Abstract
T cells move randomly (“random-walk”), a characteristic thought to be integral to their function. Using migration assays and time-lapse microscopy, we found that CD8+ T cells lacking the lymph node homing receptors CCR7 and CD62L migrate more efficiently in transwell assays, and that these same cells are characterized by a high frequency of cells exhibiting random crawling activity under culture conditions mimicking the interstitial/extravascular milieu, but not when examined on endothelial cells. To assess the energy efficiency of cells crawling at a high frequency, we measured mRNA expression of genes key to mitochondrial energy metabolism (peroxisome proliferator–activated receptor γ coactivator 1β [PGC-1β], estrogen-related receptor α [ERRα], cytochrome C, ATP synthase, and the uncoupling proteins [UCPs] UCP-2 and -3), quantified ATP contents, and performed calorimetric analyses. Together these assays indicated a high energy efficiency of the high crawling frequency CD8+ T-cell population, and identified differentially regulated heat production among nonlymphoid versus lymphoid homing CD8+ T cells.
Introduction
Phenotypic and immune-functional analyses have defined differences in the nature of CD8+ T-cell populations, differing homing being one of the key distinctions among subsets. Naive and central-memory CD8+ T cells express the lymph node homing receptors CCR7 and CD62L and screen lymphoid tissue for cognate antigen. Effector-memory CD8+ T cells, by contrast, lack expression of CCR7 and CD62L and home primarily into nonlymphoid tissues.1-5 Screening for cognate antigen via random lymphocyte motility (“random-walk” activity), an obviously energy-consuming biologic feature, has been shown to be characteristic of both lymphoid as well as nonlymphoid homing CD8+ T cells.6-11 Mimicking intravascular and extravascular conditions in vitro, we sought here to (1) characterize random lymphocyte motility among lymphoid and nonlymphoid homing CD8+ T cells, and (2) relate random lymphocyte motility and mitochondrial biogenesis/ATP synthesis.
Methods
Isolation of peripheral blood mononuclear cells, CD4+ and CD8+ T cells, cell culture, FACScan analysis, and fluorescence-activated cell sorting
Detailed procedures are outlined in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Anticoagulated blood was drawn from healthy donors after written informed consent was obtained in accordance with the Declaration of Helsinki.
Migration assay
Bulk lymphocytes or sorted CD8+ T-cell subsets were resuspended at 6 × 105 cells/mL in RPMI 1640 containing 10% fetal calf serum, 50 U/mL penicillin and 50 μg/mL streptomycin (R10; all from Invitrogen, Luzern, Switzerland) and loaded in duplicates into uncoated, fibronectin-coated (10 μg/mL; Human Plasma Fibronectin, Temecula, CA) or human umbilical vein endothelial cell (HUVEC)–coated 5-μm pore-size polycarbonate transwell inserts (Costar, Corning, NY). One milliliter R10 or endothelial cell medium (EGM-2MV; Lonza, Basel, Switzerland) for HUVECs, respectively, was added to the lower well. After 16 hours of incubation at 37°C, cells from both compartments were stained with the appropriate antibodies.
ELISpot
Detailed procedures are outlined in Document S1.
Time-lapse microscopy
CCR7−CD8+ and CCR7+CD8+ T cells were FACS sorted and resuspended in R10 and plated in uncoated 24- or 48-well cell-culture dishes (Costar) coated with fibronectin (10 μg/mL) or on HUVEC-coated plates. After an incubation period of 1 hour, pictures were taken every minute using an Olympus IX 81 motorized inverted microscope (Olympus Schweiz AG, Volketswil, Switzerland). Data were analyzed using the cell P and cell R software (professional imaging software; Olympus; http://www.microscopy.olympus.eu/microscopes/software.htm) and ImageJ software (Image Processing and Analysis in Java, National Institutes of Health, Bethesda, MD; http://rsbweb.nih.gov/ij/).
