Abstract
Expression of latent membrane protein 2 (LMP2A) during B-cell development leads to global alterations in gene transcription similar to those seen in Hodgkin Reed-Sternberg cells of Hodgkin lymphoma (HL). Along with the consistent detection of LMP2A in Epstein-Barr virus–associated HL, this implicates a role for LMP2A in the pathogenesis of HL. We have shown that LMP2A constitutively activates the Notch1 pathway to autoregulate the LMP2A promoter. To determine whether constitutive activation of the Notch pathway is important for LMP2A-mediated alterations in B-cell development in vivo, TgE-LMP2A–transgenic mice were intercrossed with mice expressing loxP-flanked Notch1 genes and Cre recombinase. B cells from TgE Notch1lox/lox-CD19+/Cre mice have an increase in immunoglobulin M and CD43 and a decrease in CD5 expression in the bone marrow compared with TgE Notch1lox/lox mice, indicating the LMP2A signal for developmental aberrations is impaired in the absence of Notch1. Real-time reverse-transcribed polymerase chain reaction analysis reveals that LMP2A requires the Notch1 pathway to alter levels of B cell–specific transcription factors, E2A and EBF. Interestingly, Notch1 appears to be important for LMP2A-mediated survival in low interleukin-7. We propose that LMP2A and the Notch1 pathway may cooperate to induce the alterations in B-cell identity seen in Hodgkin Reed-Sternberg cells.
Introduction
Epstein-Barr virus (EBV) is a ubiquitous human herpesvirus that infects greater than 90% of the adult population.1 Although most EBV infections are asymptomatic, disease arises in adolescents as infectious mononucleosis and in immunocompromised patients as lymphoproliferative disorders.1 EBV is associated with malignancies of lymphoid and epithelial origin, including Hodgkin lymphoma (HL), Burkitt lymphoma, and nasopharyngeal carcinoma.1 As is characteristic of herpesviruses, EBV is able to persist in the human host through the establishment of a lifelong latent infection. EBV establishes latency in vitro in B lymphocytes by limiting viral gene expression to a subset of genes, which includes EBV nuclear antigens 1, 2, 3A, 3B, 3C, and LP (EBNAs), latent membrane protein 1 (LMP1), and latent membrane protein 2 (LMP2A).1
The transgenic mouse model used by our laboratory has been invaluable in elucidating the function of LMP2A in vivo. In the TgE-LMP2A mice, expression of LMP2A interferes with normal B-cell development and allows B-cell receptor (BCR)–negative cells to exit the bone marrow and colonize peripheral lymphoid organs, such as the spleen.2 Normally, in the bone marrow, successful immunoglobulin (Ig) heavy-chain rearrangement is necessary for the transition from the CD19+CD43+ pre-BI stage to the CD19+CD43− pre-BII stage of development. Subsequently, the light-chain genes are rearranged and the light-chain complexes with the heavy chain to form a BCR. The BCR is expressed at the cell surface and allows the cell to transition to the IgM+ immature B-cell stage and migrate of the bone marrow into the periphery.3 The striking phenotype of the TgE-LMP2A–transgenic line is the lack of expression of surface IgM in the bone marrow and spleen along with a reduction in the total number of B cells. In addition, although TgE-LMP2A mice are unable to rearrange the heavy-chain genes necessary for expression of a pre-BCR or a BCR, they are able to transition to a CD43− stage.2,4 TgE-LMP2A mice have a defect at the pre-B stage of development; however, LMP2A is able to bypass the normal developmental requirements and allows BCR− cells to migrate of the bone marrow and colonize the peripheral lymphoid organs. This indicates that LMP2A can provide a survival signal to IgM− cells, as these cells would normally undergo apoptosis in the periphery.
The signal that is provided to LMP2A-transgenic B cells must cross a threshold for the dramatic developmental phenotype to be observed. In TgE-LMP2A mice, a high level of LMP2A expression is observed, whereas another group of LMP2A transgenic mice, designated Tg6 LMP2A mice, express lower levels of LMP2A.4 Intriguingly, Tg6 LMP2A mice do not display the alterations in B-cell development unique to the TgE-LMP2A mice.4 These data were the first indication that there is a signal strength or expression threshold that must be met for the LMP2A survival capacity. Two additional studies have extended this finding.5,6 It is thought that a strong BCR signal leads to the development of B cells belonging to the B-1 subset, whereas an intermediate strength signal drives development of the B-2 subset. B-1 cells express CD5 and are typically found in the peritoneal and pleuropericardial cavities, in contrast to B-2 cells that are CD5− and constitute the B-cell populations in spleen, lymph nodes, and peripheral blood. In TgE-LMP2A mice, the B cells in the bone marrow and spleen have a significant proportion of B cells expressing CD5, indicative of a B-1 phenotype, whereas Tg6 LMP2A mice do not.6 Similar findings have been demonstrated using LMP2A “knockin” transgenic mice.5 Therefore, it can be concluded that a signal of appropriate strength from LMP2A is required for aberrant B-cell development in the bone marrow and spleen of transgenic mice and CD5 expression can be used as a marker for LMP2A signal strength.
