAGGAGCCAGAGGATG3′ (474 bp). PCR of GAPDH with primers 5′CCACCCATGGCAAATTCCATGGCA3′ and 5′ATCTAGACGGCAGGTCAGGTCCACC3′ (598 bp) was performed as an internal control.
Detection of Fms-like tyrosine kinase 3 (FLT3) internal tandem duplication (ITD) and D835 mutation
Western blot analysis
Nuclear and whole cell lysates were prepared as previously described38 for immunoblot analysis of RUNX3 and RUNX1 expression, respectively. RUNX3 and RUNX1 proteins were detected by a rabbit polyclonal antibody against RUNX3 (Abcam, Cambridge, MA) and RUNX1 (Calbiochem, San Diego, CA). A β-actin antibody (Abcam) served as a loading control. Immunostained proteins were visualized using an enhanced chemiluminescence system (GE Healthcare).
Sodium bisulfite modification and methylation-specific PCR (MSP)
Bisulfite-modified DNA was prepared using the Methylamp DNA Modification Kit (Epigentek, Brooklyn, NY). The methylation status of 3 consecutive regions (designated as regions 1-3) within the RUNX3 P2 promoter, which was important for RUNX3 silencing,8,39 was determined by MSP as previously described.39 Regions 1 through 3 analyzed in this study were referred to as regions 6 through 8 reported in Homma et al.39 MSP products were TA-cloned and sequenced.
5′-aza-2′-deoxycytidine (5′-AZA) treatment
Kasumi-1, CMK, and Reh cells (5 × 106) were treated with 5′-AZA (Sigma-Aldrich, St Louis, MO) for 4 days before analysis. Drugs were replenished at day 2 of the treatment.
DNA constructs, transient transfection, and reporter gene assays
Human RUNX3 P1 promoter fragments (the first nucleotide [nt] of exon 1 assigned as +1) were cloned into pGL3-Basic (Promega, Madison, WI). Putative RUNX binding sites were mutated to EcoRI sites as previously described.38 pCMV-RUNX1 (encodes AML1B), pCMV-RUNX1-ETO, and pCMV-CBFβ-MYH11 expression plasmids were kindly provided by Prof S. W. Hiebert (Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, TN). pCMV-RUNX1-ETOΔ469 was prepared by cloning the corresponding coding region into pCMV. Transient transfection of HeLa and A549 cells was performed using Lipofectamine 2000 (Invitrogen) as previously described,40 with 0.5 μg of RUNX3 P1 promoter constructs, 1 μg (or otherwise indicated) pCMV expression plasmids, and 6.25 ng of Renilla luciferase plasmid pRL-CMV (Promega) for normalization of transfection efficiency. For U937, the procedures were similar except that 2.4 × 106 cells were transfected with 1.5 μg of RUNX3 promoter constructs, 1.5 μg (or otherwise indicated) pCMV expression plasmids, and 1 μg pRL-CMV in the presence of 0.5% FBS and harvested for luciferase assays 24 hours after transfection.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed in Kasumi-1, U937, and ME-1 cells according to the manufacturer's protocol (Upstate Biotechnology, Lake Placid, NY) with 5 μg anti-RUNX1 N-terminus or anti-ETO antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Immunoprecipitation with anti-IgG or without an antibody was done as controls. Primers used for RUNX3 promoter detection were 5′TGAGCTGAGGTTGGGTTGA3′ and 5′AGGCTCTGGTGGGTACGA3′ (150 bp).
Nucleofection
U937 cells (106) were suspended in 100 μL of Solution V (Amaxa, Cologne, Germany) containing 20 μg of pCMV expression plasmids and nucleofected with the X-001 program. Cells were then cultured in fresh medium containing 10% FBS for 24 hours before analysis. Transfection efficiency was monitored by flow cytometric analysis of green fluorescent protein expression from U937 cells nucleofected with the same amount of pCMS-EGFP (Clontech, Mountain View, CA).
Data analysis
To examine the relationship between RUNX3 expression levels and various clinicopathological variables, childhood AML patients (n = 44) were dichotomized at the median RUNX3 level and divided into low and high expression groups. Unpaired t and χ2 tests were used to analyze the relationship with continuous and categorical variables between groups. Event-free survival (EFS) and overall survival (OS) were analyzed using the Kaplan-Meier method and survival curves were compared by the log-rank test. EFS was measured from the date of diagnosis until failure to achieve CR, relapse, or death from any cause (whichever occurred first), censoring for those alive and event-free at last follow-up. OS was measured from the date of diagnosis until death from any cause, censoring for those alive at last follow-up. Cox regression analysis was performed to test the significance of RUNX3 expression after controlling for other potential prognostic factors including age (continuous variable), gender, presentation white blood cell (WBC) count (continuous variable), FAB subtype (M7 and non-M7), cytogenetics (favorable, intermediate, and adverse),41 and treatment received (4 courses, 5 courses, and allogeneic transplant). Two-sided P values less than .05 were considered statistically significant. Survival analysis was performed with SPSS 13.0 (SPSS, Chicago, IL).
Results
RUNX3 expression level carries prognostic implications in childhood AML
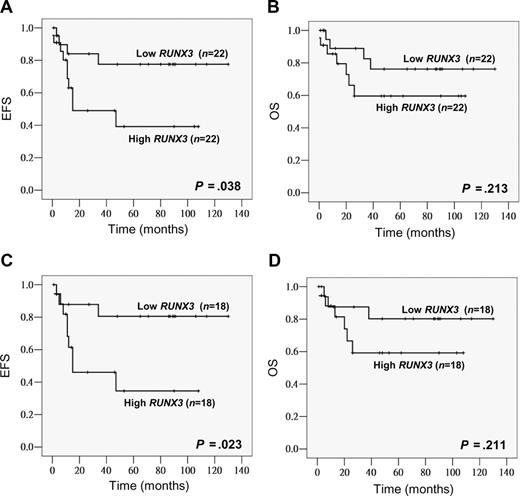
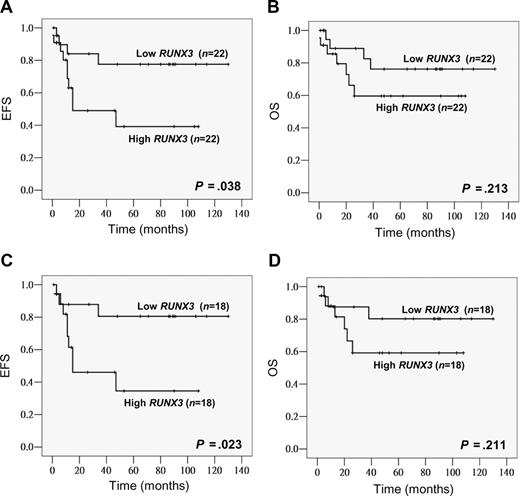
Among the 44 childhood AML patients, 42 (95%) achieved induction remission. Ten patients (23%) relapsed and 11 (25%) died. The last follow-up time was October 2007. The median follow-up period was 30 months for all patients and 53 months for the survivors (n = 33). The median RUNX3 level in the low and high RUNX3 expression groups was 0.038 (range, 0.006-0.090) and 0.204 (range, 0.092-0.677), respectively. Compared with the low expression group, high RUNX3 expressors tended to have a lower CR rate (91% vs 100%, P = .148), a higher relapse rate (35% vs 14%, P = .104; Table 1) and also a poorer EFS (P = .038, estimated 2-year EFS rates: 49% vs 84%; Figure 1A). OS also tended to be worse in high RUNX3 expressors though no statistical significance was reached (P = .213; Figure 1B). None of the potential prognostic factors including age, gender, presentation WBC count, FAB subtype, and cytogenetics alone was found to have significant impacts (P < .05) on EFS and OS. To examine the potential relationship between FLT3 mutation and RUNX3 expression as suggested by Lacayo et al,42 FLT3 mutation status was also investigated in our childhood patient cohort. FLT3 mutations were found in 8 of 44 patients (18%), an incidence similar to those reported before.43,44 Seven cases (16%) were positive for FLT3 ITD and one case (2%) for the D835E mutation. Sequence analysis revealed that all ITD mutations were in-frame and ranged from 15 bp to 69 bp. When patients were stratified by the FLT3 status, a more significant association between high RUNX3 expression and poorer EFS could be observed in the FLT3–wild-type group (n = 36, P = .023, estimated 2-year EFS rates: 46% vs 88%) (Figure 1C). The FLT3-mutant group was not analyzed because the number of cases was small. In multivariate analysis, high RUNX3 expression was an independent prognostic factor for EFS in childhood AML patients with wild-type FLT3 (P = .036, HR = 4.111, 95% CI = 1.094-15.441), and this remained marginally when all childhood patients were included (P = .052, HR = 3.191, 95% CI = 0.990-10.285).
