Abstract
Peripheral blood monocytes are plastic cells that migrate to tissues and differentiate into various cell types, including macrophages, dendritic cells, and osteoclasts. We have described the migration of cellular inhibitor of apoptosis protein 1 (cIAP1), a member of the IAP family of proteins, from the nucleus to the Golgi apparatus in monocytes undergoing differentiation into macrophages. Here we show that, once in the cytoplasm, cIAP1 is involved in the degradation of the adaptor protein tumor necrosis factor receptor–associated factor 2 (TRAF2) by the proteosomal machinery. Inhibition of cIAP1 prevents the decrease in TRAF2 expression that characterizes macrophage formation. We demonstrate that TRAF2 is initially required for macrophage differentiation as its silencing prevents Iκ-Bα degradation, nuclear factor-κB (NF-κB) p65 nuclear translocation, and the differentiation process. Then, we show that cIAP1-mediated degradation of TRAF2 allows the differentiation process to progress. This degradation is required for the macrophages to be fully functional as TRAF2 overexpression in differentiated cells decreases the c-Jun N-terminal kinase–mediated synthesis and the secretion of proinflammatory cytokines, such as interleukin-8 and monocyte chemoattractant protein 1 (MCP-1) in response to CD40 ligand. We conclude that TRAF2 expression and subsequent degradation are required for the differentiation of monocytes into fully functional macrophages.
Introduction
Tumor necrosis factor receptor (TNFR)–associated factors (TRAFs) form an evolutionarily conserved family of intracellular adaptors that bind directly or indirectly to members of the TNFR and the interleukin-1 (IL-1)/Toll-like receptor (TLR) families.1,2 They participate in the transduction of signals from these receptors to downstream events that regulate cell proliferation, differentiation, and death
The member of this family known as TRAF2 directly binds CD27, CD30, CD40, CD137, TNFR2, and receptor activator of nuclear factor-κB (RANK). TRAF2 can also bind TNFR1 indirectly, through interaction with TNFR-associated death domain protein.3
On receptor engagement, TRAF2 is recruited in a receptor-associated multiprotein complex4-6 where it contributes to stimulate specific downstream signaling pathways. Depending on cell type, differentiation stage, and stimulated receptors, these signaling pathways can involve c-jun N-terminal kinase (JNK), nuclear factor κB (NF-κB), and p38 mitogen-activated protein kinase (p38MAPK).4,5,7-9 TRAF2 is also a key regulator of TNFR1-mediated apoptosis.10-13 TRAF2 activity is regulated by its interaction with protein partners, such as TRAF1,14-16 subcellular localization,7,8,15 ubiquitylation, and degradation by the proteasome pathway.8,12,13,17-19
A yeast 2-hybrid screen of proteins able to bind TRAF2 identified a direct interaction with cIAP1 (cellular inhibitor of apoptosis protein 1, also named BIRC2, HIAP2), a member of the IAP family of proteins.20,21 Thanks to the presence of a C-terminal zinc finger domain (RING domain) that displays an E3-ubiquitin ligase activity, cIAP1 was demonstrated to promote TRAF2 ubiquitylation and to target the protein for proteasome-mediated degradation.12,13,22-24
We have previously shown that cIAP1 was required for macrophage differentiation.25 We have also shown that cIAP1 migrated from the nucleus to the cytoplasm to concentrate at the surface of the Golgi apparatus in monocytes undergoing differentiation into macrophages.26 However, the role of cIAP1 and the functional significance of its differentiation-associated redistribution remained unknown. Here we show that TRAF2 is initially required for the differentiation of monocytes into macrophages. Then, cIAP1 triggers its proteosomal degradation, which appears to be required for the normal outcome of the differentiation process. cIAP1 also maintains a low level of TRAF2 in differentiated macrophages, which favors the secretion of proinflammatory cytokines on exposure to CD40 ligand (CD40L).
Methods
Antibodies
The antihuman cIAP1 and antihuman HSC70 mouse monoclonal antibodies were obtained from BD Biosciences (Le Pont de Claix, France) and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. The following rabbit polyclonal antibodies were used: antihuman cIAP1, antihuman X-linked inhibitor of apoptosis protein (XIAP; R&D Systems, Lille, France), antihuman TRAF2 (StressGen, Victoria, BC), antihuman poly(ADP-ribose) polymerase (Santa Cruz Biotechnology), antihuman JNK/stress-activated protein kinase (SAPK), antihuman phospho-JNK/SAPK, antihuman IκBα (Cell Signaling Technology, Ozyme, Saint-Quentin-en-Yvelines, France). For immunofluorescence experiments, antihuman NF-κB p65 (Santa Cruz Biotechnology) and fluorescein isothiocyanate (FITC)–conjugated antihuman GM-130 (Transduction Laboratories, Lexington, KY; BD Biosciences, San Jose, CA) were used. For flow cytometry experiments, we used FITC or allophycocyanin (APC)–conjugated anti-CD11b or anti-CD71 antibodies (BD Biosciences PharMingen). Secondary antibodies used included goat horseradish peroxidase (HRP)–conjugated antimouse or antirabbit antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) for Western blot analysis and goat 488 or 568–Alexa Fluor antirabbit or antimouse antibodies (Molecular Probes, Eugene, OR) for immunofluorescence studies.
Chemicals
Recombinant human (rh) macrophage colony-stimulating factor (M-CSF) was obtained from PeproTech (Neuilly-sur-seine, France) and rhCD40L from Abcys (Paris, France). 12-O-Tetradecanoyl-phorbol-13-acetate (TPA) was from Sigma-Aldrich (St Quentin Fallavier, France), Smac-N7 peptide, lactacystin, and kinase inhibitor SP600125 from Calbiochem (Fontenay sous Bois, France), and Z-leu-leu-leu-H-(aldehyde) (MG132) from Peptide Institute (Osaka, Japan). Leptomycin B (LMB) was kindly provided by Dr M. Yoshida (University of Tokyo, Tokyo, Japan).
Cell culture and differentiation
U937 and THP-1 were obtained from the ATCC (Manassas, VA) and differentiated by 20 nM TPA exposure as described.27 Human peripheral blood monocytes were obtained from healthy donors with informed consent and purified using a CD14+ monocyte isolation kit with a light-scattering column according to the manufacturer's instructions (Miltenyi Biotec, Paris, France) and then incubated (0.5 × 106/mL) for up to 6 days in RPMI 1640 medium (Lonza, Verviers, Belgium), 10% fetal calf serum (Lonza), in the presence of 100 ng/mL M-CSF to trigger their differentiation into macrophages. The differentiated phenotype was identified by a flow cytometric analysis of the cell surface expression of CD11b (U937 and THP-1 cells) or CD71 (human macrophages) as described.28
Cell extracts, immunoprecipitation, cell fractionation, and immunoblot analysis
Whole-cell lysates were prepared by incubating the cells in lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% Triton X-100) in the presence of protease inhibitors and then sonicated. Extracts for phosphoprotein analysis were prepared by lysing the cells in lysis buffer (50 mM Tris-HCl, pH 7.4, 50 mM NaCl, 15 mM Na4P2O7, 5 mM NaF, 150 mM Na3VO4, 1% Triton X-100) in the presence of protease inhibitors and then centrifuged to remove cellular debris. Cell fractionation and immunoprecipitation experiments were performed as described.26,28 All extracts were stored at −80°C until analysis. Protein concentration was measured using the Bio-Rad DC Protein Assay Kit (Ivry sur Seine, France). Forty micrograms of proteins was incubated in loading buffer (125 mM Tris-HCl, pH 6.8, 10% β-mercaptoethanol, 4.6% sodium dodecyl sulfate, 20% glycerol, 0.003% bromophenol blue), separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and electroblotted to nitrocellulose membrane (Whatman, Schleicher & Schuell, Versailles, France). After blocking nonspecific binding sites for 2 hours by 2% of bovine serum albumin in Tris-buffered saline (TBS)–Tween (0.05 M Tris base, 0.9% NaCl, pH 7.6, 0.05% Tween 20), the membrane was incubated overnight at +4°C with the primary antibody, washed, incubated with horseradish peroxidase-conjugated secondary antibodies for 45 minutes at room temperature, and then washed again 3 times in TBS-Tween. The immunoblot was revealed using an enhanced chemiluminescence detection kit (GE Healthcare, Little Chalfont, United Kingdom) by autoradiography.