B-cell development from early lymphoid progenitors (ELPs) requires the concerted action of B cell–specific transcription factors E2A, early B-cell factor (EBF), and Pax-5.7 E2A and EBF play a key role in the differentiation of pro-B cells, and targeted deletion of either gene results in a complete block in B-cell development before recombination of the heavy-chain locus.7 E2A activates the expression of EBF, and these transcription factors cooperate to regulate B cell–specific gene expression, including λ5, VpreB, mb1, Rag1, Rag2, and Pax-5, genes that are necessary for the pro-B to pre-B cell transition. Pax-5 acts during the pre-B stage of development to regulate the commitment to B-cell differentiation by activating the transcription of B-cell determinants, such as mb1 and CD19.7 Whereas Pax-5 is dispensable for early B-cell differentiation, it is critical for the commitment and maintenance of the B-cell identity.8 Expression of Pax-5 during B-cell commitment causes repression of Notch1 transcription; therefore, B cells express lower levels of Notch1 compared with T cells.7 Notch1 activity in T cells leads to a down-regulation of B cell–specific transcription through the inhibition of E2A and EBF transcriptional activity as well as the degradation of E47, one of the monomers that comprises E2A.7 It is interesting to note that the changes in transcription seen when Notch1 is activated during lymphopoiesis are similar to those seen in the developing B cells of the TgE-LMP2A mice. Similarly, these transcriptional changes are also attributed to the phenotype of the Hodgkin Reed-Sternberg (HRS) cells, the malignant cells of HL. Because of changes in B cell–specific transcription factor expression and lack of B-cell markers, HRS cells are described as having an altered B-cell identity.9
Microarray analysis of B cells in TgE-LMP2A mice demonstrates that, when expressed during development, LMP2A has global effects on transcriptional regulation. Specifically, LMP2A is able to down-regulate the expression of many genes that are required for B-cell development and identity, including E2A, EBF, Pax-5, Ig-β, Ig-α, RAG-1, RAG-2, Vpre-B, λ5, and TdT.10,11 The changes in B-cell development seen in the TgE-LMP2A mice are probably the outcome of such transcriptional alterations. In addition, the decrease in B cell–specific gene transcription with a concomitant increase in genes that play a role in Notch signaling, such as RBP-Jκ, Delta-like1, and Hes-1, indicate that LMP2A may constitutively activate the Notch pathway in vivo. Indeed, we have shown that LMP2A expression leads to the constitutive activation of Notch in B cells and epithelial cells.12 Interestingly, the changes in transcriptional regulation seen in TgE-LMP2A mice are similar to those seen in an HRS cell in HL, linking LMP2A to the development of this malignancy.10 Recently, aberrant activity of Notch1 has been linked to the down-regulation of E2A and EBF in HRS cells.13 Given the effects of Notch1 activity in cell fate decisions during lymphopoiesis, we hypothesized that constitutive activation of Notch1 in the TgE-LMP2A mice may also contribute to the developmental phenotype seen in these mice. Notch1 and LMP2A are both highly expressed in HL, and the identity of the HRS cells is altered compared with a normal germinal center (GC) B cell.9,14,15 To determine whether LMP2A activation of the Notch1 pathway contributes to the alterations in B-cell development in the TgE-LMP2A mice, we have crossed them with Notch1lox/lox mice generated by Freddy Radtke (Lausanne, Switzerland) and CD19+/Cre mice to mediate B cell–specific deletion of the Notch1 locus.16,17 Deletion of Notch1 in developing B cells expressing LMP2A decreases the signal strength of LMP2A indicating that LMP2A uses the Notch1 pathway to alter the development of B cells in the bone marrow of TgE-LMP2A mice. These results imply a role for LMP2A and Notch1 in the altered identity of HRS cells in HL.
Methods
Mice
Notch1lox/lox mice were obtained from Freddy Radtke.17 CD19+/Cre mice, originally generated by Robert Rickert,16 were backcrossed to a C57BL/6 background by Geoff Kansas (Chicago, IL). Notch1lox/lox mice were crossed with TgE and CD19+/Cre mice to generate Notch1lox/lox TgE and Notch1lox/lox CD19+/Cre mice. These mice were intercrossed to generate the genotypes specified. Notch1lox/lox mice were considered wild-type controls. Experiments were performed using mice 3 to 4 weeks old. Mice were maintained in the barrier facility of the Feinberg School of Medicine at Northwestern University in accordance with university animal welfare guidelines.
PCR genotyping of mice
Genomic DNA was isolated from mouse tail snips and analyzed by polymerase chain reaction (PCR). TgE mice were genotyped as described.2 The floxed Notch1 allele was detected as described.17 To detect Cre, CD19, and interleukin-2 (IL-2) (PCR internal control), the following primer sequences were obtained from The Jackson Laboratory (Bar Harbor, ME) and used according to their specified protocols: IMR1084 Cre, IMR1085 Cre, IMR0042 IL-2, IMR0043 IL-2, CD19Cre 1589, CD19Cre 1590.
PCR assay to detect Notch1 gene deletion
DNA was isolated from CD19+ bone marrow and spleen cells. The PCR assay was performed as described except that reactions were performed separately.18
Bone marrow and spleen cell isolation
Total bone marrow was flushed from the tibia and femur from both hind legs of mice using RPMI media. Red blood cells were lysed using 155 mM ammonium chloride solution. Cells were resuspended in 10 mL phosphate-buffered saline (PBS) with 10% fetal bovine serum (FBS) and filtered through nytex filter mesh to eliminate debris. Spleens were dissociated between frosted slides in RPMI and processed as described for bone marrow.
Methylcellulose cultures
A total of 106 cells were plated in 3 mL methylcellulose containing IL-7 (MethoCult M3630; StemCell Technologies, Vancouver, BC) in a 6-well dish and incubated in a humidified incubator at 37°C for 7 days. Colonies were counted using an inverted light microscope and photographed using a Nikon SMZ1000 stereoscope (Nikon Instruments, Melville, NY). To harvest the cells, cultures were washed thoroughly with PBS.
Flow cytometry
A total of 2 × 106 cells were plated in a V-bottom 96-well dish and washed with fluorescence-activated cell sorter (FACS) buffer (PBS plus 1% FBS). Cells were stained with fluorochrome-conjugated antibodies and analyzed using the LSRII flow cytometer and FACS Diva software (BD Biosciences, San Jose, CA). All antibodies were obtained from BD Biosciences.
Survival assay
A total of 100 000 cells/well in complete opti-MEM media were plated in a 48-well dish in high and low IL-7 as described.19 Survival was measured at 48 hours by staining with annexin V according to product instructions and analyzing by flow cytometry.
Real-time RT-PCR
Total RNA from CD19+ selected cells was isolated using the QIAGEN RNeasy Kit with on-column DNase digestion (QIAGEN, Valencia, CA). RNA was reverse-transcribed using the Superarray Bioscience RT2 First Strand Kit according to kit instructions. The RT2 qPCR Primer Assay-SYBR Green Kit was used with primers specific for murine EBF, E2A, and GAPDH (Superarray Bioscience, Frederick, MD). PCR was carried out using the Bio-Rad iQ5 (Bio-Rad, Hercules, CA) according to kit instructions. Cycle threshold (Ct) values were normalized to GAPDH levels. Fold change was calculated using the ΔΔCt method.