High RUNX3 expression was associated with poor EFS in childhood AML. Kaplan-Meier analysis of EFS and OS based on RUNX3 expression level (low vs high) in all childhood AML patients (total, n = 44) (A,B) or patients with wild-type FLT3 (total, n = 36; C,D).
High RUNX3 expression was associated with poor EFS in childhood AML. Kaplan-Meier analysis of EFS and OS based on RUNX3 expression level (low vs high) in all childhood AML patients (total, n = 44) (A,B) or patients with wild-type FLT3 (total, n = 36; C,D).
RUNX3 expression levels were reduced in t(8;21)- and inv(16)-CBF AML
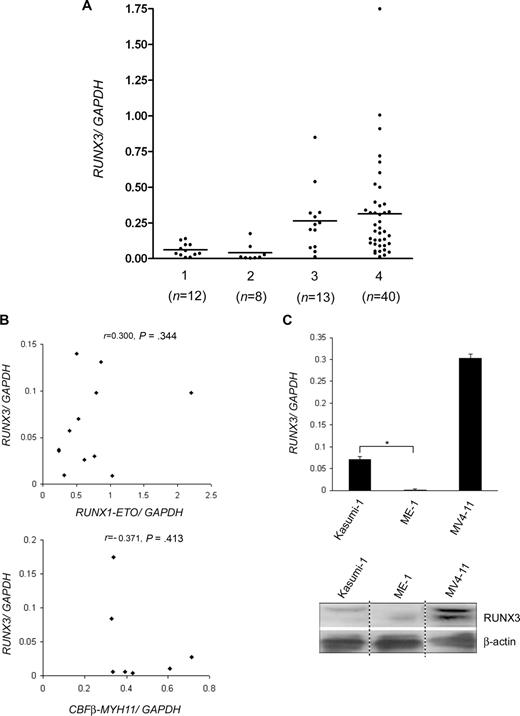
The relationship between RUNX3 expression levels and other clinicopathological parameters was summarized in Table 1. There was no statistical difference in the percentage of BM blasts between the 2 expression groups (median: 73% [low RUNX3] vs 83% [high RUNX3]). RUNX3 expression levels did not correlate with patient's age, sex, presentation WBC count, and the presence of FLT3 mutation. However, reduced RUNX3 levels correlated significantly with the presence of t(8;21) and inv(16) translocations (50% vs 14%, P = .01). In the low expression group, 6 patients had t(8;21) and 5 had inv(16), while in the high expression group, 2 patients had t(8;21) and one had inv(16). Concordantly, low RUNX3 levels were found more frequently in FAB M2 and M4 patients (59% vs 14%, P = .03). To increase the sample size for comparison of RUNX3 expression levels among patient subgroups, we measured RUNX3 mRNA levels in 29 additional diagnostic BM from adult AML patients (total n = 73; Figure 2A). The mean ratio of RUNX3/GAPDH mRNA levels was 0.062 for t(8;21)-positive cases (n = 12) and 0.041 for inv(16)-positive cases (n = 8), and the ratios were not statistically different (P = .387). No significant correlation was found between RUNX3 mRNA levels and the levels of RUNX1-ETO and CBFβ-MYH11 fusion transcripts in the CBF-AML patients (Figure 2B). RUNX3 mRNA levels in the t(8;21)-positive (P = .006) and inv(16)-positive (P = .041) cases were markedly lower than t(8;21)- and inv(16)-negative M2 and M4 cases, which had a mean ratio of 0.265 (n = 13). No statistical difference in RUNX3 levels was found between t(8;21)- and inv(16)-negative M2/M4 and non-M2/M4 cases (n = 40, mean ratio = 0.313). Accordingly, RUNX3 mRNA and protein levels were remarkably lower in t(8;21)-positive Kasumi-1 and inv(16)-positive ME-1 cells than MV4-11 cells, which were derived from a FAB M5 patient lacking the 2 translocations (Figure 2C). Collectively, these data indicate that RUNX3 expression is specifically repressed in t(8;21)– and inv(16)–CBF-AML.
RUNX3 expression levels in AML patients and cell lines. (A) RUNX3 mRNA levels in 73 diagnostic BM from AML patients were determined by real-time RT-PCR and normalized using GAPDH. Patients were divided into 4 subgroups: (1) t(8;21)-positive; (2) inv(16)-positive; (3) t(8;21)- and inv(16)-negative M2/M4; and (4) t(8;21)- and inv(16)-negative non-M2/M4. The number of patients in each subgroup is shown. The horizontal line indicates the mean RUNX3/GAPDH ratio. (B) Correlation analysis between RUNX3 mRNA levels and the levels of RUNX1-ETO and CBFβ-MYH11 fusion transcripts. RUNX1-ETO and CBFβ-MYH11 levels in the t(8;21) and inv(16)-positive patients were determined by real-time RT-PCR and normalized using GAPDH. Of the 8 inv(16)-positive patients, 7 cases express the common type A transcript (also expressed by ME-1), and 1 case expresses the type D transcript,32 which was not amplified by the type A primer set and was excluded from the analysis. r indicates Pearson correlation coefficient. (C) Real-time RT-PCR (top panel) and immunoblot (bottom panel) analysis of RUNX3 expression in Kasumi-1, ME-1, and MV4-11 cell lines, which represent different subgroups of AML. For real-time RT-PCR, results are expressed as mean plus or minus SD from triplicate assays. The mean RUNX3/GAPDH mRNA ratio was 0.002 for ME-1. *P < .001. For immunoblot, the image was assembled from different lanes in the same film.