Plasmid constructs, siRNAs, and cell transfection
Six-day–differentiated human blood monocytes and 48-hour–differentiated THP-1 cells were transfected with pBABE-TRAF2 wt (kindly provided by Olivier Micheau, Inserm UMR 866, Dijon, France) or related control vector (3 μg/106 cells) using Lipofectamine LTX reagent (Invitrogen) or with siRNA (2 μg/106 cells) using Oligofectamine reagent (Invitrogen). Undifferentiated THP-1 and U937 cells and human blood monocytes were nucleoporated with small interfering RNA (siRNA) using Amaxa nucleofector kit (Amaxa Biosystems, Gaithersburg, MD) following the manufacturer's instructions. The siRNA sequences were cIAP1 siRNA sequence 1 (forward: 5′-GGCCAAGAGUUUGUUGAUtt-3′; reverse: 5′-AUCAACAAACUCUUGGCCtt-3′; Eurogentec, Seraing, Belgium); cIAP1 sequence 2 (121288) designed and purchased from Ambion (Courtaboeuf, France), TRAF2 siRNA sequence 1 (Hs_TRAF2_4) and sequence 2 (Hs_TRAF2_5) both designed and provided by QIAGEN (Courtaboeuf, France).
Retroviral supernatant production and transduction protocol
A mutated form of TRAF2 within the RING and the fourth zinc finger domain (TRAF2*) (kindly provided by Ze'ev Ronai, Ruttenberg Cancer Center, New York, NY), cIAP1 wt, and cIAP1-ΔRING (deleted for the RING domain) cDNAs were inserted into a bicistronic retroviral pMX-IG vector (kindly provided by Toshio Kitamura, Advanced Clinical Research Center, Institute of Medical Science, Tokyo, Japan) that harbors the internal ribosomal entry site and simultaneously expresses green fluorescent protein (GFP) with the protein of interest. HEK 293T cells (ATCC) were grown in Dulbecco modified Eagle medium (DMEM)–10% fetal calf serum to 70% confluence in a 6-well culture plates and transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol with plasmid DNA of interest in the presence of MLV gag-pol and ecotropic envelope encoding plasmids. Viral particles containing supernatant were collected every day for 3 days. THP-1 and U937 cells were incubated in viral supernatant in the presence of 4 μg/mL polybrene (Sigma-Aldrich). Cells were subjected to 3 runs of virus infection at a 1-day interval. The efficiency of infection is checked by analysis of GFP-positive cells by flow cytometry. We observed a 1.4- to 1.6-fold increase in the mean of fluorescence intensity of whole cell population in pMX-IG compared with control-transduced cells.
Phagocytic assay
To evaluate the phagocytic activity of monocytic cells, U937 and THP-1 cells were differentiated in a 24-well dish during 48 to 72 hours by 20 nM TPA treatment and incubated with FITC-conjugated Escherichia coli BioParticles (Invitrogen) in fresh medium at 37°C. After incubation, cells were immediately put on ice to stop phagocytosis and washed 3 times in ice-cold Dulbecco phosphate-buffered saline (DPBS; Lonza). The fluorescence of FITC-conjugated E coli BioParticles, which were absorbed onto the cell surface, was faded by incubated cells for 1 minute in 0.25 mg/mL Trypan blue solution. Cells were then washed and slightly scraped in DPBS. The FITC-conjugated E coli BioParticles engulfed were evaluated by a flow cytometry analysis (LSRII, BD Biosciences). A negative control was performed by incubating cells with E coli BioParticles at 4°C.
Cytokine antibody blot array and ELISA
Monocyte-derived macrophages were first transfected with DNA constructs as previously described. Twenty hours after, cells were washed and incubated in fresh medium containing 500 ng/mL CD40L. The cell culture supernatant was collected 24 hours after CD40L stimulation and stored at −80°C until analysis. The presence of cytokines in macrophage supernatant was assessed using human cytokine antibody arrays panel A (R&D Systems) following the manufacturer's instructions. The autoradiography films were digitalized and analyzed by densitometry software ImageJ (http://rsb.info.nih.gov/ij/index.html). Monocyte chemoattractant protein 1 (MCP-1) and IL-8 were quantified using an enzyme-linked immunosorbent assay (ELISA) kit from eBioscience and Biosource (Biosource Europe, Nivelles, Belgium), respectively, following the manufacturer's instructions.
RNA purification and quantitative real-time PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA was reverse transcribed by Moloney murine leukemia virus (MMLV) reverse transcriptase with oligo(dT) primers (Promega, Madison, WI). Specific cDNAs were amplified on a 7500 FAST thermocycler (Applied Biosystems, Foster City, CA) using the SyBr Green detection protocol as outlined by the manufacturer (Applied Biosystems). Specific forward and reverse primers were: cyclophilin, 5′-GTCGACGGCGAGCCC-3′ and 5′-TCTTTGGGACCTTGTCTGCAA-3′ used as a standardizing control; TNF-α, 5′-GGAGAAGGGTGACCGACTCA-3′ and 5′-TGCCCAGACTCGGCAAAG-3′; MCP-1, 5′-TCTCTGCCGCCCTTCTGT-3′ and 5′-GCATCTGGCTGAGCGAGC-3′; IL-8, 5′-CTGGCCGTGGCTCTCTTG-3′ and 5′-CTTGGCAAAACTGCACCTTCA-3′; TRAF2 5′-TGGCTGGCCGCATACC-3′ and 5′-TGTAGCCGTACCTGCTGGTGTA-3′; and cIAP1 5′-CCTGGAGATAGGGTAGCCTGC-3′ and 5′-TGACATAGCATCATCCTTTGGTTC-3′ (Eurogentec).
Immunofluorescence studies
Cells were fixed in paraformaldehyde (PFA; 2%) for 10 minutes at room temperature, washed twice, saturated in DPBS containing 0.1% saponin and 5% nonfat milk, and incubated overnight at room temperature in the presence of primary antibody diluted in DPBS containing 0.1% saponin and 0.5% bovine serum albumin (BSA). After washing, cells were incubated for 30 minutes with secondary antibody and washed again with DPBS. Nuclei were stained by Hoechst 33342 (Sigma-Aldrich). To rule out nonspecific signal, cells were incubated in the presence of an irrelevant mouse or rabbit immunoglobulin and a specific secondary antibody. Analysis was performed using either a fluorescence (Nikon Eclipse 80i; Nikon, Champigny, France) or a confocal (Leica TCS SP2; Leica, Bron, France) microscope (objective 50×; original magnification ×500). The images were captured by a 3 CCD (charge-coupled device) color video camera (Sony, Paris, France), digitally saved using Archimed-Pro software (Microvision Instruments, Evry, France), and further processed using Photoshop software (Adobe Systems France, Paris, France).
Electrophoretic mobility shift assay
Nuclear fractions were obtained by incubating the cells in lysis buffer (10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid [HEPES], pH 7.8, 10 mM KCl, 0.1 mM ethylenediaminetetraacetic acid [EDTA], 0.1 mM ethyleneglycoltetraacetic acid [EGTA], 1 mM dithiothreitol [DTT], 0.6% Nonidet P40) in the presence of protease inhibitors. Cell lysate was centrifuged at 1200g for 10 minutes, and the pellet was washed once in lysis buffer and then resuspended in a buffer containing 20 mM HEPES, pH 7.8, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, in the presence of complete protease inhibitor mixture for 30 minutes on ice. Nuclear extracts were cleared by centrifugation at 20 000g for 30 minutes; then 5 μg was incubated with 100 000 cpm γ32P–end-labeled NF-κB (5′-AGTTGAGGGGCTTTCCCAGGC-3′) consensus oligonucleotide (Promega) in a reaction buffer containing 5 μL HNB (0.5 M sucrose, 15 mM Tris, pH 7.5, 60 mM KCl, 0.25 mM EDTA, pH 8, 0.125 mM EGTA, pH 5, 0.15 mM spermin, 0.5 mM spermidine, 1 mM DTT), 2 μL MgSp (10 mM MgCl2, 80 mM spermidine), 1.5 μL NaPi (10 mM NaPi, 1 mM EDTA), 10 mM DTT, and 0.2 μg poly(dI-dC). After 30 minutes, the DNA-protein complexes were separated from free oligonucleotides by electrophoresis in a 4% polyacrylamide gel and detected by a PhosphorImager.