Results
To determine the role of Notch1 signaling for LMP2A function in vivo, we took advantage of Notch1lox/lox mice generated previously.17 To delete Notch1 only in B cells, we used CD19+/Cre mice that express Cre recombinase under the control of the CD19 promoter to induce efficient knockdown in B cells.16 Intercrossing these 2 lines with our TgE-LMP2A–transgenic mice yielded the following genotypes: Notch1lox/lox (wild-type), Notch1lox/loxCD19+/Cre, TgE Notch1lox/lox, and TgE Notch1lox/loxCD19+/Cre. None of the mice displayed any gross abnormalities and the development of T cells was unaffected, as expected (data not shown).20 To detect Notch1 gene deletion, DNA was isolated from CD19+ selected B cells and analyzed by PCR. Only mice expressing Cre recombinase have a deleted Notch1 gene (data not shown).18 Protein lysates generated from B cells isolated from bone marrow and spleen for each genotype were analyzed for LMP2A expression by Western blot. Mice possessing the LMP2A TgE transgene express LMP2A from the Ig heavy-chain promoter and enhancer; as a result, the absence of Notch1 does not affect the expression of LMP2A (data not shown).
TgE-LMP2A mice have decreased numbers of B cells in the spleen, and the majority of the B cells are IgM−.2 Whereas approximately half of the spleen of a Notch1lox/lox (wild-type) and a Notch1lox/loxCD19+/Cre mouse is composed of B220+ B cells, this is reduced to approximately 20% in TgE Notch1lox/lox and TgE Notch1lox/loxCD19+/Cre mice (Figure 1B). Interestingly, however, deletion of Notch1 in the TgE mice results in a statistically significant increase in the percentage of IgM+ cells in the spleen (Figure 1C, compare 12% B220+IgM+ cells in TgE Notch1lox/lox to 21% in TgE Notch1lox/loxCD19+/Cre). When expressed as the total number of spleen cells that are IgM+, there is a statistically significant 2-fold increase in IgM+ cells in the spleens of TgE Notch1lox/loxCD19+/Cre mice compared with TgE Notch1lox/lox mice (4.2% vs 2%, respectively).
Loss of Notch1 expression in TgE mice results in increased IgM+ B cells in the spleen. (A) Total cells isolated from the spleen of each indicated genotype were stained with B220 and IgM fluorochrome-conjugated antibodies and analyzed by flow cytometry. Live cell populations, designated by forward and side scatter properties, were plotted with IgM and B220, and a representative plot is shown for each genotype. CD19-Cre–mediated deletion has been demonstrated previously to have an efficiency of approximately 80% in the bone marrow and 90% to 95% in the spleen.16 (B) The average percent of B cells based on B220 staining for each genotype is shown plus or minus SE. (C) The average percentage of B cells that have IgM on the cell surface (%B220) as well as the percentage of total spleen cells that are IgM+ (% total) for each genotype is shown plus or minus SE. The percentage of B220 cells expressing IgM (%B220) is calculated to account for differences in total B cells between the genotypes. Results from 4 independent experiments are shown. *P = .04, **P = .05 by t test. Notch1lox/lox, n = 5 mice; Notch1lox/lox CD19+/Cre, n = 4; TgE Notch1lox/lox, n = 7; TgE Notch1lox/lox CD19+/Cre, n = 8.
Loss of Notch1 expression in TgE mice results in increased IgM+ B cells in the spleen. (A) Total cells isolated from the spleen of each indicated genotype were stained with B220 and IgM fluorochrome-conjugated antibodies and analyzed by flow cytometry. Live cell populations, designated by forward and side scatter properties, were plotted with IgM and B220, and a representative plot is shown for each genotype. CD19-Cre–mediated deletion has been demonstrated previously to have an efficiency of approximately 80% in the bone marrow and 90% to 95% in the spleen.16 (B) The average percent of B cells based on B220 staining for each genotype is shown plus or minus SE. (C) The average percentage of B cells that have IgM on the cell surface (%B220) as well as the percentage of total spleen cells that are IgM+ (% total) for each genotype is shown plus or minus SE. The percentage of B220 cells expressing IgM (%B220) is calculated to account for differences in total B cells between the genotypes. Results from 4 independent experiments are shown. *P = .04, **P = .05 by t test. Notch1lox/lox, n = 5 mice; Notch1lox/lox CD19+/Cre, n = 4; TgE Notch1lox/lox, n = 7; TgE Notch1lox/lox CD19+/Cre, n = 8.
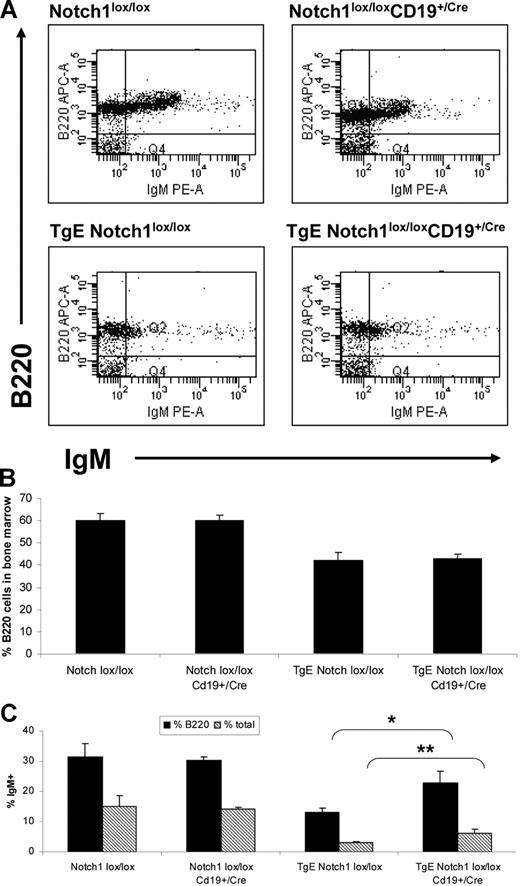
Similar to the spleen, deletion of Notch1 does not affect total B-cell numbers in the bone marrow of TgE Notch1lox/lox mice (Figure 2A,B) but does result in a statistically significant increase in the expression of IgM (Figure 2C, compare 13% B220+IgM+ in TgE Notch1lox/lox to 22% in TgE Notch1lox/lox CD19+/Cre). Interestingly, the increase of IgM+ cells in the TgE Notch1lox/lox CD19+/Cre mice makes this population nearly comparable with the B220+IgM+ B-cell population in the Notch1lox/lox mouse (31%). The increase of IgM+ cells in TgE Notch1lox/lox CD19+/Cre compared with TgE Notch1lox/lox mice is also expressed as a percentage of total bone marrow in Figure 2C, 6% versus 2.9%, respectively.