RUNX3 expression levels in AML patients and cell lines. (A) RUNX3 mRNA levels in 73 diagnostic BM from AML patients were determined by real-time RT-PCR and normalized using GAPDH. Patients were divided into 4 subgroups: (1) t(8;21)-positive; (2) inv(16)-positive; (3) t(8;21)- and inv(16)-negative M2/M4; and (4) t(8;21)- and inv(16)-negative non-M2/M4. The number of patients in each subgroup is shown. The horizontal line indicates the mean RUNX3/GAPDH ratio. (B) Correlation analysis between RUNX3 mRNA levels and the levels of RUNX1-ETO and CBFβ-MYH11 fusion transcripts. RUNX1-ETO and CBFβ-MYH11 levels in the t(8;21) and inv(16)-positive patients were determined by real-time RT-PCR and normalized using GAPDH. Of the 8 inv(16)-positive patients, 7 cases express the common type A transcript (also expressed by ME-1), and 1 case expresses the type D transcript,32 which was not amplified by the type A primer set and was excluded from the analysis. r indicates Pearson correlation coefficient. (C) Real-time RT-PCR (top panel) and immunoblot (bottom panel) analysis of RUNX3 expression in Kasumi-1, ME-1, and MV4-11 cell lines, which represent different subgroups of AML. For real-time RT-PCR, results are expressed as mean plus or minus SD from triplicate assays. The mean RUNX3/GAPDH mRNA ratio was 0.002 for ME-1. *P < .001. For immunoblot, the image was assembled from different lanes in the same film.
Absence of RUNX3 P2 promoter methylation in AML
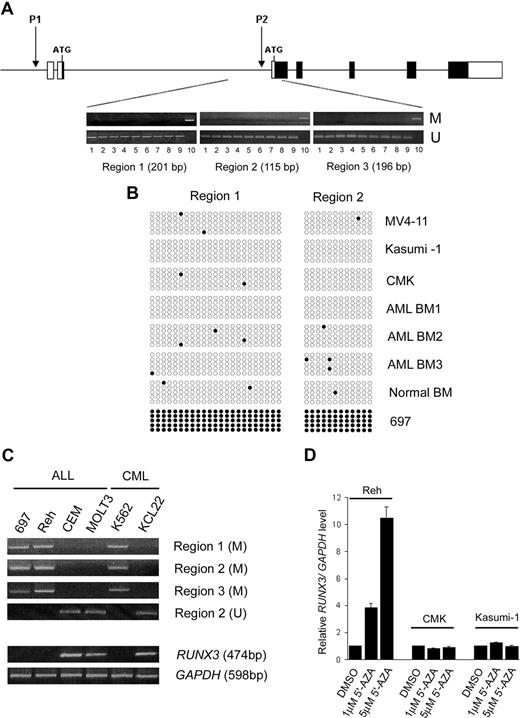
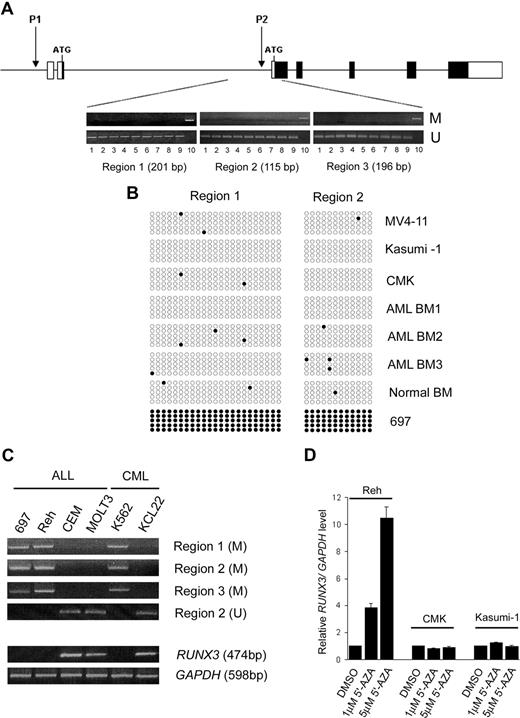
RUNX3 P2 methylation is a major mechanism causing RUNX3 silencing in solid tumors. To examine whether the reduced RUNX3 expression found in t(8;21)- and inv(16)-positive AML was due to promoter methylation, 3 consecutive regions within the RUNX3 P2 promoter were analyzed by MSP. The 3 promoter regions were found to be unmethylated in all the AML samples examined, including 7 AML cell lines (Kasumi-1, THP-1, MV4-11, CMK, NB4, ME-1, and HL-60) and 68 diagnostic BM from AML patients (39 childhood and 29 adult; Figure 3A). The promoter regions were also unmethylated in 5 normal PB and BM samples. Sequence analysis revealed that almost all CpG dinucleotides in regions 1 and 2 were unmethylated in the AML samples (Figure 3B). In contrast, the RUNX3 P2 promoter regions were methylated in two fourths of acute lymphoblastic leukemia (ALL; 697, Reh, MOLT-3, and CEM) and one half of chronic myeloid leukemia (CML) cell lines (K562 and KCL22; Figure 3C). In 697 cells, all the CpG dinucleotides in regions 1 and 2 were methylated (Figure 3B). RT-PCR revealed that RUNX3 P2 methylation was associated with RUNX3 silencing in these cell lines (Figure 3C). In addition, treatment of the methylated Reh cell line with 5′-AZA drastically induced RUNX3 mRNA levels (Figure 3D). In contrast, RUNX3 levels were not induced by the demethylating agent in the unmethylated Kasumi-1 and CMK (RUNX3/GAPDH ratio = 0.009) cell lines, which expressed very low levels of RUNX3 transcripts. Collectively, these findings suggest that promoter methylation is not involved in controlling RUNX3 expression in AML.
RUNX3 P2 promoter was unmethylated in AML. (A) Schematic diagram of the human RUNX3 gene structure. ■ and □ represent coding and noncoding regions, respectively. The location of the RUNX3 P1 and P2 promoters is shown. Below the diagram are representative results of MSP analysis of 3 consecutive regions in the RUNX3 P2 promoter in AML samples. The size of each analyzed region is indicated. Lanes: 1, MV4-11; 2, Kasumi-1; 3, CMK; 4, ME-1; 5-6, 2 diagnostic childhood AML BM; 7-8, 2 diagnostic adult AML BM; 9, normal BM; 10, Reh (positive control). M and U represent PCR using primers specific for the methylated and unmethylated sequences, respectively. (B) Results of nucleotide sequencing of MSP products from regions 1 and 2 in 3 AML cell lines (MV4-11, Kasumi-1, and CMK), 3 diagnostic childhood AML BM, a normal BM, and the 697 cell line. Each row of circles represents one PCR clone. Open and filled circles indicate unmethylated and methylated CpG dinucleotides, respectively. (C) MSP (top panel) and RT-PCR (bottom panel) analysis of RUNX3 P2 methylation and expression in 4 ALL and 2 CML cell lines. Note that RUNX3 P2 methylation is associated with RUNX3 silencing in these leukemic cell lines. (D) Treatment with the DNA demethylating agent 5′-AZA induced RUNX3 mRNA expression in the methylated Reh cell line but not in unmethylated Kasumi-1 and CMK cell lines. RUNX3 mRNA levels were determined by real-time RT-PCR and normalized using GAPDH. Results are presented as relative RUNX3/GAPDH level by comparing the normalized RUNX3 level in treatment groups with that in the respective control group (DMSO treated).