Results
Monocyte differentiation into macrophages is associated with a decrease in TRAF2 expression
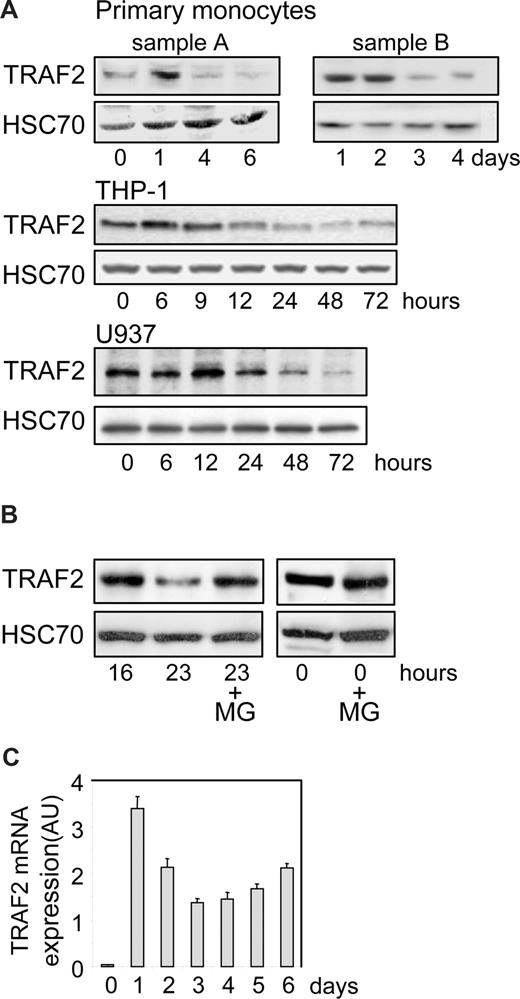
We performed an immunoblot analysis of TRAF2 expression in human peripheral blood monocytes undergoing differentiation into macrophages on exposure to M-CSF. We observed an increase in TRAF2 expression at day 1, then a time-dependent decrease in the protein level (Figure 1A, 2 independent samples). A similar observation was made in THP-1 and U937 cell lines, 2 monoblastic cell lines whose differentiation into macrophages was induced by exposure to the phorbol ester TPA. In these cell lines, TRAF2 expression increased during the first hours of the differentiation process and then progressively decreased (Figure 1A). In THP-1 cells (Figure 1B), addition of the proteasome inhibitor MG132 to the culture medium 16 hours after the beginning of TPA exposure prevented the subsequent decrease in TRAF2 expression as observed 7 hours later, that is, 23 hours after initiation of the differentiation. A similar inhibition of TRAF2 protein level decrease was obtained by inhibiting the proteasome with MG132 in TPA-treated U937 cells (data not shown). A quantitative real-time PCR analysis demonstrated that exposure of peripheral blood monocytes to M-CSF induced a rapid and strong increase in TRAF2 mRNA level. Then, this mRNA remained highly expressed compared with nondifferentiated cells while the protein was progressively degraded (Figure 1C).
TRAF2 expression decreases along differentiation into macrophages. (A) Immunoblot analysis of TRAF2 expression in peripheral blood monocytes exposed to 100 ng/mL M-CSF and in THP-1 and U937 cells exposed to 20 nM TPA for the indicated times (sample A and sample B represent 2 independent experiments). HSC70 indicates loading control. (B) Immunoblot analysis of TRAF2 expression in THP-1 cells exposed for the indicated times to 20 nM TPA. In indicated samples (+MG), 30 μM MG132 was added 7 hours before preparing extracts and immunoblot analysis, ie, 16 hours after initiation of the differentiation by TPA. HSC70 indicates loading control. (C) Real-time PCR analysis of mRNA expression of TRAF2 in peripheral blood monocytes exposed to 100 ng/mL M-CSF (mean ± SD of independent triplicates). AU indicates arbitrary units.
TRAF2 expression decreases along differentiation into macrophages. (A) Immunoblot analysis of TRAF2 expression in peripheral blood monocytes exposed to 100 ng/mL M-CSF and in THP-1 and U937 cells exposed to 20 nM TPA for the indicated times (sample A and sample B represent 2 independent experiments). HSC70 indicates loading control. (B) Immunoblot analysis of TRAF2 expression in THP-1 cells exposed for the indicated times to 20 nM TPA. In indicated samples (+MG), 30 μM MG132 was added 7 hours before preparing extracts and immunoblot analysis, ie, 16 hours after initiation of the differentiation by TPA. HSC70 indicates loading control. (C) Real-time PCR analysis of mRNA expression of TRAF2 in peripheral blood monocytes exposed to 100 ng/mL M-CSF (mean ± SD of independent triplicates). AU indicates arbitrary units.
cIAP1 is involved in the differentiation-associated TRAF2 degradation
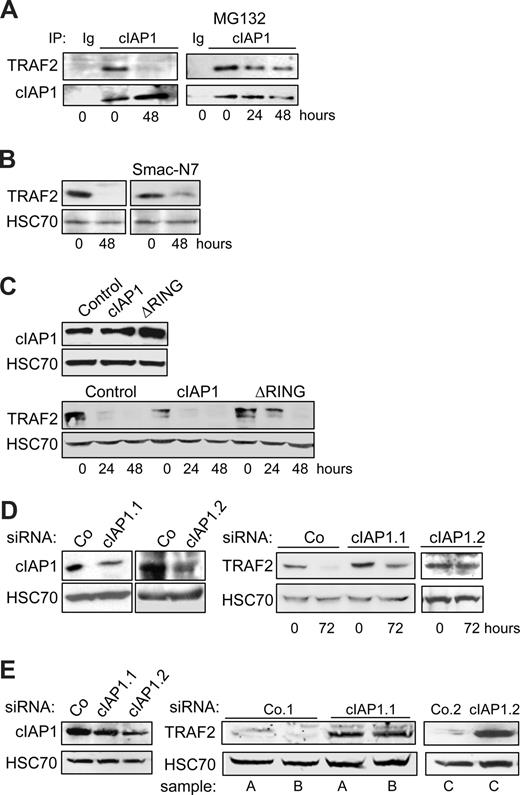
The RING domain containing protein cIAP1 was demonstrated to be an E3 enzyme in the ubiquitylation process that leads to TRAF2 degradation by the proteasome machinery.12,13,23 Coimmunoprecipitation experiments demonstrated that cIAP1 interacted with TRAF2 in undifferentiated cells. This interaction was no more detected in terminally differentiated cells (Figure 2A), in accordance with the decreased expression of TRAF2 protein (Figure 1A). Addition of the proteasome inhibitor MG132 to the culture medium 7 hours before the coimmunoprecipitation experiment permitted the TRAF2/cIAP1 interaction to be partially recovered (Figure 2A). To determine whether cIAP1 was involved in the degradation of TRAF2 that accompanies the macrophagic differentiation, we first exposed the cells to the IAP inhibitor Smac-N7, a cell-permeable peptide made of the 7 N-terminal amino acids of the endogenous IAP inhibitor, Smac. This inhibitory peptide was shown to prevent cIAP1-TRAF2 interaction.29 Its addition to the culture medium partially prevented the differentiation-associated degradation of TRAF2 (Figure 2B). We also used a retrovirus vector to introduce in THP-1 cells an E3-defective mutant of cIAP1 in which the RING domain has been deleted (cIAP1-ΔRING). The expression of this cIAP1 dominant-negative mutant12 delayed the degradation of TRAF2 in THP-1 cells undergoing differentiation into macrophages (Figure 2C). In contrast, retrovirus-mediated overexpression of wild-type cIAP1 decreased the basal level of TRAF2 expression in undifferentiated cells (Figure 2C). Lastly, the use of 2 cIAP1-targeting siRNAs that specifically decreased cIAP1 expression (Figure 2D left panel) prevented the differentiation-associated TRAF2 depletion (Figure 2D right panel). Interestingly, when introduced into macrophages obtained by ex vivo differentiation of peripheral blood monocytes by M-CSF, the 2 cIAP1 siRNAs induced a drastic increase in TRAF2 level (Figure 2E), suggesting that cIAP1 was also required for maintaining a low level of TRAF2 protein in differentiated macrophages.