Loss of Notch1 expression in TgE mice results in increased IgM+ B cells in the bone marrow. Total cells isolated from the bone marrow of each indicated genotype were stained with B220 and IgM-conjugated antibodies and analyzed by flow cytometry. Live cell populations, designated by forward and side scatter properties, were plotted with IgM and B220, and a representative plot for each genotype is shown (A). (B) The average percentage of B220+ cells in the bone marrow for each genotype is indicated plus or minus SE. (C) The average percentage of B cells expressing IgM in the bone marrow from 4 independent experiments is shown. Data are represented as the percentage of B220+ cells that express IgM (%B220) as well as the percentage of total bone marrow that is B220+IgM+ (percentage of total). *P = .04, **P = .03 by t test. Notch1lox/lox, n = 4 mice; Notch1lox/lox CD19+/Cre, n = 4; TgE Notch1lox/lox, n = 4; TgE Notch1lox/lox CD19+/Cre, n = 6.
Loss of Notch1 expression in TgE mice results in increased IgM+ B cells in the bone marrow. Total cells isolated from the bone marrow of each indicated genotype were stained with B220 and IgM-conjugated antibodies and analyzed by flow cytometry. Live cell populations, designated by forward and side scatter properties, were plotted with IgM and B220, and a representative plot for each genotype is shown (A). (B) The average percentage of B220+ cells in the bone marrow for each genotype is indicated plus or minus SE. (C) The average percentage of B cells expressing IgM in the bone marrow from 4 independent experiments is shown. Data are represented as the percentage of B220+ cells that express IgM (%B220) as well as the percentage of total bone marrow that is B220+IgM+ (percentage of total). *P = .04, **P = .03 by t test. Notch1lox/lox, n = 4 mice; Notch1lox/lox CD19+/Cre, n = 4; TgE Notch1lox/lox, n = 4; TgE Notch1lox/lox CD19+/Cre, n = 6.
The signal strength of LMP2A is important for the development of CD5+ B-1 cells, as transgenic mice expressing lower levels of LMP2A do not develop B-1 cells in the bone marrow.5,6 Therefore, we wanted to determine whether the signal strength of LMP2A would be affected by a lack of Notch1 expression. In the bone marrow of TgE Notch1lox/lox mice, approximately 46% of the B220+ B cells express CD5, indicating their B-1 identity (Figure 3). In TgE Notch1lox/loxCD19+/Cre mice, this percentage is decreased to 35%, suggesting that the strength of the LMP2A signal is partially dependent upon Notch1. Notch1lox/lox and Notch1lox/loxCD19+/Cre mice did not develop B-1 cells in the bone marrow, as expected.6
The signal strength of LMP2A is decreased in the bone marrow of TgE Notch1lox/loxCD19+/Cre mice. Total cells isolated from the bone marrow of each indicated genotype were stained with B220 and CD5 fluorochrome-conjugated antibodies and analyzed by flow cytometry. Live cell populations, designated by forward and side scatter properties, were gated to examine CD5 expression and a representative plot is shown for each genotype (A). (B) The average results from 4 independent experiments are shown plus or minus SE. Populations are represented as the percentage of total bone marrow that is CD5 positive (percentage of total) as well as the percentage of B cells that are CD5 positive (%B220). *P = .03, **P = .02 by t test. Notch1lox/lox, n = 5 mice; Notch1lox/lox CD19+/Cre, n = 5; TgE Notch1lox/lox, n = 7; TgE Notch1lox/lox CD19+/Cre, n = 9.
The signal strength of LMP2A is decreased in the bone marrow of TgE Notch1lox/loxCD19+/Cre mice. Total cells isolated from the bone marrow of each indicated genotype were stained with B220 and CD5 fluorochrome-conjugated antibodies and analyzed by flow cytometry. Live cell populations, designated by forward and side scatter properties, were gated to examine CD5 expression and a representative plot is shown for each genotype (A). (B) The average results from 4 independent experiments are shown plus or minus SE. Populations are represented as the percentage of total bone marrow that is CD5 positive (percentage of total) as well as the percentage of B cells that are CD5 positive (%B220). *P = .03, **P = .02 by t test. Notch1lox/lox, n = 5 mice; Notch1lox/lox CD19+/Cre, n = 5; TgE Notch1lox/lox, n = 7; TgE Notch1lox/lox CD19+/Cre, n = 9.