RUNX3 P2 promoter was unmethylated in AML. (A) Schematic diagram of the human RUNX3 gene structure. ■ and □ represent coding and noncoding regions, respectively. The location of the RUNX3 P1 and P2 promoters is shown. Below the diagram are representative results of MSP analysis of 3 consecutive regions in the RUNX3 P2 promoter in AML samples. The size of each analyzed region is indicated. Lanes: 1, MV4-11; 2, Kasumi-1; 3, CMK; 4, ME-1; 5-6, 2 diagnostic childhood AML BM; 7-8, 2 diagnostic adult AML BM; 9, normal BM; 10, Reh (positive control). M and U represent PCR using primers specific for the methylated and unmethylated sequences, respectively. (B) Results of nucleotide sequencing of MSP products from regions 1 and 2 in 3 AML cell lines (MV4-11, Kasumi-1, and CMK), 3 diagnostic childhood AML BM, a normal BM, and the 697 cell line. Each row of circles represents one PCR clone. Open and filled circles indicate unmethylated and methylated CpG dinucleotides, respectively. (C) MSP (top panel) and RT-PCR (bottom panel) analysis of RUNX3 P2 methylation and expression in 4 ALL and 2 CML cell lines. Note that RUNX3 P2 methylation is associated with RUNX3 silencing in these leukemic cell lines. (D) Treatment with the DNA demethylating agent 5′-AZA induced RUNX3 mRNA expression in the methylated Reh cell line but not in unmethylated Kasumi-1 and CMK cell lines. RUNX3 mRNA levels were determined by real-time RT-PCR and normalized using GAPDH. Results are presented as relative RUNX3/GAPDH level by comparing the normalized RUNX3 level in treatment groups with that in the respective control group (DMSO treated).
RUNX1-ETO repressed the RUNX3 P1 promoter through 2 conserved RUNX binding sites
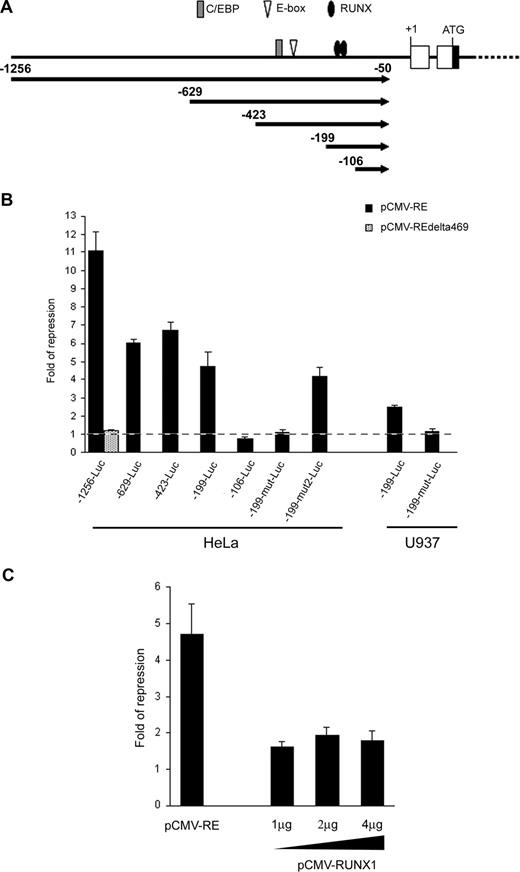
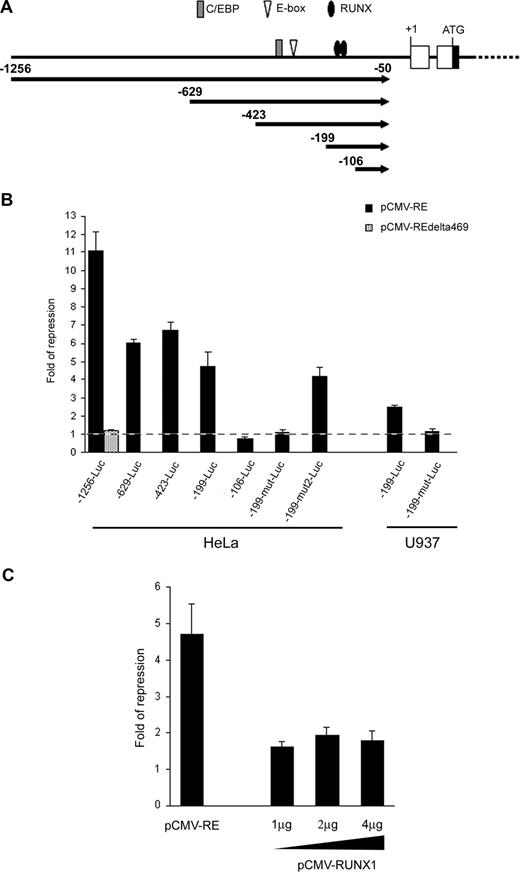
Sequence analysis revealed that the RUNX3 P1 promoter contained a number of binding sites for RUNX, E proteins and CCAAT/enhancer-binding protein (C/EBP) transcription factors which are potential targets for RUNX1-ETO repression (Figure 4A).20,22,45 To test whether RUNX1-ETO could repress the RUNX3 P1 promoter, we cloned a 1.2-kb P1 promoter fragment containing these transcription factor binding sites into pGL3-Basic and cotransfected the construct (-1256-Luc) with the t(8;21) expression plasmid into HeLa cells. Ectopic expression of RUNX1-ETO repressed the P1 promoter by 11-fold (Figure 4B). However, deletion of the C-terminal region of RUNX1-ETO (RUNX1-ETOΔ469), which is required for oligomerization and corepressor recruitment,18 ablated the repression. To identify the RUNX1-ETO response region, we performed 5′-deletion mapping on the RUNX3 P1 promoter. Removal of sequences from nt -1256 to -199 did not abolish the RUNX1-ETO effect as revealed by the persistence of 4.6-fold repression on -199-Luc (Figure 4B). Mutation or deletion of the putative E-box and C/EBP sites had no effect on both basal and RUNX1-ETO-regulated promoter activity (data not shown). However, further deletion to nt -106, which involved the 2 conserved RUNX binding sites at -157/-152 and -149/-144, completely abolished the RUNX1-ETO repression (Figure 4B). Likewise, concurrent mutation of these RUNX sites (-199-mut-Luc) abolished the RUNX1-ETO effect. In contrast, mutation of a RUNX-like site (TGAGGT; -199-mut2-Luc) at -194/-189 did not affect the RUNX1-ETO effect (Figure 4B). These data thus indicate that RUNX1-ETO repressed the RUNX3 P1 promoter via the 2 conserved RUNX sites at -157/-152 and -149/-144. Similar results were obtained in U937 myeloid cells (Figure 4B). We next investigated whether RUNX1 could regulate the RUNX3 P1 promoter by cotransfecting -199-Luc with various amounts of a RUNX1 expression plasmid into HeLa cells, which do not express RUNX1.20 Overexpression of RUNX1 had a weak repressive effect on the P1 promoter, as compared with cotransfection with the same amount of a RUNX1-ETO expression plasmid (Figure 4C). Increasing the amount of the RUNX1 expression plasmid did not augment the repression (Figure 4C).