cIAP1 is responsible for differentiation-associated TRAF2 depletion. (A) Coimmunoprecipitation of cIAP1 and TRAF2 in U937 cells exposed for the indicated times to 20 nM TPA. (Right panel) MG132 was added during the last 7 hours of exposure to TPA (IP with an anti-cIAP1 antibody; immunoblot with anti-TRAF2 and anti-cIAP1 antibodies; Ig indicates irrelevant immunoglobulin). (B) U937 cells were left untreated or treated with 20 nM TPA for 48 hours in the absence or presence of 50 μM Smac-N7 peptide before immunoblot analysis of TRAF2. (C) U937 cells were transduced with murine stem cell virus–based retroviral vector, either empty (Control) or encoding cIAP1 or ΔRING (a cIAP1 construct deleted of the C-terminal E3-ubiquitin ligase domain), and cIAP1 expression was checked by immunoblotting (top panel) before inducing cell differentiation by exposure to 20 nM TPA for the indicated times and analyzing TRAF2 expression by immunoblot (bottom panel). (D) THP-1 cells were transfected with either negative-control (Co) or 2 cIAP1 targeting siRNA sequences (1 and 2), 48 hours before exposing the cells to 20 nM TPA. (Left panel) Immunoblot analysis of cIAP1, 48 hours after siRNA treatment. (Right panel) Immunoblot analysis of TRAF2 at indicated times after TPA exposure (20 nM). (E) Macrophages were obtained by exposure of human blood monocytes for 6 days to M-CSF (100 ng/mL) and then transfected with either negative-control (Co) or cIAP1 targeting siRNAs (1 and 2). cIAP1 (left panel) and TRAF2 (right panel) expression was studied by immunoblotting, 48 hours after siRNA transfection in independent samples (A-C). (B-E) HSC70 used as loading control.
cIAP1 is responsible for differentiation-associated TRAF2 depletion. (A) Coimmunoprecipitation of cIAP1 and TRAF2 in U937 cells exposed for the indicated times to 20 nM TPA. (Right panel) MG132 was added during the last 7 hours of exposure to TPA (IP with an anti-cIAP1 antibody; immunoblot with anti-TRAF2 and anti-cIAP1 antibodies; Ig indicates irrelevant immunoglobulin). (B) U937 cells were left untreated or treated with 20 nM TPA for 48 hours in the absence or presence of 50 μM Smac-N7 peptide before immunoblot analysis of TRAF2. (C) U937 cells were transduced with murine stem cell virus–based retroviral vector, either empty (Control) or encoding cIAP1 or ΔRING (a cIAP1 construct deleted of the C-terminal E3-ubiquitin ligase domain), and cIAP1 expression was checked by immunoblotting (top panel) before inducing cell differentiation by exposure to 20 nM TPA for the indicated times and analyzing TRAF2 expression by immunoblot (bottom panel). (D) THP-1 cells were transfected with either negative-control (Co) or 2 cIAP1 targeting siRNA sequences (1 and 2), 48 hours before exposing the cells to 20 nM TPA. (Left panel) Immunoblot analysis of cIAP1, 48 hours after siRNA treatment. (Right panel) Immunoblot analysis of TRAF2 at indicated times after TPA exposure (20 nM). (E) Macrophages were obtained by exposure of human blood monocytes for 6 days to M-CSF (100 ng/mL) and then transfected with either negative-control (Co) or cIAP1 targeting siRNAs (1 and 2). cIAP1 (left panel) and TRAF2 (right panel) expression was studied by immunoblotting, 48 hours after siRNA transfection in independent samples (A-C). (B-E) HSC70 used as loading control.
Differentiation-associated TRAF2 degradation occurs in the Golgi apparatus–associated compartment
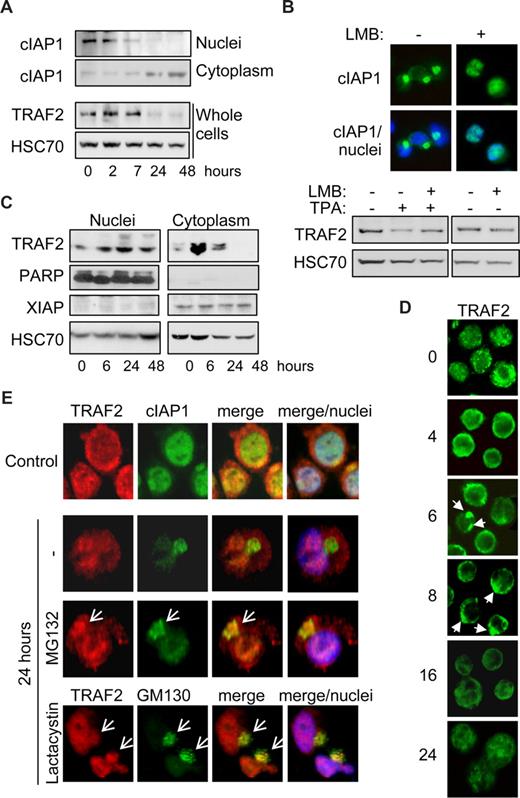
We previously showed that, in cells undergoing differentiation, cIAP1 migrated from the nucleus to the cytoplasm through a nuclear export signal (NES)–chromosome maintenance region 1 (CRM1)-dependent mechanism, to concentrate at the Golgi apparatus surface,26 which was confirmed in Figure 3A,E. The expression of cIAP1 in the cytoplasm is inversely correlated with TRAF2 expression (Figure 3A). Leptomycin B blocked the nuclear export of cIAP1 (Figure 3B top panel) and prevented the decrease in TRAF2 protein expression (Figure 3B) associated with differentiation. In undifferentiated cells, TRAF2 is expressed both in the nuclear and in the cytoplasm-enriched fractions (Figure 3C). Along macrophage formation, TRAF2 protein expression first increases in the cytoplasm (Figure 3C), which is in accordance with results obtained in whole cell extracts (Figure 1A), then decreases when cIAP1 accumulates in this cellular compartment. Conversely, nuclear TRAF2 expression remains unchanged (Figure 3C). Fluorescence microscopy analyses confirmed that, in undifferentiated cells, TRAF2 was expressed in both the cytoplasm and the nucleus (Figure 3D,E). During the first hours of TPA-induced differentiation, TRAF2 accumulated at the cytoplasm periphery and formed clusters (Figure 3D). Then, TRAF2 labeling gets progressively less bright, coinciding with TRAF2 depletion in the cytoplasm. After 24 hours of TPA treatment, TRAF2 was mainly detected in the nuclear compartment (Figure 3D,E). Inhibition of the proteasome machinery by addition of MG132 for 7 hours or lactacystin for 4 hours before immunofluorescence staining revealed that the adaptor protein colocalized with cIAP1 and the Golgi apparatus marker GM130 in a cytoplasm perinuclear structure (Figures 3E, S1 for controls, available on the Blood website; see the Supplemental Materials link at the top of the online article). These results suggest that TRAF2 is degraded in the Golgi apparatus–associated compartment by a cIAP1 and proteasome-dependent process.
TRAF2 is degraded in the cytoplasm. (A) Immunoblot analysis of cIAP1 in nucleus and cytoplasm-enriched fractions and TRAF2 in whole-cell extracts from THP-1 cells undergoing differentiation on TPA exposure (20 nM for the indicated times). HSC70 indicates loading control. (B) THP-1 cells were induced to differentiate into macrophages by exposure to 20 nM TPA for 24 hours, in the absence or presence of 100 nM leptomycin B (LMB) before analyzing cIAP1 localization by fluorescence microscopy (top panel, c-IAP1 in green). Nuclei were stained using Hoechst blue. (Bottom panel) Immunoblot analysis of TRAF2 expression. HSC70 indicates loading control. (C) Immunoblot analysis of TRAF2 in nucleus and cytoplasm-enriched fractions from TPA-treated THP-1 cells (20 nM for the indicated times). Poly(ADP-ribose) polymerase was used as a nucleus fraction marker and X-linked inhibitor of apoptosis protein as a cytoplasm fraction marker. HSC70 indicates loading control. (D) Fluorescence microscopic analysis of TRAF2 expression (green) in THP-1 cells exposed for the indicated times to 20 nM TPA (original magnification ×400). (E) Confocal fluorescence microscopic analysis of TRAF2 (red) and cIAP1 (green) or TRAF2 (red) and GM130 (green) expression in undifferentiated (Control) and TPA-differentiated (24 hours) THP-1 cells, in the absence or presence of 30 μM MG132 or 10 μM lactacystin added for the 7 or 4 last hours of treatment, respectively. Nuclei were stained using Hoechst blue (original magnification ×500).