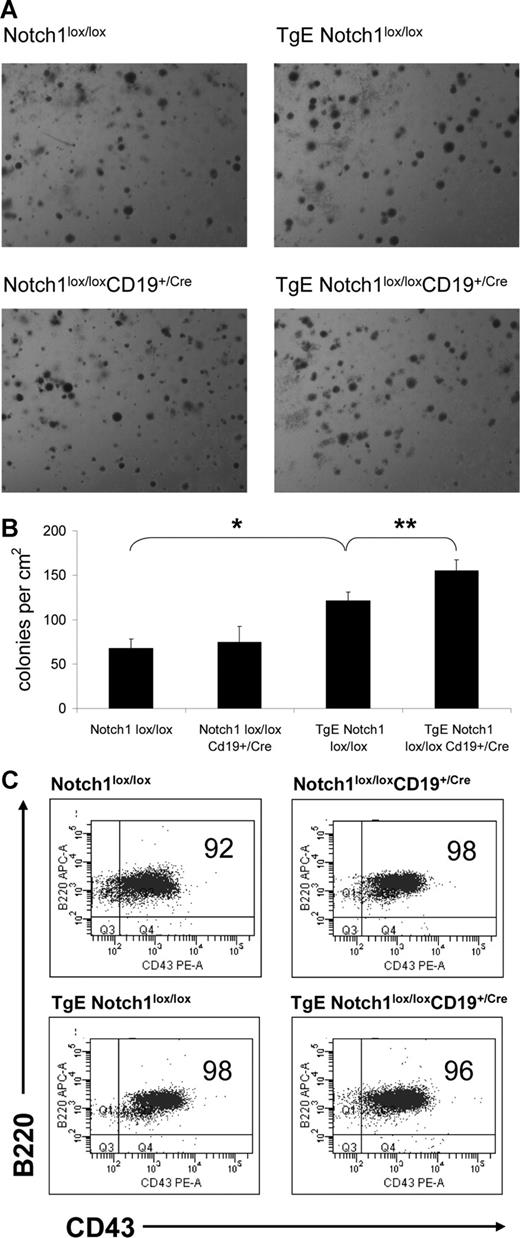
In the bone marrow, the transition from CD43+ pre-B1 stage to CD43− pre-BII stage of development is mediated by the expression and activity of the pre-BCR. In the absence of a pre-BCR, CD43+ B cells accumulate in the bone marrow and are unable to transition to a CD43− stage, as seen in a Rag−/− mouse.21,22 Expression of LMP2A allows B cells to bypass the developmental checkpoints and progress to the CD43− pre-BII stage, albeit with less efficiency than a wild-type mouse. The expression of CD43 was analyzed on B cells from the bone marrow to determine whether Notch1 was required for LMP2A to provide a surrogate pre-BCR signal and mediate progression from the CD43+ to CD43− pre-B cell stages (Figure 4A). There is an increase in the number of CD43+ B cells in TgE Notch1lox/lox mice compared with Notch1lox/lox mice (compare 40% to 63%, Figure 4B), a phenotype that has been described previously.4 In TgE Notch1lox/loxCD19+/Cre mice where Notch1 is deleted, there is a greater accumulation of CD43+ B cells and 72% of B cells are CD43+ (Figure 4A,B). Approximately 23% of the total bone marrow cells in a TgE Notch1lox/lox mouse express CD43, compared with 30% in the TgE Notch1lox/loxCD19+/Cre mice, an increase that is statistically significant. The accumulation of CD43+ B cells in the bone marrow of TgE Notch1lox/loxCD19+/Cre mice suggests that LMP2A depends on the expression of Notch1 to alter B-cell development and indicates further that Notch1 is an important component for the signal strength of LMP2A.
CD43+ B cells accumulate in the bone marrow of TgE Notch1lox/lox CD19+/Cre mice. Total cells isolated from the bone marrow of each indicated genotype were stained with B220 and CD43 fluorochrome-conjugated antibodies and analyzed by flow cytometry. Live cell populations, designated by forward and side scatter properties, were gated and B220 and CD43 expression was examined. A representative dot plot for each genotype is shown (A). (B) The average percentage of B cells in the bone marrow that express CD43 is shown plus or minus SE. Populations are represented as the percentage of total bone marrow that is CD43+ (% total) as well as the percentage of B cells that are CD43+ (%B220). *P = .05, **P = .04 by t test. Notch1lox/lox, n = 5 mice; Notch1lox/lox CD19+/Cre, n = 5; TgE Notch1lox/lox, n = 8; TgE Notch1lox/lox CD19+/Cre, n = 11.
CD43+ B cells accumulate in the bone marrow of TgE Notch1lox/lox CD19+/Cre mice. Total cells isolated from the bone marrow of each indicated genotype were stained with B220 and CD43 fluorochrome-conjugated antibodies and analyzed by flow cytometry. Live cell populations, designated by forward and side scatter properties, were gated and B220 and CD43 expression was examined. A representative dot plot for each genotype is shown (A). (B) The average percentage of B cells in the bone marrow that express CD43 is shown plus or minus SE. Populations are represented as the percentage of total bone marrow that is CD43+ (% total) as well as the percentage of B cells that are CD43+ (%B220). *P = .05, **P = .04 by t test. Notch1lox/lox, n = 5 mice; Notch1lox/lox CD19+/Cre, n = 5; TgE Notch1lox/lox, n = 8; TgE Notch1lox/lox CD19+/Cre, n = 11.
Bone marrow B cells from TgE-LMP2A mice are capable of growth in methylcellulose medium containing IL-7 and typically form twice the number of colonies than for wild-type mice.23 The increase in colony formation is probably because of the increase in CD43+ cells in the bone marrow of TgE mice, as these cells are the most responsive to growth in IL-7.24 Because TgE Notch1lox/loxCD19+/Cre mice displayed a statistically significant increase in the accumulation of CD43+ cells in the bone marrow compared with TgE Notch1lox/lox mice, we sought to determine whether the bone marrow B cells in these mice have an altered ability to form colonies in methylcellulose. Figure 5A shows representative images of the colonies formed in methylcellulose containing IL-7 after 7 days. Notch1lox/lox and Notch1lox/loxCD19+/Cre bone marrow cells form similar numbers of colonies with an average of 68 plus or minus 10 and 74 plus or minus 18, respectively (Figure 5B). As expected, almost twice as many colonies formed from the bone marrow of TgE Notch1lox/lox mice (121 ± 10). Bone marrow from TgE Notch1lox/loxCD19+/Cre mice have an average colony count of 155 plus or minus 12, which is a statistically significant increase compared with TgE Notch1lox/lox mice. The cells recovered from the methylcellulose cultures were analyzed by flow cytometry. Figure 5C is a representative dot plot for each genotype, which shows that the majority of cells are CD43+ B cells (B220+), regardless of the genotype.
Bone marrow from TgE Notch1lox/lox CD19+/Cre mice forms more colonies in methylcellulose than TgE Notch1lox/lox mice. Bone marrow cells were plated in methylcellulose with IL-7. After 7 days, the numbers of colonies in 1 cm2 were counted in triplicate wells for each genotype. (A) Representative images of colonies formed from bone marrow cells for each genotype. Methylcellulose cultures were viewed with a Nikon SMZ1000 stereomicroscope with an HR Plan Apo 1× WD54 objective (Nikon Instruments, Melville, NY). Images were acquired using a Polaroid Digital Camera model PDM02 and DMC le version V1.25 software (Polaroid, Concord MA) and processed using Adobe Photoshop version 7.0 software (Adobe Systems, San Jose, CA). (B) The number of colonies in 1 cm2 of the culture dish was counted in triplicate for each genotype and the average number of colonies for 3 independent experiments is shown plus or minus SE. *P = .001, **P = .03 by t test. (C) Cells from methylcellulose cultures were stained with B220 and CD43 antibodies and analyzed by flow cytometry. The live cell population, designated by forward and side scatter properties, was gated to examine expression of B220 and CD43. A representative plot for each genotype is shown with percentages for each population. Typically, the majority of cells are B220+CD43+ B cells.