RUNX1-ETO repressed the RUNX3 P1 promoter through 2 conserved RUNX binding sites. (A) Schematic diagram of the RUNX3 P1 promoter showing the consensus RUNX, E-box, and C/EBP binding sites along with the promoter deletions analyzed in subsequent panels. (B) Wild-type and mutant RUNX3 P1 promoter constructs were cotransfected with pCMV-RUNX1-ETO and pRL-CMV into HeLa and U937 cells. The -1256-Luc was also cotransfected with pCMV-RUNX1-ETOΔ469 to examine the effect of C-terminal deletion on RUNX3 repression. (C) The -199-Luc promoter construct was cotransfected with 1 μg of pCMV-RUNX1-ETO or increasing amounts (1 μg, 2 μg, and 4 μg) of pCMV-RUNX1 together with pRL-CMV into HeLa cells. In all experiments, cotransfection with the same amount of empty pCMV was done in parallel. Transfection efficiency was normalized according to the cotransfected pRL-CMV Renilla luciferase activity. Results are presented as fold of repression by comparing the normalized firefly luciferase activity of the construct cotransfected with the expression plasmids to that cotransfected with empty pCMV. Results are expressed as mean plus or minus SE from triplicate assays. pCMV-RE indicates pCMV-RUNX1-ETO.
RUNX1-ETO repressed the RUNX3 P1 promoter through 2 conserved RUNX binding sites. (A) Schematic diagram of the RUNX3 P1 promoter showing the consensus RUNX, E-box, and C/EBP binding sites along with the promoter deletions analyzed in subsequent panels. (B) Wild-type and mutant RUNX3 P1 promoter constructs were cotransfected with pCMV-RUNX1-ETO and pRL-CMV into HeLa and U937 cells. The -1256-Luc was also cotransfected with pCMV-RUNX1-ETOΔ469 to examine the effect of C-terminal deletion on RUNX3 repression. (C) The -199-Luc promoter construct was cotransfected with 1 μg of pCMV-RUNX1-ETO or increasing amounts (1 μg, 2 μg, and 4 μg) of pCMV-RUNX1 together with pRL-CMV into HeLa cells. In all experiments, cotransfection with the same amount of empty pCMV was done in parallel. Transfection efficiency was normalized according to the cotransfected pRL-CMV Renilla luciferase activity. Results are presented as fold of repression by comparing the normalized firefly luciferase activity of the construct cotransfected with the expression plasmids to that cotransfected with empty pCMV. Results are expressed as mean plus or minus SE from triplicate assays. pCMV-RE indicates pCMV-RUNX1-ETO.
CBFβ-MYH11 repressed the RUNX3 P1 promoter through cooperation with RUNX1 and the same RUNX binding sites that mediated RUNX1-ETO repression
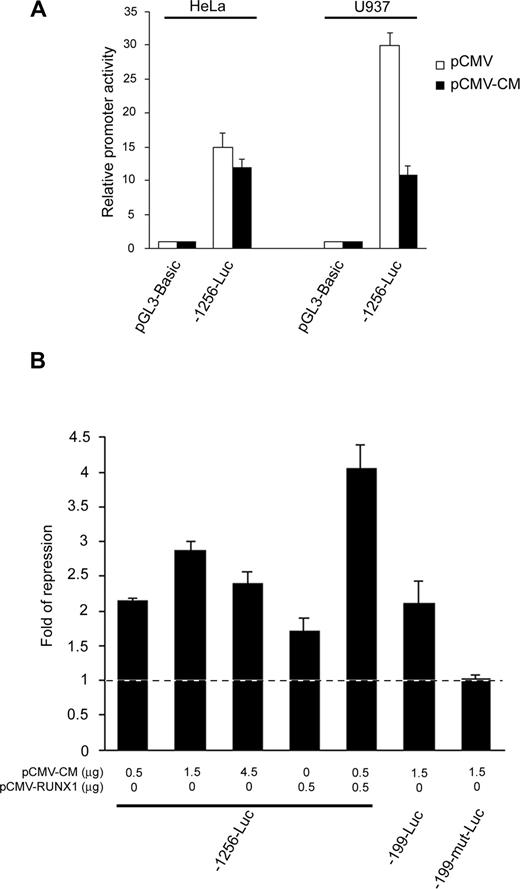
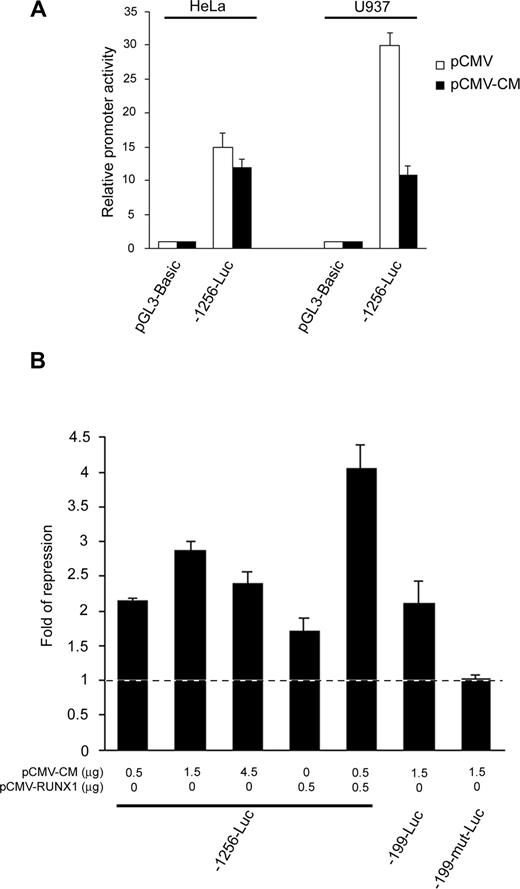
Given that RUNX1 cooperates with CBFβ-MYH11 to repress transcription,26 we sought to investigate whether they repressed the RUNX3 P1 promoter in a cooperative manner. Ectopic expression of CBFβ-MYH11 repressed the 1.2-kb P1 promoter fragment in RUNX1-expressing U937 cells but not in RUNX1-nonexpressing HeLa cells (Figure 5A), suggesting the requirement of RUNX1 in mediating the inv(16) repression. To further illustrate the cooperation, we cotransfected -1256-Luc with increasing amounts of the inv(16) expression plasmid into U937 cells. While the repressive effect increased from 0.5 μg to 1.5 μg of expression plasmid, it did not enhance further when more plasmid (4.5 μg) was used (Figure 5B), indicating that other factors may be limiting at high CBFβ-MYH11 concentrations. Thus, we next cotransfected -1256-Luc and the inv(16) expression plasmid in the presence or absence of the RUNX1 expression plasmid into the myeloid cells. CBFβ-MYH11 and RUNX1 alone moderately repressed the P1 promoter. However, an additive repressive effect was observed when both CBFβ-MYH11 and RUNX1 were overexpressed in the cells (Figure 5B). These findings thus confirm that CBFβ-MYH11 cooperates with RUNX1 to repress the RUNX3 P1 promoter. Because RUNX1 DNA binding is required for the CBFβ-MYH11/RUNX1 cooperative action,26 we speculated that the 2 conserved RUNX sites at nt -157/-152 and -149/-144 might mediate the inv(16) repression. In U937 cells, CBFβ-MYH11 repressed both -1256-Luc and -199-Luc, which retain the 2 conserved RUNX sites. However, concurrent mutation of these sites (-199-mut-Luc) abolished the repression (Figure 5B). Thus, CBFβ-MYH11/RUNX1 repressed the RUNX3 P1 promoter through the same RUNX binding sites that also mediated the t(8;21) repression.