TRAF2 is degraded in the cytoplasm. (A) Immunoblot analysis of cIAP1 in nucleus and cytoplasm-enriched fractions and TRAF2 in whole-cell extracts from THP-1 cells undergoing differentiation on TPA exposure (20 nM for the indicated times). HSC70 indicates loading control. (B) THP-1 cells were induced to differentiate into macrophages by exposure to 20 nM TPA for 24 hours, in the absence or presence of 100 nM leptomycin B (LMB) before analyzing cIAP1 localization by fluorescence microscopy (top panel, c-IAP1 in green). Nuclei were stained using Hoechst blue. (Bottom panel) Immunoblot analysis of TRAF2 expression. HSC70 indicates loading control. (C) Immunoblot analysis of TRAF2 in nucleus and cytoplasm-enriched fractions from TPA-treated THP-1 cells (20 nM for the indicated times). Poly(ADP-ribose) polymerase was used as a nucleus fraction marker and X-linked inhibitor of apoptosis protein as a cytoplasm fraction marker. HSC70 indicates loading control. (D) Fluorescence microscopic analysis of TRAF2 expression (green) in THP-1 cells exposed for the indicated times to 20 nM TPA (original magnification ×400). (E) Confocal fluorescence microscopic analysis of TRAF2 (red) and cIAP1 (green) or TRAF2 (red) and GM130 (green) expression in undifferentiated (Control) and TPA-differentiated (24 hours) THP-1 cells, in the absence or presence of 30 μM MG132 or 10 μM lactacystin added for the 7 or 4 last hours of treatment, respectively. Nuclei were stained using Hoechst blue (original magnification ×500).
TRAF2 is a positive regulator of macrophage differentiation
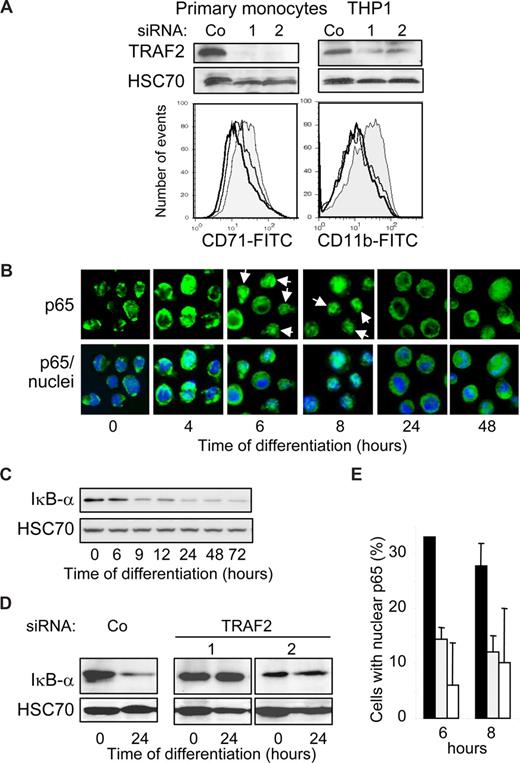
The initial increase in TRAF2 mRNA and protein expression (Figure 1) and its clustering at the cell periphery (Figure 3D) suggested that TRAF2 might play a role in the first phases of the differentiation process. This TRAF2 expression increase was efficiently prevented by the use of 2 independent and specific siRNAs in human blood monocytes and THP-1 cells (Figure 4A top panel). TRAF2-targeting siRNAs prevented their differentiation induced by M-CSF (CD71 expression) and TPA (CD11b expression) exposure, respectively (Figure 4A bottom panel). We28 and others30 previously showed that macrophage differentiation required a moderate and transient activation of NF-κB transcription factor. Here we show that macrophage differentiation is associated with a nuclear translocation of p65 NF-κB subunit (Figure 4B) and a decrease in Iκ-Bα expression (Figure 4C). The 2 TRAF2-targeting siRNAs prevent both Iκ-Bα decrease (Figure 4D) and p65 nuclear redistribution (Figure 4E).
TRAF2 is required for macrophage differentiation. (A,B,D,E) THP-1 cells or human monocytes were transfected with either negative-control (Co) or TRAF2 targeting siRNAs (sequences 1 and 2), 48 hours before exposing the cells to differentiating agents (20 nM TPA for THP-1 and 100 ng/mL M-CSF for human monocytes). (A) (Top panel) Immunoblot analysis of TRAF2, 48 hours after siRNA treatment. (Bottom panel) Flow cytometry analysis of CD11b or CD71 membrane expression, 48 hours after addition of differentiation inducers. Gray histogram represents negative control siRNA; white histograms, TRAF2-targeting siRNA sequence 1 (black line) and 2 (gray line). One representative of 3 experiments is shown. (B) Fluorescence microscopy analysis of NF-κB p65 subunit (green) in THP-1 cells exposed for the indicated times to 20 nM TPA (original magnification ×400). Nuclei were stained using Hoechst (blue). (C) Immunoblot analysis of Iκ-Bα in THP-1 cells exposed for the indicated times to 20 nM TPA. (D) Immunoblot analysis of Iκ-Bα in THP-1 cells exposed for the indicated times to 20 nM TPA, 48 hours after transfection with indicated siRNAs. (E) Percentage of cells with nuclear p65 quantified by immunofluorescence in THP-1 cells exposed for the indicated times to 20 nM TPA, 48 hours after treatment with scramble (black) or TRAF2-specific (gray represents sequence 1; white, sequence 2) siRNAs. Data are mean plus or minus SD of 3 independent experiments.
TRAF2 is required for macrophage differentiation. (A,B,D,E) THP-1 cells or human monocytes were transfected with either negative-control (Co) or TRAF2 targeting siRNAs (sequences 1 and 2), 48 hours before exposing the cells to differentiating agents (20 nM TPA for THP-1 and 100 ng/mL M-CSF for human monocytes). (A) (Top panel) Immunoblot analysis of TRAF2, 48 hours after siRNA treatment. (Bottom panel) Flow cytometry analysis of CD11b or CD71 membrane expression, 48 hours after addition of differentiation inducers. Gray histogram represents negative control siRNA; white histograms, TRAF2-targeting siRNA sequence 1 (black line) and 2 (gray line). One representative of 3 experiments is shown. (B) Fluorescence microscopy analysis of NF-κB p65 subunit (green) in THP-1 cells exposed for the indicated times to 20 nM TPA (original magnification ×400). Nuclei were stained using Hoechst (blue). (C) Immunoblot analysis of Iκ-Bα in THP-1 cells exposed for the indicated times to 20 nM TPA. (D) Immunoblot analysis of Iκ-Bα in THP-1 cells exposed for the indicated times to 20 nM TPA, 48 hours after transfection with indicated siRNAs. (E) Percentage of cells with nuclear p65 quantified by immunofluorescence in THP-1 cells exposed for the indicated times to 20 nM TPA, 48 hours after treatment with scramble (black) or TRAF2-specific (gray represents sequence 1; white, sequence 2) siRNAs. Data are mean plus or minus SD of 3 independent experiments.