ATP quantification
Flourescence-activated cell sorter (FACS)–sorted CD8+ T-cell subsets were resuspended at 106 cells/mL in purified water, transferred to cryotubes (Nunc A/S, Roskilde, Denmark) and snap-frozen in liquid nitrogen. After thawing, cells were incubated for 10 minutes in boiling water and centrifuged for 5 minutes at 20 000g. The supernatant was kept on ice until ATP measurements were performed according to the manufacturer's protocol (FL-AA; Sigma-Aldrich, Steinheim, Germany).
Gene expression analysis
Detailed procedures are outlined in Document S1. Primers are listed in Table S1.
Calorimetry
Bulk lymphocytes and FACS-sorted CCR7− and CCR7+CD8+ T cells were resuspended at 1.2 × 106 or 2.4 × 106 cells/mL in R10. After a calibration period of 24 hours, heat production of bulk and sorted lymphocytes was recorded every minute during 12 hours using an isothermal calorimeter (Thermal Activity Monitor 3102 TAM III; TA Instruments, New Castle, DE).
Statistical analysis
Results were tested for normality using the D'Agostino-Pearson omnibus normality test. Student t tests, Mann-Whitney U tests, and Wilcoxon signed rank tests were performed using Prism 4 software (GraphPad Software, San Diego, CA). P values less than .05 were considered statistically significant. Results are given as mean plus or minus standard deviation (SD) or median and range, as appropriate.
Results and discussion
Random cellular movement of CD8+ T cells
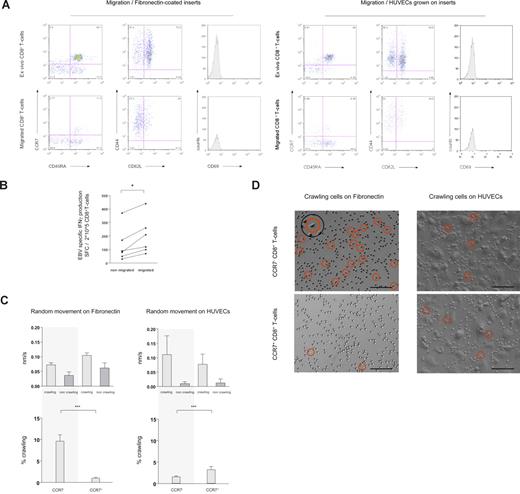
Distinct anatomical compartments are screened for cognate antigen by specialized subsets of T cells.1-5 Antigen-screening efficiency of T cells in both lymphoid and nonlymphoid organs is thought to be increased by random cellular movement (“random-walk”).6,7,10,12-15 Aiming at assigning random cellular movement activity to distinct cell populations we first compared the phenotype of CD8+ T cells ex vivo with that of CD8+ T cells that, in the absence of a chemotactic gradient, migrated across 5-μM pore inserts coated with either fibronectin (Figure 1A left panel) or HUVECs (Figure 1A right panel). Irrespective of whether CD8+ T cells were interacting with fibronectin or with HUVECs, the phenotype of transmigrating CD8+ T cells became skewed toward cells lacking expression of the lymph node homing receptors CCR7 and CD62L (Figure 1A and Table S2). CD8+ T cells were also stained for expression of the maturation marker CD45RA, the cell-surface glycoprotein CD44 (involved in lymphocyte homing), and the activation marker CD69.4,16-20 Neither of these molecules was differentially expressed on nonmigrating versus migrating CD8+ T cells (Figure 1A). We also used CD27 and CD28, 2 phenotypic markers of CD8+ T-cell differentiation,1 to characterize cells accumulating in migration assays. As for expression of CCR7 and CD62L, also according to the expression of CD27 and CD28 accumulation of cells with an effector-memory phenotype (CD27−CD28− “late differentiated” cells) was observed (data not shown). We then quantified migration of sorted CCR7− and CCR7+CD8+ T cells, by relating the starting number of cells to the number of cells that had migrated after 16 hours of incubation. Indicating their higher intrinsic random movement activity, the percentage of migrating cells was almost 3-fold higher among sorted CCR7−CD8+ T cells (median percentage of cells migrating after 16 hours: CCR7− 27.7% [range, 23.7%-49.0%], n = 5; CCR7+ 10.1% [range, 1.3%-14.4%], n = 6; P = .004) Lastly, we compared the size (forward scatter histogram) of migrating versus nonmigrating cells, thus testing and dismissing the hypothesis that accumulation of CD8+ T cells with an effector-memory phenotype merely reflected selection for smaller cells (data not shown).