CBFβ-MYH11 cooperated with RUNX1 to repress the RUNX3 P1 promoter through the same conserved RUNX binding sites. (A) The -1256-Luc promoter construct was cotransfected with pCMV-CBFβ-MYH11 or empty pCMV together with pRL-CMV into HeLa and U937 cells. (B) Wild-type and mutant RUNX3 P1 promoter constructs were cotransfected with the indicated amount of pCMV-CBFβ-MYH11 and/or pCMV-RUNX1 together with pRL-CMV into U937 cells. Cotransfection with the same amount of empty pCMV was done in parallel. In all experiments, transfection efficiency was normalized according to the cotransfected pRL-CMV Renilla luciferase activity. Results are presented as relative promoter activity by comparing the normalized firefly luciferase activity of the construct to that of pGL3-Basic or as fold of repression by comparing the normalized firefly luciferase activity of the construct cotransfected with the expression plasmids to that cotransfected with empty pCMV. Results are expressed as mean plus or minus SE from triplicate assays. pCMV-CM indicates pCMV-CBFβ-MYH11.
CBFβ-MYH11 cooperated with RUNX1 to repress the RUNX3 P1 promoter through the same conserved RUNX binding sites. (A) The -1256-Luc promoter construct was cotransfected with pCMV-CBFβ-MYH11 or empty pCMV together with pRL-CMV into HeLa and U937 cells. (B) Wild-type and mutant RUNX3 P1 promoter constructs were cotransfected with the indicated amount of pCMV-CBFβ-MYH11 and/or pCMV-RUNX1 together with pRL-CMV into U937 cells. Cotransfection with the same amount of empty pCMV was done in parallel. In all experiments, transfection efficiency was normalized according to the cotransfected pRL-CMV Renilla luciferase activity. Results are presented as relative promoter activity by comparing the normalized firefly luciferase activity of the construct to that of pGL3-Basic or as fold of repression by comparing the normalized firefly luciferase activity of the construct cotransfected with the expression plasmids to that cotransfected with empty pCMV. Results are expressed as mean plus or minus SE from triplicate assays. pCMV-CM indicates pCMV-CBFβ-MYH11.
RUNX1-ETO and RUNX1 bound the RUNX3 P1 promoter
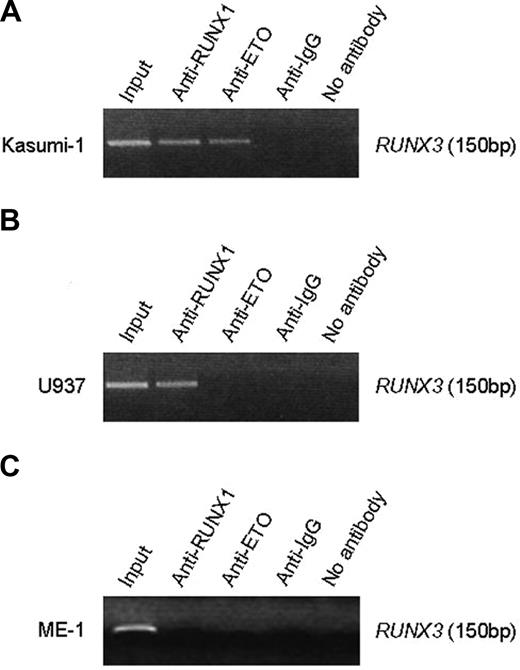
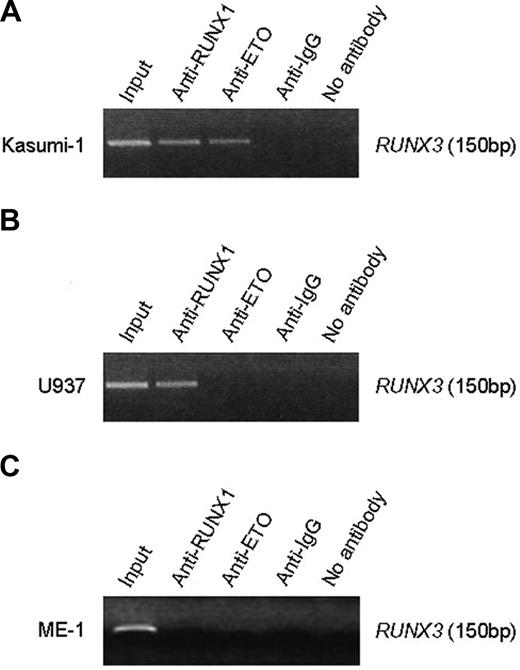
ChIP assays showed that a 150-bp DNA fragment (nt -199/-50) flanking the 2 conserved RUNX sites in the RUNX3 P1 promoter was amplified from Kasumi-1 cells when chromatin was immunoprecipitated with the anti-RUNX1 N-terminus antibody (Figure 6A). Because Kasumi-1 cells also express RUNX1, we performed ChIP with anti-ETO antibody to distinguish RUNX1-ETO from wild-type RUNX1. The RUNX3 promoter sequence was also detected with anti-ETO in Kasumi-1 cells (Figure 6A), indicating that RUNX1-ETO associates with the P1 promoter. To determine whether RUNX1 associates with the P1 promoter in the absence of RUNX1-ETO, we carried out ChIP assays in U937 cells that express RUNX1 but lack the fusion protein. The RUNX3 promoter sequence was only detected with anti-RUNX1 but not anti-ETO antibody (Figure 6B). These findings thus indicate that both RUNX1-ETO and RUNX1 bind the RUNX3 P1 promoter in vivo. Indeed, it was shown using gel shift assays that RUNX1 also bound the 2 conserved RUNX sites in the RUNX1 P1 promoter.29 Interestingly, we failed to detect the RUNX3 promoter with anti-RUNX1 in ME-1 cells, which express CBFβ-MYH11 (Figure 6C). The failure might be possibly due to masking of RUNX1 epitope or sequestration of RUNX1 from the promoter by the inv(16) fusion protein.