TRAF2 degradation is required for terminal differentiation of macrophages
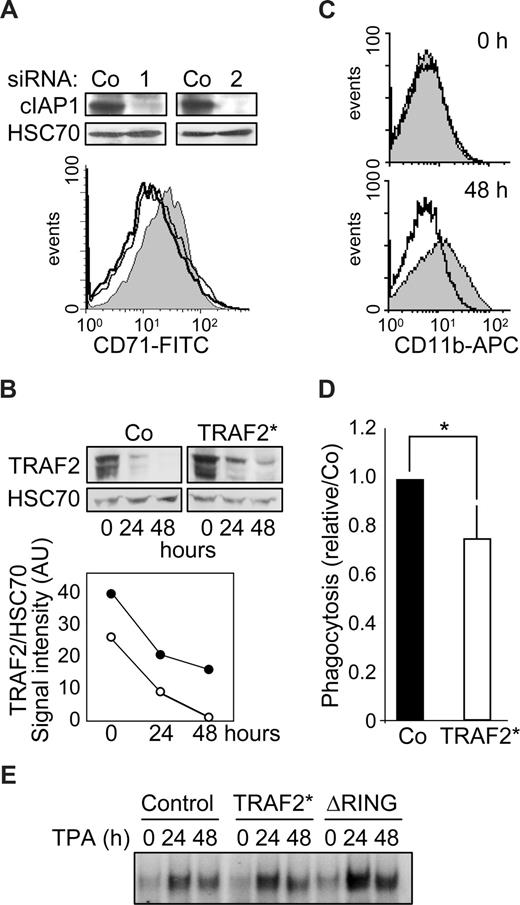
cIAP1 knockdown by 2 specific siRNAs blocks TPA-induced differentiation of THP-1 cells (not shown) and M-CSF–induced differentiation of human peripheral blood monocytes (Figure 5A). Monocytic cells were transduced with a murine stem cell virus-based retroviral vector encoding simultaneously TRAF2 and GFP thanks to the presence of an internal ribosome entry site, and then induced to differentiate by TPA exposure. A flow cytometric analysis of GFP-positive cells indicated that approximately 70% of the cells had been efficiently transduced (Figure S2). To reduce the differentiation-associated degradation of exogenous TRAF2, we used a mutated form of the protein (TRAF2*) that was shown to resist more efficiently than wild-type protein to proteasome-mediated degradation.7 Retrovirus-mediated expression of TRAF2* induced an approximately 1.5-fold increase in TRAF2 protein expression compared with cells transduced with the empty vector (Figure 5B). Although degradation of endogenous TRAF2 occurred along the differentiation, TRAF2 expression always remained higher in TRAF2*-transduced cells (Figure 5B), and this overexpression was sufficient to prevent macrophage differentiation, as evidenced by studying CD11b expression at the cell surface (Figure 5C). TRAF2 overexpression also induced an approximately 20% decrease in the ability of cells to engulf bacteria, a differentiation-associated property (Figure 5D). An electrophoretic mobility shift assay (EMSA) also suggested that overexpression of TRAF2* or the cIAP1-ΔRING construct partially prevented the down-regulation of NF-κB activity that is required for the differentiation into macrophages to progress (Figure 5E).
TRAF2 degradation is required for macrophage differentiation. (A) (Top panel) Immunoblot analysis of c-IAP1 expression in human monocytes transfected 48 hours before with control (Co) or cIAP1-specific (1 and 2) siRNAs. (Bottom panel) Flow cytometry analysis of CD71 expression at the surface of human monocytes transfected with control (gray area) or cIAP1 targeting siRNA sequences 1 (black line) and 2 (dark gray line) and exposed 48 hours later to 100 ng/mL M-CSF for 48 hours. (B-E) THP-1 cells were transduced with an empty retroviral vector (Co) or a vector encoding a proteasome-resistant TRAF2 mutant (TRAF2*) or a vector encoding a cIAP1ΔRING mutant (ΔRING), then induced to differentiate into macrophages by exposure to 20 nM TPA for the indicated times. (B) Immunoblot analysis of TRAF2 expression in transduced cells exposed for the indicated times to 20 nM TPA. HSC70 indicates loading control. (Bottom panel) TRAF2/HSC70 ratio of signal intensities quantified by densitometry (○ indicates control vector; and ●, TRAF2*). (C) Transduced cells were left untreated (top panel) or exposed to 20 nM TPA for 48 hours (bottom panel) before flow cytometry analysis of CD11b membrane expression. Gray area represents control vector; black lines, TRAF2*-transfected cells. (D) Engulfment activity of labeled bacteria E coli was measured in transduced cells exposed for 48 hours to 20 nM TPA. Results were normalized to control cells (mean ± SD of 3 independent experiments). *Statistically significant differences (P < .05, Student test). (E) NF-kB DNA-binding activity was assessed by EMSA in THP-1-transduced cells exposed for the indicated times (hours) to 20 nM TPA. A representative experiment is shown.
TRAF2 degradation is required for macrophage differentiation. (A) (Top panel) Immunoblot analysis of c-IAP1 expression in human monocytes transfected 48 hours before with control (Co) or cIAP1-specific (1 and 2) siRNAs. (Bottom panel) Flow cytometry analysis of CD71 expression at the surface of human monocytes transfected with control (gray area) or cIAP1 targeting siRNA sequences 1 (black line) and 2 (dark gray line) and exposed 48 hours later to 100 ng/mL M-CSF for 48 hours. (B-E) THP-1 cells were transduced with an empty retroviral vector (Co) or a vector encoding a proteasome-resistant TRAF2 mutant (TRAF2*) or a vector encoding a cIAP1ΔRING mutant (ΔRING), then induced to differentiate into macrophages by exposure to 20 nM TPA for the indicated times. (B) Immunoblot analysis of TRAF2 expression in transduced cells exposed for the indicated times to 20 nM TPA. HSC70 indicates loading control. (Bottom panel) TRAF2/HSC70 ratio of signal intensities quantified by densitometry (○ indicates control vector; and ●, TRAF2*). (C) Transduced cells were left untreated (top panel) or exposed to 20 nM TPA for 48 hours (bottom panel) before flow cytometry analysis of CD11b membrane expression. Gray area represents control vector; black lines, TRAF2*-transfected cells. (D) Engulfment activity of labeled bacteria E coli was measured in transduced cells exposed for 48 hours to 20 nM TPA. Results were normalized to control cells (mean ± SD of 3 independent experiments). *Statistically significant differences (P < .05, Student test). (E) NF-kB DNA-binding activity was assessed by EMSA in THP-1-transduced cells exposed for the indicated times (hours) to 20 nM TPA. A representative experiment is shown.
TRAF2 modulates the response of mature macrophages to CD40L
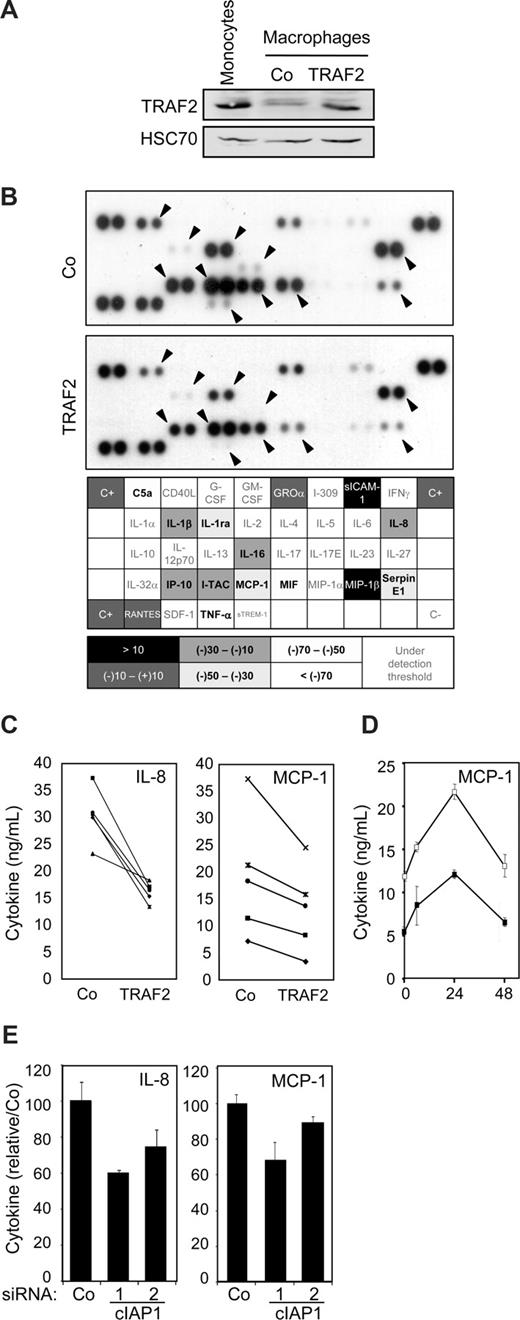
As low level of TRAF2 expression was maintained through a cIAP1-dependent mechanism in macrophages (Figure 2E), we wondered whether this regulation was involved in macrophage-specific functional properties. Macrophages obtained by a 6-day exposure of peripheral blood monocytes to M-CSF were transiently transfected with an empty vector or a wild-type TRAF2 encoding vector. In these conditions, TRAF2-protein level in transfected macrophages was almost similar to that observed in primary monocytes (Figure 6A). These cells were stimulated for 24 hours with 500 ng/mL CD40L, and cell supernatants were analyzed by the use of a cytokine antibody array (Figure 6B). Of the 36 cytokines analyzed, 15 were detected in CD40L-stimulated macrophage supernatants. The level of 11 of them, including C5a, IL-1β, IL-1ra, IL-8, IL-16, IP-10, I-TAC, MCP-1, MIF, Serpin E1, and TNF-α, was lower in the supernatant of TRAF2-overexpressing macrophages than in the supernatant of macrophages transfected with the empty vector (Figure 6B). An ELISA quantification confirmed that TRAF2 restoration inhibited MCP-1 (∼30%) and IL-8 (∼35%) secretion (Figure 6C), and this effect was maintained over time (Figure 6D). siRNA-mediated decrease in cIAP1 expression also decreased the secretion of IL-8 and MCP-1 (Figure 6E).