Bone marrow from TgE Notch1lox/lox CD19+/Cre mice forms more colonies in methylcellulose than TgE Notch1lox/lox mice. Bone marrow cells were plated in methylcellulose with IL-7. After 7 days, the numbers of colonies in 1 cm2 were counted in triplicate wells for each genotype. (A) Representative images of colonies formed from bone marrow cells for each genotype. Methylcellulose cultures were viewed with a Nikon SMZ1000 stereomicroscope with an HR Plan Apo 1× WD54 objective (Nikon Instruments, Melville, NY). Images were acquired using a Polaroid Digital Camera model PDM02 and DMC le version V1.25 software (Polaroid, Concord MA) and processed using Adobe Photoshop version 7.0 software (Adobe Systems, San Jose, CA). (B) The number of colonies in 1 cm2 of the culture dish was counted in triplicate for each genotype and the average number of colonies for 3 independent experiments is shown plus or minus SE. *P = .001, **P = .03 by t test. (C) Cells from methylcellulose cultures were stained with B220 and CD43 antibodies and analyzed by flow cytometry. The live cell population, designated by forward and side scatter properties, was gated to examine expression of B220 and CD43. A representative plot for each genotype is shown with percentages for each population. Typically, the majority of cells are B220+CD43+ B cells.
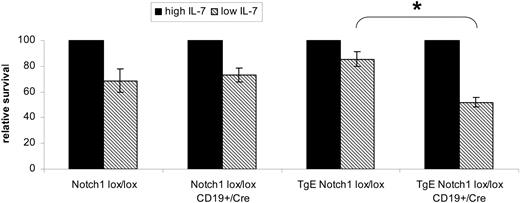
We have shown that bone marrow B cells from TgE-LMP2A mice survive in low concentrations of IL-7 similar to wild-type cells, despite the lack of pre-BCR expression for TgE-LMP2A cells.19 These data suggest that LMP2A imparts a surrogate pre-BCR signal in B cells that allows for survival in low concentrations of IL-7. Thus far, the data suggest that Notch1 signaling is important for LMP2A-mediated signaling. Therefore, we examined whether deletion of Notch1 would affect the surrogate pre-BCR signal provided by LMP2A. B cells recovered from methylcellulose cultures were analyzed for their ability to survive in response to high and low concentrations of IL-7 (Figure 6). After 48 hours, 70% to 85% of cells remained alive in the cultures grown with low IL-7 for Notch1lox/lox, Notch1lox/loxCD19+/Cre, and TgE Notch1lox/lox mice. In contrast, only 50% of TgE Notch1lox/loxCD19+/Cre cells were alive after 48 hours in low IL-7. This result is similar to what is seen when Rag−/− B cells are placed in low IL-7 for 48 hours and suggests that the deletion of Notch1 decreases the ability for LMP2A to provide a surrogate pre-BCR signal for survival.19
Bone marrow B cells TgE Notchlox/lox CD19+/Cre mice have a decreased survival capacity in low IL-7. Equal numbers of B220+ cells harvested from methylcellulose cultures were plated in either high IL-7 or low IL-7. At 48 hours, cells were analyzed by flow cytometry to determine the cell recovery. The percentage of cells in the live cell gate, based on forward and side scatter properties, was recorded and graphed as relative survival compared with high IL-7 for each genotype. We have demonstrated previously that relative survival inversely correlates with apoptosis as measured by annexin V binding.19 Results are the average of 3 independent experiments after 48 hours plus or minus SE. *P = .002 by t test. Notch1lox/lox, n = 4 mice; Notch1lox/lox CD19+/Cre, n = 3; TgE Notch1lox/lox, n = 5; TgE Notch1lox/lox CD19+/Cre, n = 6.
Bone marrow B cells TgE Notchlox/lox CD19+/Cre mice have a decreased survival capacity in low IL-7. Equal numbers of B220+ cells harvested from methylcellulose cultures were plated in either high IL-7 or low IL-7. At 48 hours, cells were analyzed by flow cytometry to determine the cell recovery. The percentage of cells in the live cell gate, based on forward and side scatter properties, was recorded and graphed as relative survival compared with high IL-7 for each genotype. We have demonstrated previously that relative survival inversely correlates with apoptosis as measured by annexin V binding.19 Results are the average of 3 independent experiments after 48 hours plus or minus SE. *P = .002 by t test. Notch1lox/lox, n = 4 mice; Notch1lox/lox CD19+/Cre, n = 3; TgE Notch1lox/lox, n = 5; TgE Notch1lox/lox CD19+/Cre, n = 6.
LMP2A expression during B-cell development leads to a down-regulation of B cell–specific gene transcription.10 Notch1 activity during lymphopoiesis similarly affects B cell–specific gene transcription, and recent data demonstrate that aberrant Notch1 activity in HRS cells leads to a decrease in E2A and EBF mRNA.13,25,26 Therefore, we wanted to determine whether the levels of E2A and EBF in TgE Notch1lox/loxCD19+/Cre mice would revert to wild-type levels. As shown in Figure 7, quantitative real-time RT-PCR demonstrates that, in TgE Notch1lox/loxCD19+/Cre mice, E2A and EBF levels are increased compared with TgE Notch1lox/lox mice. Changes in mRNA levels are specific to mice expressing the TgE transgene, as there is no difference between Notch1lox/lox and Notch1lox/loxCD19+/Cre mice. These results indicate that Notch1 plays a role in the developmental aberrations seen in the TgE Notch1lox/lox mice. That TgE Notch1lox/loxCD19+/Cre B cells have increased IgM surface expression compared with TgE Notch1lox/lox B cells is probably an outcome of LMP2A-mediated repression of E2A and EBF transcription being relieved in the absence of Notch1.