RUNX1-ETO and RUNX1 associated with the RUNX3 P1 promoter. (A) ChIP analysis of the RUNX3 P1 promoter in Kasumi-1 cells using an antibody directed to the N-terminus of RUNX1, ETO, or IgG. Immunoprecipitated chromatin was analyzed by PCR of a 150-bp RUNX3 P1 promoter fragment. (B) ChIP analysis of the RUNX3 P1 promoter in U937 cells, which express RUNX1 but not RUNX1-ETO. (C) ChIP analysis of the RUNX3 P1 promoter in ME-1 cells, which express CBFβ-MYH11. The same antibodies described in panel A were used for immunoprecipitation in panels B and C. Note that no PCR product was obtained when anti-IgG or no antibody was used for immunoprecipitation.
RUNX1-ETO and RUNX1 associated with the RUNX3 P1 promoter. (A) ChIP analysis of the RUNX3 P1 promoter in Kasumi-1 cells using an antibody directed to the N-terminus of RUNX1, ETO, or IgG. Immunoprecipitated chromatin was analyzed by PCR of a 150-bp RUNX3 P1 promoter fragment. (B) ChIP analysis of the RUNX3 P1 promoter in U937 cells, which express RUNX1 but not RUNX1-ETO. (C) ChIP analysis of the RUNX3 P1 promoter in ME-1 cells, which express CBFβ-MYH11. The same antibodies described in panel A were used for immunoprecipitation in panels B and C. Note that no PCR product was obtained when anti-IgG or no antibody was used for immunoprecipitation.
RUNX1-ETO and CBFβ-MYH11 repressed endogenous RUNX3 expression
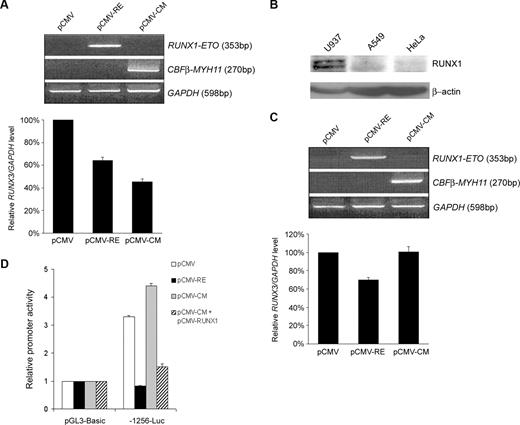
The ability of RUNX1-ETO and CBFβ-MYH11 to repress RUNX3 promoter suggested that the fusion proteins might inhibit endogenous RUNX3 expression. To address this issue, the U937 cell line that endogenously expresses high levels of RUNX3 was selected for nucleofection.10 Under our conditions, a transfection efficiency of 54% plus or minus 6% (mean ± SD from 3 independent experiments) was achieved. Ectopic expression of RUNX1-ETO and CBFβ-MYH11 in U937 cells reduced RUNX3 mRNA levels by 36% and 55%, respectively (Figure 7A). Because the inv(16)-mediated RUNX3 repression required RUNX1, we tested whether CBFβ-MYH11 failed to inhibit endogenous RUNX3 expression in A549 lung cancer cells that lacked RUNX1 as revealed by immunoblot analysis (Figure 7B). Similar to U937 cells, ectopic expression of RUNX1-ETO suppressed RUNX3 mRNA levels by 30% in A549 cells (Figure 7C). In contrast, no significant change in RUNX3 expression could be detected when CBFβ-MYH11 was expressed. Concordantly, we found that RUNX1-ETO but not CBFβ-MYH11 could repress the RUNX3 P1 promoter in the lung cancer cells (Figure 7D). Cotransfection with RUNX1 rescued the ability of CBFβ-MYH11 to repress the P1 promoter.
RUNX1-ETO and CBFβ-MYH11 inhibited endogenous RUNX3 expression. (A) U937 cells were nucleofected with 20 μg of pCMV, pCMV-RUNX1-ETO, or pCMV-CBFβ-MYH11. Expression of fusion products was validated by RT-PCR (upper panel), and RUNX3 mRNA levels were determined by real-time RT-PCR and normalized using GAPDH (lower panel) 24 hours after nucleofection. Results are presented as relative RUNX3/GAPDH level by comparing the normalized RUNX3 level transfected with pCMV-RUNX1-ETO or pCMV-CBFβ-MYH11 to that transfected with empty pCMV. (B) Immunoblot analysis of RUNX1 and β-actin (loading control) expression in U937, A549, and HeLa cells. (C) A549 cells were transfected with 10 μg of pCMV, pCMV-RUNX1-ETO, or pCMV-CBFβ-MYH11 using lipofectamine 2000. The transfection efficiency from 3 independent experiments was 53% plus or minus 4%. Validation of fusion product expression (top panel) and measurement of RUNX3 mRNA levels (bottom panel) were done as described in panel A 48 hours after transfection. (D) RUNX3 P1 promoter construct -1256-Luc was cotransfected with pCMV-RUNX1-ETO, pCMV-CBFβ-MYH11, pCMV-CBFβ-MYH11 and pCMV-RUNX1 (0.5 μg each), or empty pCMV together with pRL-CMV into A549 cells. Transfection efficiency was normalized according to the cotransfected pRL-CMV Renilla luciferase activity. Results are presented as relative promoter activity by comparing the normalized firefly luciferase activity of the construct to that of pGL3-Basic. Results are expressed as mean plus or minus SE from triplicate assays.
RUNX1-ETO and CBFβ-MYH11 inhibited endogenous RUNX3 expression. (A) U937 cells were nucleofected with 20 μg of pCMV, pCMV-RUNX1-ETO, or pCMV-CBFβ-MYH11. Expression of fusion products was validated by RT-PCR (upper panel), and RUNX3 mRNA levels were determined by real-time RT-PCR and normalized using GAPDH (lower panel) 24 hours after nucleofection. Results are presented as relative RUNX3/GAPDH level by comparing the normalized RUNX3 level transfected with pCMV-RUNX1-ETO or pCMV-CBFβ-MYH11 to that transfected with empty pCMV. (B) Immunoblot analysis of RUNX1 and β-actin (loading control) expression in U937, A549, and HeLa cells. (C) A549 cells were transfected with 10 μg of pCMV, pCMV-RUNX1-ETO, or pCMV-CBFβ-MYH11 using lipofectamine 2000. The transfection efficiency from 3 independent experiments was 53% plus or minus 4%. Validation of fusion product expression (top panel) and measurement of RUNX3 mRNA levels (bottom panel) were done as described in panel A 48 hours after transfection. (D) RUNX3 P1 promoter construct -1256-Luc was cotransfected with pCMV-RUNX1-ETO, pCMV-CBFβ-MYH11, pCMV-CBFβ-MYH11 and pCMV-RUNX1 (0.5 μg each), or empty pCMV together with pRL-CMV into A549 cells. Transfection efficiency was normalized according to the cotransfected pRL-CMV Renilla luciferase activity. Results are presented as relative promoter activity by comparing the normalized firefly luciferase activity of the construct to that of pGL3-Basic. Results are expressed as mean plus or minus SE from triplicate assays.