Influence of TRAF2 on CD40-mediated cytokine secretion in macrophages. Macrophages were obtained by exposure of human blood monocytes for 6 days to M-CSF (100 ng/mL) and then transfected with an empty vector (Co) or a TRAF2-expressing vector (TRAF2). Twenty-four hours after transfection, macrophages were treated with 500 ng/mL CD40L. (A) Immunoblot analysis of TRAF2 expression, 24 hours after transfection. Peripheral blood monocytes were used as a control. (B) (Top panel) Cytokines were detected in the culture supernatant of control (Co) and TRAF2-transfected cells treated with CD40L for 24 hours using an antibody array. (Bottom panel) Spots were quantified by densitometry analysis and reported to internal control (C+). The ratio between cytokine quantities detected in control and TRAF2-transfected macrophage supernatants was quantified. (C-E) ELISA quantification of IL-8 and MCP-1 secretion in (C) control (Co) and TRAF2-transfected cells (TRAF2) treated with CD40L for 24 hours (each line indicates an independent sample) or (D) control (□) and TRAF2-transfected cells (■) treated with CD40L for the indicated times (hours). Results are the mean plus or minus SD of 3 measurements in one sample. (E) Control (Co) and cIAP1 siRNA-transfected cells (2 sequences 1 and 2) treated with CD40L for 24 hours (mean ± SD of 3 independent samples normalized to the control).
Influence of TRAF2 on CD40-mediated cytokine secretion in macrophages. Macrophages were obtained by exposure of human blood monocytes for 6 days to M-CSF (100 ng/mL) and then transfected with an empty vector (Co) or a TRAF2-expressing vector (TRAF2). Twenty-four hours after transfection, macrophages were treated with 500 ng/mL CD40L. (A) Immunoblot analysis of TRAF2 expression, 24 hours after transfection. Peripheral blood monocytes were used as a control. (B) (Top panel) Cytokines were detected in the culture supernatant of control (Co) and TRAF2-transfected cells treated with CD40L for 24 hours using an antibody array. (Bottom panel) Spots were quantified by densitometry analysis and reported to internal control (C+). The ratio between cytokine quantities detected in control and TRAF2-transfected macrophage supernatants was quantified. (C-E) ELISA quantification of IL-8 and MCP-1 secretion in (C) control (Co) and TRAF2-transfected cells (TRAF2) treated with CD40L for 24 hours (each line indicates an independent sample) or (D) control (□) and TRAF2-transfected cells (■) treated with CD40L for the indicated times (hours). Results are the mean plus or minus SD of 3 measurements in one sample. (E) Control (Co) and cIAP1 siRNA-transfected cells (2 sequences 1 and 2) treated with CD40L for 24 hours (mean ± SD of 3 independent samples normalized to the control).
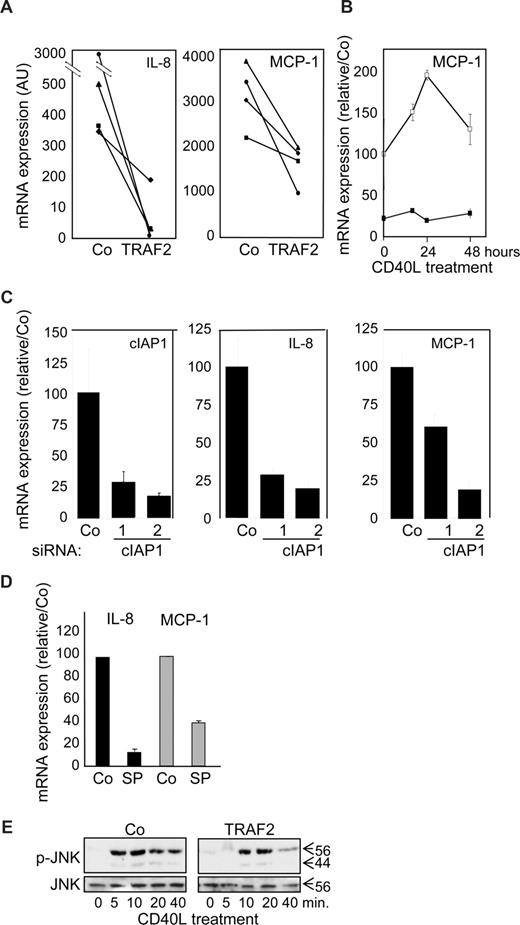
A quantitative real-time–PCR analysis showed that TRAF2 overexpression (Figure 7A) as well as siRNA-mediated decrease in cIAP1 expression (Figure 7C) decreased the expression of IL-8 and MCP-1 mRNA, which was confirmed with time (Figure 7B). JNK was shown to be one of the key pathways that control the cytokine secretion in response to CD40L in macrophages.31 Inhibition of JNK by SP600123 abolished CD40L-induced increase in MCP-1, and IL-8 mRNA (Figure 7D). In this setting, TRAF2 silencing did not interfere with CD40-induced JNK activation (not shown), but TRAF2 overexpression delayed and rapidly down-regulated JNK activation (Figure 7E). Altogether, these data suggest that TRAF2, although being required for optimal differentiation at the beginning of the process, must be subsequently down-regulated for an optimal cytokine response of macrophages to CD40L and c-IAP1 is involved in this down-regulation.
Influence of TRAF2 on CD40-mediated cytokine mRNA expression in macrophages. Macrophages were obtained by exposure of human blood monocytes for 6 days to M-CSF (100 ng/mL) and then transfected with an empty vector (Co) or a TRAF2-expressing vector (TRAF2) or cIAP1 siRNA sequence 1 and 2 and treated with CD40L 24 hours later. (A-D) Real-time PCR analyses of mRNA expression of MCP-1 and IL-8 (A) in Co and TRAF2-transfected macrophages treated for 24 hours with 500 ng/mL CD40L (each line represent an independent sample). (B) Control (open symbols) and TRAF2-transfected (closed symbols) macrophages treated for the indicated times with 500 ng/mL CD40L. (C) Control and c-IAP1 targeting siRNAs macrophages treated for 24 hours with 500 ng/mL CD40L (mean ± SD of 3 independent experiments normalized to controls). (D) Macrophages treated by 500 ng/mL CD40L for 24 hours in the absence (Co) or presence of 10 μM SP600123 (SP) (mean ± SD of 3 independent experiments normalized to controls). (E) Immunoblot analysis of JNK and phosphorylated JNK (p-JNK) in control (Co) and TRAF2-overexpressing macrophages exposed to CD40L for the indicated times. One representative of 3 independent experiments is shown.
Influence of TRAF2 on CD40-mediated cytokine mRNA expression in macrophages. Macrophages were obtained by exposure of human blood monocytes for 6 days to M-CSF (100 ng/mL) and then transfected with an empty vector (Co) or a TRAF2-expressing vector (TRAF2) or cIAP1 siRNA sequence 1 and 2 and treated with CD40L 24 hours later. (A-D) Real-time PCR analyses of mRNA expression of MCP-1 and IL-8 (A) in Co and TRAF2-transfected macrophages treated for 24 hours with 500 ng/mL CD40L (each line represent an independent sample). (B) Control (open symbols) and TRAF2-transfected (closed symbols) macrophages treated for the indicated times with 500 ng/mL CD40L. (C) Control and c-IAP1 targeting siRNAs macrophages treated for 24 hours with 500 ng/mL CD40L (mean ± SD of 3 independent experiments normalized to controls). (D) Macrophages treated by 500 ng/mL CD40L for 24 hours in the absence (Co) or presence of 10 μM SP600123 (SP) (mean ± SD of 3 independent experiments normalized to controls). (E) Immunoblot analysis of JNK and phosphorylated JNK (p-JNK) in control (Co) and TRAF2-overexpressing macrophages exposed to CD40L for the indicated times. One representative of 3 independent experiments is shown.