Phenotypic and functional characteristics of randomly moving of CD8+ T cells. (A) In transwell migration experiments, migration of CD8+ T cells through fibronectin-coated 5-μM pore membranes (left panel) and through HUVECs grown on 5-μM pore membranes (right panel) was assessed in absence of a chemotactic gradient. Ex vivo expression of CCR7 and CD45RA, CD44 and CD62L, and the activation marker CD69 was compared with expression on migrating cells (16 hours migration). Migrating CD8+ T cells were skewed toward a CCR7− and CD62L− phenotype. (B) As assessed in transwell migration experiments, the frequency of EBV-specific CD8+ T cells producing IFNγ was higher amid migrating than nonmigrating cells. *P < .05. (C) Using time-lapse microscopy, the movement pattern of sorted CCR7− and CCR7+CD8+ T cells on fibronectin-coated cell-culture plates and on HUVECs was visualized. The velocity of crawling and noncrawling cells was analyzed assessing 1-minute–spaced pictures. The average speed of crawling cells was similar among subsets on both fibronectin and HUVECs, whereas noncrawling cells moved slightly more rapid on fibronectin (top panel). Crawling and noncrawling cells were enumerated on 5-minute–spaced pictures throughout a 1-hour observation period, and the mean percentage of crawling cells compared among CCR7− and CCR7+CD8+ T cells. Crawling was a prominent feature of CCR7−CD8+ T cells on fibronectin (∼10% of all cells). On HUVECs the frequency of crawling CD8+ T cells was higher among the CCR7+ subset, but always remained less than 5% (bottom panel). ***P < .001 (D) On representative time-lapse video pictures crawling CD8+ T cells on fibronectin and HUVECs are marked with a circle. The insert shows the typical features of a crawling cell at higher magnification. Scale bar represents 0.1 mm.
Phenotypic and functional characteristics of randomly moving of CD8+ T cells. (A) In transwell migration experiments, migration of CD8+ T cells through fibronectin-coated 5-μM pore membranes (left panel) and through HUVECs grown on 5-μM pore membranes (right panel) was assessed in absence of a chemotactic gradient. Ex vivo expression of CCR7 and CD45RA, CD44 and CD62L, and the activation marker CD69 was compared with expression on migrating cells (16 hours migration). Migrating CD8+ T cells were skewed toward a CCR7− and CD62L− phenotype. (B) As assessed in transwell migration experiments, the frequency of EBV-specific CD8+ T cells producing IFNγ was higher amid migrating than nonmigrating cells. *P < .05. (C) Using time-lapse microscopy, the movement pattern of sorted CCR7− and CCR7+CD8+ T cells on fibronectin-coated cell-culture plates and on HUVECs was visualized. The velocity of crawling and noncrawling cells was analyzed assessing 1-minute–spaced pictures. The average speed of crawling cells was similar among subsets on both fibronectin and HUVECs, whereas noncrawling cells moved slightly more rapid on fibronectin (top panel). Crawling and noncrawling cells were enumerated on 5-minute–spaced pictures throughout a 1-hour observation period, and the mean percentage of crawling cells compared among CCR7− and CCR7+CD8+ T cells. Crawling was a prominent feature of CCR7−CD8+ T cells on fibronectin (∼10% of all cells). On HUVECs the frequency of crawling CD8+ T cells was higher among the CCR7+ subset, but always remained less than 5% (bottom panel). ***P < .001 (D) On representative time-lapse video pictures crawling CD8+ T cells on fibronectin and HUVECs are marked with a circle. The insert shows the typical features of a crawling cell at higher magnification. Scale bar represents 0.1 mm.
In parallel, migration experiments were performed using isolated CD4+ T cells. Increased migration of CD4+ T cells with an effector-memory phenotype, analogous to CD8+ T cells, was observed, and (also analogous to CD8+ T cells) no difference in the expression of CD45RA, CD44, or CD69 was seen (data not shown).