Notch1 expression is required for LMP2A to alter mRNA levels of B cell–specific transcription factors in TgE mice. Real-time RT-PCR gene expression analysis was performed using total RNA isolated from B220+ bone marrow B cells harvested from methylcellulose cultures of each genotype. Primers specific for E2A and EBF were used along with GAPDH as a control for RNA levels. Fold change in gene expression was calculated using the ΔΔCt method, and fold change is shown relative to wild-type (Notch1lox/lox) mice. Results are the average of at least 3 independent experiments plus or minus SE. Notch1lox/lox, n = 6 mice; Notch1lox/lox CD19+/Cre, n = 4; TgE Notch1lox/lox, n = 3; TgE Notch1lox/lox CD19+/Cre, n = 6.
Notch1 expression is required for LMP2A to alter mRNA levels of B cell–specific transcription factors in TgE mice. Real-time RT-PCR gene expression analysis was performed using total RNA isolated from B220+ bone marrow B cells harvested from methylcellulose cultures of each genotype. Primers specific for E2A and EBF were used along with GAPDH as a control for RNA levels. Fold change in gene expression was calculated using the ΔΔCt method, and fold change is shown relative to wild-type (Notch1lox/lox) mice. Results are the average of at least 3 independent experiments plus or minus SE. Notch1lox/lox, n = 6 mice; Notch1lox/lox CD19+/Cre, n = 4; TgE Notch1lox/lox, n = 3; TgE Notch1lox/lox CD19+/Cre, n = 6.
Discussion
Microarray analysis demonstrated that LMP2A induces global alterations in gene transcription that are similar to those seen in HRS cells of HL, cells that have lost their B-cell identity.9,10 Notably, it appeared that LMP2A may constitutively activate the Notch pathway in vivo, a finding that has been confirmed in B cells and epithelial cells expressing LMP2A.12 Because of the critical role that Notch signaling plays during B/T lineage specification, we sought to explore the possibility that LMP2A-mediated activation of Notch signaling in vivo may be responsible for the developmental changes seen in the B cells of TgE-LMP2A mice. By crossing our TgE-LMP2A mice with Notch1lox/loxCD19+/Cre mice, we were able to demonstrate that LMP2A uses Notch1 signaling in vivo. Abrogation of Notch1 expression in B cells decreases the strength of the LMP2A signal, resulting in a decreased capacity to alter B-cell development. These results indicate that Notch1 may cooperate with LMP2A to provide developmental and survival signals to LMP2A-expressing B cells.
In lymphocytes, Notch1 activity is usually associated with the development of T cells during lymphopoiesis as well as mature T-lineage specification, αβ vs γδ.27 Studies of inducible knockout of either Notch1 or RBP-Jκ does not affect the development of B cells, whereas T-cell development is severely impaired.17,28 Therefore, as expected, the deletion of Notch1 in B cells did not affect T-cell or B-cell development in the Notch1lox/lox or Notch1lox/loxCD19+/Cre mice. Although Notch1 does not appear to be important for proper B-cell development, both Notch1 and Notch2 have been demonstrated to be expressed in B220+ murine bone marrow cells.29 In addition, the presence of Hes-1 transcript in early pre-B cells as well as expression of the Notch ligand Jagged1 on bone marrow stromal cells strongly suggest that Notch signaling occurs in both human and mouse B cells.29-31 We have observed that, in LMP2A-expressing B cells, Notch1 is important for LMP2A function. The result of deleting the Notch1 gene in TgE mice presents a novel mechanism whereby LMP2A exploits a signaling pathway not normally attributed to a B cell.
In studies where mice were reconstituted with bone marrow transduced with constitutively active Notch1, it has been demonstrated that T cells develop in the bone marrow at the expense of B-cell development.32 This in conjunction with our observations in TgE mice raises an important question. If LMP2A constitutively activates Notch1 in the bone marrow of mice, why is B-cell development able to proceed at all? Although the precise mechanism of action is unknown, signal strength through Notch1 can have dramatic effects on the lineage commitment of an ELP. The function of Notch1 is known to be affected by a 2-fold change in expression, as Notch1+/− hematopoietic stem cells are unable to efficiently contribute to thymocyte development.33 Experiments using immobilized Delta1 ligand demonstrated that stem cells exposed to high concentrations of ligand differentiated into T cells, whereas cells exposed to low concentrations committed to B-cell differentiation.34 Complimenting these results, experiments using CD34+ progenitor cells seeded in low and high concentrations of the γ-secretase inhibitor DAPT (N-[N-3,5-difluorophenacetyl-l-alanyl]-S-phenylglycine t-butyl ester) differentiated into T cells and B cells, respectively, with intermediate concentrations of DAPT yielded a less efficient skewing toward B-cell differentiation.35 Altogether, these results indicate that a high strength of Notch1 signaling is required for T-lineage commitment. This dose-response or strength of signal phenomenon for Notch1 is the most plausible explanation as to why T cells are not developing in the bone marrow of TgE-LMP2A mice. The altered B-cell identity seen in TgE-LMP2A mice is probably the result of a weak or intermediate strength of Notch1 activity that is sufficient to induce alterations in transcriptional regulation and B-cell development but not strong enough to reach the threshold of Notch1 activity necessary for T-cell development.
There is no difference in B-1 B-cell development in Notch1lox/loxCD19+/Cre mice compared with Notch1lox/lox wild-type mice, indicating that the changes seen in bone marrow of TgE Notch1lox/lox and TgE Notch1lox/loxCD19+/Cre mice are specifically the result of the expression of LMP2A. CD5 expression on B cells is an indicator of signal strength, either through the BCR or through the surrogate BCR signal provided by LMP2A. CD5+ B-1 cells do not develop in the bone marrow of mice expressing low levels of LMP2A or an LMP2A-signaling mutant.5,6 The observation that TgE Notch1lox/loxCD19+/Cre mice have decreased CD5 expression in the bone marrow compared with TgE Notch1lox/lox mice suggests that, in the absence of Notch1, the signal strength of LMP2A is diminished.