Discussion
Once in the tissues, peripheral blood monocytes evolve into a variety of terminally differentiated cells, depending on signals received from the microenvironment. The molecular mechanisms that sustain the differentiation processes and confer to differentiated cells their specific properties are only partially depicted. Here we show that the adaptor molecule TRAF2 is both a positive (initial phase) and a negative (secondary phase) regulator of the differentiation of monocytes into macrophages. The protein must be degraded by a proteasome-dependent mechanism that involves cIAP1 for terminal differentiation of monocytes into macrophages and for mature macrophages to optimally respond to stimulation with CD40L.
TRAF2 is an important regulator of cell signaling response to TNFR-mediated extracellular stimuli, leading to cell proliferation, cell activation, and cytokine secretion.2 Ex vivo studies have revealed that TRAF2 could also have a role in the regulation of differentiation in 2 specific cell types. First, TRAF2 was proposed to modulate B-cell maturation by mediating CD40-mediated immunoglobulin class switching.32,33 Second, TRAF2 plays an important role in RANK and TNFRs signals that trigger osteoclast differentiation.34 Here we show that TRAF2 is also a regulator of macrophage differentiation and activity. TRAF2 acts as an adaptor molecule that connects several plasma membrane receptors to downstream signaling pathways.2 TRAF2 was shown to be recruited to membrane complexes and translocated to raft microdomains to participate to signaling events before being degraded by the proteasome system.6,8,14,19 Here we show that, in monocytes induced to differentiate into macrophages, TRAF2 mRNA rapidly increases and the protein forms clusters at the periphery of the cell. This redistribution of TRAF2 protein coincides with Iκ-Bα degradation and the nuclear translocation of NF-κB p65 subunit, 2 events that are prevented by TRAF2 silencing. These results suggest that TRAF2 takes parts in the canonical NF-κB activating pathway during initial phase of monocyte differentiation into macrophages.
Later on in the differentiation process, TRAF2 is degraded by the proteasome system, which was demonstrated to be an important regulatory event in various signaling pathways.8,11,13,17,19 Overexpression of a TRAF2 mutant known to resist to proteasome-mediated degradation prevent the progression of macrophage differentiation and interfere with the down-regulation of NF-κB activity. Altogether, our results suggest that initial accumulation of TRAF2 and its subsequent degradation by a cIAP1-dependent process play a role in the time-dependent modulation of NF-κB activity along the differentiation process.28,30
TRAF2 is targeted for proteolytic degradation by ubiquitylation. The molecule is composed of an N-terminal RING domain35 that characterizes E3 ubiquitin ligase activity36 and was proposed to promote its own ubiquitylation.19 Siah2 protein is also a RING-containing protein that can mediate TRAF2 ubiquitylation to target the protein for degradation under stressful conditions.17 cIAP1 is another RING-containing ubiquitin ligase that can trigger TRAF2 ubiquitylation and proteasome-dependent degradation, eg, on engagement of TNFR2.8,12,13 We have previously shown that cIAP1 is required for optimal differentiation of monocytes into macrophages.25 We show here that cIAP1 is one of the E3 ligases involved in the proteasome-dependent degradation of TRAF2 along this differentiation process. Because the effect of cIAP1 inhibition on differentiation-associated TRAF2 down-regulation was only partial, we cannot rule out a function for TRAF2 itself, Siah2, or other E3 in this event.
We also observed that the degradation of TRAF2 in monocytes undergoing macrophage differentiation was associated with the translocation of the protein to a perinuclear structure. By inhibiting the proteasome, we demonstrated that TRAF2 colocalizes with cIAP1 and proteins of the Golgi apparatus-associated compartment.26 TRAF2 was shown also to colocalize with endoplasmic reticulum markers, including the cognate E2 enzyme Ubc6. Interestingly, Ubc6 behaves as the E2 enzyme for cIAP1 E3 activity, leading to TRAF2 degradation.8 Altogether, these observations indicate that cIAP1 interaction with TRAF2 that leads to its proteasome-mediated degradation along macrophage formation might occur in the Golgi-associated compartment.
TRAF2 depletion sensitizes specific cells to TNF-α-induced apoptosis.6,11,14,18,19 The premature death of TRAF2-deficient mice was related to an increased sensitivity of thymocytes and hematopoietic progenitors to TNF-induced cell death.10 Accordingly, cIAP1-mediated TRAF2 degradation, for example, in response to TNFR2 stimulation, sensitizes specific cells to TNFR1-mediated death.12,13 Interestingly, cIAP1-mediated TRAF2 degradation observed in monocytes undergoing macrophage differentiation did not sensitize these cells to TNF-induced apoptosis (data not shown) but was required for an optimal response of differentiated macrophages to CD40L. CD40 is expressed on antigen-presenting cells that include macrophages, whereas the expression of its ligand is almost restricted to activated CD4+ T cells. T cell–dependent stimulation of CD40 in macrophages induces their activation, enhances their survival,37 up-regulates the expression of costimulatory molecules,31,38,39 and regulates their antimicrobial activity.40 It was previously shown that trimeric CD40 cytoplasmic domain could form multiprotein complexes that contained TRAF2 and cIAP1, together with other TRAFs.41,42 Association of cIAP1 with the CD40 cytoplasmic domain complex was dependent on the presence of an intact TRAF1/2/3-binding site, but the role of cIAP1 in this complex remained undetermined.41,42 We demonstrate that cIAP1-mediated TRAF2 depletion associated with macrophage differentiation facilitates the cytokine secretion in response to CD40 stimulation. Accordingly, an elevated level of TNF-α has been observed in sera from TRAF2-deficient mice10 that is responsible for the premature death of the animals.43
The TRAF-dependent signaling pathway activated by CD40L depends on the cell type and the differentiation stage.44 In macrophages, CD40 proinflammatory signals involve the adaptor TRAF6 and the ERK and JNK-dependent pathways.31,39,45 Macrophages that harbor a CD40 mutant unable to bind TRAF2 still produced cytokine in response to CD40L showing that TRAF2 is dispensable for its function.31,46 The present results show that TRAF2 actually negatively regulates this pathway by preventing JNK activation and downstream production of MCP-1 and IL-8 in response to CD40L. This negative effect of TRAF2 on macrophage response to CD40L could possibly be mediated by heterodimerization with another TRAF, such as TRAF6, as it has been observed in epithelial cells.9,16,47
Although TRAF2 is a positive regulator of the differentiation of monocytes into macrophages, its down-regulation by cIAP1 in mature macrophages may be part of a coordinated response that prepares these cells to their various functions, ie, appropriate response to CD40 ligand. When associated with the Golgi apparatus, cIAP1 could regulate the expression of other signaling regulators that include NEMO/IKKγ,48 RIP1,49 ASK1,50 and NIK,51 which were demonstrated to be cIAP1 ubiquitin targets. Further studies will determine whether these kinases are cIAP1 targets in this differentiation setting and how their modulations contribute to the macrophage functions.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Arlette Hammann and Lydie Desoches for technical assistance, Nathalie Droin for efficient help in real-time PCR analysis, Minoru Yoshida for providing LMB, Toshio Kitamura for pMXs-IG expressing retroviral vector, Ze'ev Ronai for TRAF2 mutant cDNA, and Olivier Micheau for wt TRAF2 expressing vector.
This work was supported by grants from the Ligue Nationale Contre le Cancer (Equipe Labellisee), the Comité de Côte d'Or of the Ligue Nationale Contre le Cancer, the Association Centpoursanglavie, the Agence Nationale de la Recherche, and the National Institute of Cancer.
Authorship
Contribution: A.D. performed experiments and analyzed data; J.C. performed experiments; S.C. performed immunoprecipitation experiments; R.F. performed EMSA experiments; E.S. supervised the research group and corrected the paper; and L.D.D. supervised the experiments and research team, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Laurence Dubrez-Daloz, Institut National de la Santé et de la Recherche Médicale, Unite Mixte de Recherche 866, Faculté de Médecine, 7 boulevard Jeanne d'Arc, 21079 Dijon cedex, France; e-mail: Laurence.Dubrez-Daloz@u-bourgogne.fr.