To monitor random migration at the antigen-specific level we then compared the frequency of Epstein-Barr virus (EBV)–specific interferon γ (IFNγ)–producing CD8+ T cells among nonmigrating and migrating populations. EBV-specific CD8+ T cells were readily detected both among nonmigrating and migrating cells, and, on each occasion, accumulation of effector-memory cells among the migrating cells (as shown in Figure 1A) was reflected by an increase in the frequency of cells releasing the effector-cytokine IFNγ (Figure 1B).
We next aimed at visualizing random migration of CD8+ T cells on fibronectin, mimicking the extravascular/interstitial milieu, and on HUVECs. Both on fibronectin and on HUVECs, at any given time point crawling and noncrawling CD8+ T cells could readily be discerned (crawling is defined here as flattening of the cell stoma with extension of pseudopodia; see insert in Figure 1D). The average speed of crawling cells (CCR7− and CCR7+) was similar on fibronectin and on HUVECs, whereas noncrawling cells (CCR7− and CCR7+) moved slightly more rapidly on fibronectin, possibly reflecting a moving restraint imposed by the bold relief of HUVECs (Figure 1C top panels). Intriguingly, comparing the percentage of crawling versus noncrawling CD8+ T cells on fibronectin versus HUVECs, a striking difference was observed between cells expressing versus not expressing the lymph node–homing chemokine receptor CCR7. While for each recorded time point on fibronectin, approximately 10% of all CCR7−CD8+ T cells exhibited a crawling phenotype, the frequency of CCR7+CD8+ T cells crawling on fibronectin was always less than 2%. By contrast, on HUVECs the percentage of crawling cells was higher among CCR7+CD8+ T cells, yet it always remained less than 5% for both subsets (Figure 1C bottom panels). Representative live microscopy images of CCR7− and CCR7+CD8+ T cells on fibronectin and HUVECs are shown in Figure 1D. Time-lapse movies are available as Videos S1 through S4.
Together these experiments identified robust differences between phenotypically distinct subsets of CD8+ T cells with regards to their random movement activity, and the frequency distribution of crawling cells. These differences may imply that stochastic detection of cognate antigen in the extravascular milieu at a given rate requires higher crawling activity in structurally more variable nonlymphoid tissues than within anatomically highly defined secondary lymph nodes.
ATP content and energy efficiency of CCR7− versus CCR7+CD8+ T cells
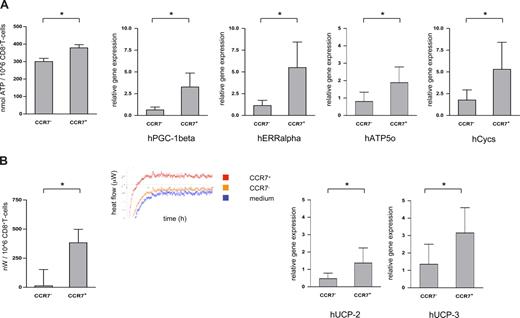
Cellular locomotion relies on transient interactions of motor proteins with microtubules or actin filaments, which then activates the binding and hydrolysis of ATP.21 Using a robust luciferin-based assay, ATP contents were found to be consistently lower in CCR7− compared with CCR7+CD8+ T cells (CCR7−CD8+ T cells: median ATP content 299 ± 62 nmol/106 cells; CCR7+CD8+ T cells: median ATP content 378 ± 62 nmol/106 cells, n = 10; P = .025; Figure 2A left panel). Intriguingly, however, lower ATP contents did not simply reflect increased ATP consumption by crawling cells, since messenger RNA of peroxisome proliferator–activated receptor γ coactivator 1β (PGC-1β; a strong activator of mitochondrial biogenesis/ATP synthesis), estrogen-related receptor α (ERRα; an important partner of PGC-1), cytochrome C (Cycs), and ATP synthase (Atp5o) were all expressed at a higher level in the CCR7+CD8+ T-cell subset as well (Figure 2A right panels).22 CCR7−CD8+ T cells thus seem able to maintain a higher crawling frequency despite lower overall ATP generation.