Discussion
In this study, we showed that RUNX3 expression was an independent prognostic factor in childhood AML. High RUNX3 expression was associated with worse EFS and this association was independent of the FLT3 mutation status. A previous study by Lacayo et al also showed that a high RUNX3/ATRX expression ratio was associated with a worse EFS in childhood AML, but only in the FLT3 mutant group.42 Worthy of note is that they analyzed primarily on the ratio of RUNX3 to ATRX expression and in a cohort with a disproportionately large number (28%) of patients refractory to induction and a much higher frequency (45%) of FLT3 mutations. These differences may likely contribute to the differential observations in the apparent link between FLT3 mutation and RUNX3 expression in these 2 studies. On the other hand, low RUNX3 expression associated with better EFS was overrepresented by patients with t(8;21) and inv(16), a known prognostically favorable subgroup of AML.41 Analysis of the 35 identically treated patients also revealed a similar trend toward poorer EFS with high RUNX3 (P = .271). The trend was more obvious (P = .109) when the 2 patients who received extra chemotherapy and had high RUNX3 were included in the analysis (data not shown). Larger prospective studies are needed to further analyze the prognostic use of RUNX3 in childhood AML.
Despite conferring a better prognosis, t(8;21) and inv(16) AML exhibit substantial clinical and biologic heterogeneity such that some patients develop relapse with shortened survival. The heterogeneity is also reflected by the existence of different clinical subsets that could be defined by distinct gene expression profiles.46 Accordingly, we found that 3 pediatric patients with t(8;21) or inv(16) had high RUNX3 expression (Table 1). Importantly, 2 of these 3 patients (67%) relapsed early in 11 months as compared with 18% (2 of 11) of CBF-AML patients in the low RUNX3 expression group did in an average of 23 months from presentation. However, these 2 CBF-AML subgroups were found to be similar with respect to their age, gender, presentation WBC count, as well as the distribution of genetic aberrations including FLT3 and KIT exons 8 and 17 (our unpublished data, August 2007) mutations, which were reported to affect the prognosis of CBF-AML.46-48 Thus, it suggests that RUNX3 may be a useful marker that helps discriminate prognostically distinct CBF-AML subgroups for future risk-adapted therapies. Why RUNX3 not repressed by RUNX1-ETO and CBFβ-MYH11 in these 3 patients is currently unknown but might be due to the presence of other deregulated pathways that interfere with RUNX3 repression by the fusion proteins. The identification of these pathways may open novel therapeutic approaches to high-risk CBF-AML patients.
It has been shown that t(8;21) and inv(16) are associated with locus-specific promoter methylation leading to transcriptional silencing.49,50 However, none of the AML specimens examined, including CBF-AML, showed RUNX3 P2 methylation (Figure 3). Interestingly, methylation-associated RUNX3 silencing was detected in half of the ALL and CML cell lines, suggesting that RUNX3 methylation occurs in certain types of hematological malignancies.
Most importantly, in this study, we uncovered a new mode of RUNX3 inactivation by showing that the gene was transcriptionally repressed by the t(8;21) and inv(16) fusion proteins in AML. Both forms of the repression were mediated by the same RUNX binding sites in the P1 promoter and were presumably sufficient to reduce RUNX3 expression in the absence of P2 methylation. However, the t(8;21) repression involved direct binding of RUNX1-ETO to the RUNX3 promoter. For inv(16), our findings indicated a cooperative action between CBFβ-MYH11 and RUNX1 and that the repression occurred at the level of RUNX1 DNA binding because mutation of its cognate binding sites abolished the effect. This coincides with previous findings that a RUNX1 mutant that failed to bind DNA did not cooperate with inv(16) to repress transcription.26 CBFβ-MYH11 can recruit the mSin3A corepressor and HDAC8, and mSin3A itself was shown to associate with HDAC1 and HDAC2.25 In addition, CBFβ-MYH11 can multimerize through its myosin tail, which may further enhance corepressor binding.25 However, the recruitment of multiple corepressors and formation of a highly multimerized structure might potentially have precluded our detection of CBFβ-MYH11 on the endogenous RUNX3 promoter in ME-1 cells using ChIP with an anti-CBFβ antibody (data not shown). It should be noted that unlike most of the genes repressed by RUNX1-ETO and CBFβ-MYH11,19-21,51 RUNX1 did not activate but rather modestly repressed the RUNX3 promoter (Figures 4,5). Our findings thus support the notion that t(8;21) and inv(16) convert RUNX1 into a constitutive repressor.27,52
The transforming growth factor β (TGFβ) signaling pathway is a major antiproliferative and differentiation signal for hematopoietic progenitors.53 In AML, Smad4 mutations and inactivation of cytoplasmic PML, a critical TGFβ regulator, by PML-RARα have been described.54,55 Interestingly, RUNX3 has also been regarded as a TGFβ mediator because of its ability to cooperate with Smads to regulate transcription.56 The specific repression of RUNX3 transcription by RUNX1-ETO and CBFβ-MYH11 suggests that these fusion proteins may contribute to leukemogenesis in part by disruption of the TGFβ/RUNX3/Smad pathway.
c-Jun is a target of the myeloid master regulator PU.1 and plays a crucial role in AML development.57 Interestingly, we found that mutation of a proximal AP-1 motif dramatically reduced RUNX3 promoter activity and c-Jun could transactivate the promoter (our unpublished data, July 2007). These findings suggest RUNX3 as an indirect PU.1 target. It is possible that loss of PU.1 activity or other mechanisms may reduce c-Jun expression, and thereby hinder RUNX3 transcription. This may be a possible explanation for the decreased RUNX3 expression in some AML patients lacking t(8;21) and inv(16) (Figure 2).
The RUNX1 gene is also disrupted by the t(12;21) translocation in childhood B-cell ALL, resulting in the production of the TEL-RUNX1 fusion protein.58 Although TEL-RUNX1 is similar to RUNX1-ETO in that it can also block RUNX1-dependent transactivation and recruit corepressors, these fusion proteins have distinct promoter specificities.59 Moreover, unlike in AML, RUNX3 was epigenetically silenced by promoter methylation in t(12;21)-positive Reh cells (Figure 3). Whether RUNX3 is also transcriptionally repressed by TEL-RUNX1 awaits further investigation.
In conclusion, we showed that RUNX3 was a common transcriptional target for repression by the t(8;21) and inv(16) fusion proteins in AML. High RUNX3 expression was associated with a poorer outcome and might serve as a marker to stratify prognostically distinct CBF-AML subgroups. Thus, RUNX3 has emerged as an important target in CBF-AML and characterization of its transcriptome will help elucidate the pathogenesis of this specific leukemia subtype and identify novel targets for therapeutic intervention.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Prof S. W. Hiebert for providing the RUNX1, RUNX1-ETO, and CBFβ-MYH11 expression plasmids; Dr K. W. Lo for providing the HeLa and A549 cell lines; and Dr K. S. Tsang for technical help in preparing and storing some patient samples.
This work was supported in part by a grant from the Research Council of the Hong Kong SAR, China (Project No. CUHK CERG 4415/05M).
Authorship
Contribution: C.K.C. designed and performed research and wrote the manuscript; L.L. and S.H.C. performed research; K.M.L., N.P.C., R.S.W., M.M.S., and C.K.L. collected and analyzed data; and M.H.N. designed research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof Margaret H. L. Ng, Department of Anatomical and Cellular Pathology, The Chinese University of Hong Kong, Prince of Wales Hospital, Shatin, New Territories, Hong Kong SAR, China; e-mail: margaretng@cuhk.edu.hk.