The efficient transition through the pro-B to pre-B checkpoint is known to largely be controlled by signals delivered through the pre-BCR, indicating that lack of CD45 results in a weak pre-BCR signal.36 We have shown that LMP2A provides a surrogate pre-BCR signal to developing B cells that allows cells to proliferate and survive in low concentrations of IL-7 as well as to transition through the CD43+ to CD43− checkpoint, albeit less efficiently than a wild-type pre-BCR signal.4,19 Our analysis suggests that, in the absence of Notch1, the pre-BCR signal provided by LMP2A is weakened and CD43+ cells accumulate because of an inefficient transition though the CD43+ to CD43− checkpoint. The further accumulation of CD43+ pro-B cells in TgE Notch1lox/loxCD19+/Cre mice provides evidence that Notch1 signaling is an important component for the strength of the LMP2A signal. Additional support was provided by analyzing the survival capacity of bone marrow B cells from each genotype. TgE Notch1lox/lox mice display a survival capacity in low IL-7 that is similar to Notch1lox/lox wild-type mice, again demonstrating that LMP2A can provide a surrogate pre-BCR signal to bone marrow B cells. In contrast, bone marrow B cells from TgE Notch1lox/loxCD19+/Cre mice are unable to survive in low concentrations of IL-7. In the same conditions, approximately 60% of Rag-deficient bone marrow B cells undergo apoptosis after 48 hours.19 Only half of the cells from TgE Notch1lox/loxCD19+/Cre survive after 48 hours, indicating that, in the absence of Notch1, LMP2A-expressing bone marrow B cells revert to a survival phenotype that resembles pre-BCR− B cells. B cells from Notch1lox/loxCD19+/Cre mice survive as well as wild-type, indicating that these results are not the result of a lack of Notch1 expression in the B cells but are specific to cells expressing LMP2A.
HRS cells are thought to arise from GC B cells that, despite acquiring somatic mutations that result in a crippled Ig, have been rescued from apoptosis.9,37 Between 30% and 50% of Hodgkin lymphomas are EBV-associated, and it has been shown recently that in vitro infection of primary GC cells with EBV can rescue those with mutated Ig genes from undergoing apoptosis.38,39 The 2 primary candidates for mediating rescue from apoptosis during the GC reaction are LMP1 and LMP2A, as these viral proteins provide a surrogate CD40 and BCR signal, respectively.2,40 Most recently, it has been demonstrated that LMP2A is required for EBV to rescue Iglow and Ig− cells from apoptosis, suggesting that the strong LMP2A signal is required to maintain the survival of cells with weak or no basal survival signal from the BCR.41 These results underline the importance of signal strength from the BCR for B-cell survival, and the in vivo data provided by the TgE Notch1lox/loxCD19+/Cre mice may extend to this concept. We have shown that the strength of the LMP2A signal is partially dependent on Notch1 expression. Deletion of the Notch1 gene in TgE-LMP2A mice diminishes the signal strength of LMP2A, as evidenced by a decrease in CD5 along with an increase in CD43 and IgM expression and E2A and EBF transcription in the bone marrow of TgE Notch1lox/loxCD19+/Cre mice. Because a weak BCR signal is unable to maintain B-cell survival, it is conceivable that a weakened LMP2A signal may be insufficient to maintain B-cell survival. Therefore, in the absence of Notch1, the strength of the LMP2A signal may not be sufficient to rescue B cells with crippling Ig mutations and would not contribute to EBV-associated oncogenesis.
The hallmark of classic HL is the lack of BCR expression and an overall loss of B-cell identity.42 Until studies identified clonally rearranged Ig genes via PCR techniques indicating that HRS cells are indeed B cells, the origin of these cells had remained elusive.43 Whereas several mechanisms that may be responsible for the lack of a functional BCR in these cells exist, the most intriguing possibility involves down-regulation of B cell–specific transcription factors that normally regulate the commitment to the B-cell lineage, such as E2A, EBF, Pax-5, PU.1, mb-1, and CD19, because these genes are similarly found to be down-regulated in the B cells of TgE-LMP2A mice.10,15,44-47 Interestingly, Notch1 is found to be highly expressed in HRS cells, with increased expression of Notch ligands such as Jagged1 found in the nonmalignant cells surrounding HRS cells.48 Activation of Notch1 in HRS cells by adjacent cells could account for the changes in B cell–specific transcription factors, as during lymphopoiesis Notch1 activity is responsible for down-regulating the B-cell lineage to allow ELPs to progress to a T-cell fate.27 This idea is supported by a recent manuscript demonstrating that Notch1 activity in HRS cell lines is responsible for decreased transcription of E2A and EBF.13 Alternatively, we have demonstrated that LMP2A can constitutively activate the Notch pathway.12 We now show Notch1 is important for LMP2A to alter the identity of the B cells in transgenic mice. Although the effects of LMP2A on B cell–specific transcription seem modest, there is considerable evidence that small changes in gene transcription can have profound effects on lymphocyte development.13,49 B cells from TgE Notch1lox/loxCD19+/Cre mice do not exhibit a dramatic reduction in B cell–specific transcription factor expression, indicating that Notch1 expression is necessary for LMP2A to alter the B-cell identity. Despite the limitations in interpreting data from a mouse model of B-cell development in the context of Hodgkin lymphoma pathogenesis, these studies provide a clear link between LMP2A, Notch1, and HL. It will be interesting to explore this link further when human primary B cells amenable to such analysis become available. Because LMP2A is highly expressed in EBV+ HRS cells and can activate the Notch pathway, we propose a mechanism whereby LMP2A-mediated activation of Notch1 leads to changes in transcriptional regulation that result in loss of B-cell identity, all the while providing a survival signal to Ig− HRS cells.50
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Freddy Radtke for providing us with Notch1lox/lox mice and Geoff Kansas for providing CD19+/Cre mice.
R.L. is a Stohlman Scholar of the Leukemia & Lymphoma Society of America and was supported by the National Cancer Institute (Public Health Service grants CA62234 and CA73507) and the National Institute of Allergy and Infectious Disease (grant AI067048). L.J.A. was supported by the Training Program in Viral Replication (T32 AI060523), National Institute of Allergy and Infectious Disease.
National Institutes of Health
Authorship
Contribution: L.J.A. designed the research, performed the experiments, analyzed the resulting data, and wrote the manuscript; and R.L. aided in the experimental design and interpretation of the resulting data and helped write the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard Longnecker, Department of Microbiology and Immunology, Northwestern University, Ward 6-260, 303 E Chicago Avenue, Chicago, IL 60611; e-mail: r-longnecker@northwestern.edu.