ATP content and energy efficiency of CCR7− versus CCR7+CD8+ T cells. (A) ATP contents of sorted CCR7− and CCR7+CD8+ T cells were measured in a luminescence-based assay (left panel); mRNA levels of PGC-1β, ERRα, cytochrome C (Cycs), and ATP synthase (Atp5o) were quantified using RT-PCR technology (right panels). CCR7−CD8+ T cells contained less ATP and expressed less mRNA for PGC-1β, ERRα, Cycs, and Atp5o than their CCR7+ counterparts. *P < .05. (B) In calorimetric analyses heat production of sorted CCR7− and CCR7+CD8+ T cells was quantified. After a calibration period of 24 hours, heat production was quantified every minute for 12 hours. The insert shows the heat flow diagram of a representative experiment (left panel). mRNA levels of the uncoupling proteins 2 and 3 of the respiratory chain (UCP-2 and UCP-3) were quantified by RT-PCR (right panels). CCR7−CD8+ T cells contained significantly less mRNA for both UCP-2 and UCP-3 than their CCR7+ counterparts. *P < .05.
ATP content and energy efficiency of CCR7− versus CCR7+CD8+ T cells. (A) ATP contents of sorted CCR7− and CCR7+CD8+ T cells were measured in a luminescence-based assay (left panel); mRNA levels of PGC-1β, ERRα, cytochrome C (Cycs), and ATP synthase (Atp5o) were quantified using RT-PCR technology (right panels). CCR7−CD8+ T cells contained less ATP and expressed less mRNA for PGC-1β, ERRα, Cycs, and Atp5o than their CCR7+ counterparts. *P < .05. (B) In calorimetric analyses heat production of sorted CCR7− and CCR7+CD8+ T cells was quantified. After a calibration period of 24 hours, heat production was quantified every minute for 12 hours. The insert shows the heat flow diagram of a representative experiment (left panel). mRNA levels of the uncoupling proteins 2 and 3 of the respiratory chain (UCP-2 and UCP-3) were quantified by RT-PCR (right panels). CCR7−CD8+ T cells contained significantly less mRNA for both UCP-2 and UCP-3 than their CCR7+ counterparts. *P < .05.
To further investigate the energy efficiency of CCR7−CD8+ T cells, we next compared the heat flow of sorted CCR7− and CCR7+CD8+ T cells using microcalorimetry assays.23 In line with a postulated higher energy efficiency, CCR7−CD8+ T cells produced a median of only 68 nW/106 cells (range, 0-530 nW; n = 8), whereas CCR7+CD8+ T cells produced a median of 390 nW/106 cells (range, 2-1050 nW; n = 8; Figure 2B left panel). Surprisingly, at least part of the higher energy efficiency of CCR7−CD8+ T cells might be brought about via up-regulation of uncoupling proteins of the oxidative chain (UCP-2 und UCP-3) in the CCR7+CD8+ T-cell subset (Figure 2B right panels).
As any biologically costly system, immunity has evolved to minimize energy expenditure in performing its task,24,25 and screening for cognate antigen within lymphoid and nonlymphoid/peripheral tissues is bound to comply with this principle. Our data regarding nonlymphoid homing CD8+ T cells recognizably comply with that principle; regulated uncoupling of the respiratory chain via up-regulation of uncoupling proteins, on the other hand, seems counterintuitive and suggests as yet unrecognized properties of lymph node homing CD8+ T cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Emmanouil Kyriakakis and Emmanuel Traunecker for technical assistance and Juerg A. Schifferli for critical review of the manuscript.
C.H. was supported by the Swiss National Science Foundation (SNF professorship grant PP00B-114850). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authorship
Contribution: G.Z. designed and performed most experiments, analyzed data, and helped write the report; P.G., P.E., O.G., and A.S. designed, performed, and analyzed experiments; A.T. designed and supervised experiments; C. Handschin designed, performed, supervised, and analyzed experiments; A.D.L. designed and supervised experiments and helped write the report; and C. Hess initiated the study, designed and supervised the research, analyzed data, and wrote the report.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christoph Hess, MD, PhD, Immunobiology Laboratory, Department of Biomedicine, University Hospital Basel, 20 Hebelstrasse, CH-4031 Basel; e-mail: chess@uhbs.ch.