Abstract
The default pathway of cell-surface T-cell receptor (TCR) complex formation, and the subsequent transport to the membrane, is thought to entail endoplasmic reticulum (ER) localization followed by proteasome degradation of the unassembled chains. We show herein an alternative pathway: short, incomplete peptide versions of TCRβ naturally occur in the thymus. Such peptides, which have minimally lost the leader sequence or have been massively truncated, leaving only the very C terminus intact, are sorted preferentially to the mitochondrion. As a consequence of the mitochondrial localization, apoptotic cell death is induced. Structure function analysis showed that both the specific localization and induction of apoptosis depend on the transmembrane domain (TMD) and associated residues at the COOH-terminus of TCR. Truncated forms of TCR, such as the short peptides that we detected in the thymus, may be products of protein degradation within thymocytes. Alternatively, they may occur through the translation of truncated mRNAs resulting from unfruitful rearrangement or from germline transcription. It is proposed that mitochondria serve as a subcellular sequestration site for incomplete TCR molecules.
Introduction
T lymphocytes bear a cell-surface T-cell receptor (TCR) complex consisting of TCRα/β chains along with CD3 components. The TCRα and the TCRβ genes code for 2 mRNA populations of a higher and a lower molecular weight forms. The higher molecular-weight species are functionally rearranged and constitute variable-(diversity)–joining-constant (V-(D)-J-C) transcripts. Most of the lower size transcripts contain J-C but lack V region sequences.1-4 These truncated nonrearranged germline transcripts initiate upstream to J, D, or C regions5-8 and are expressed early in murine ontogeny in lymphoid cells9-12 and also in nonlymphoid tissues.13-15
Some of these partial transcripts encode proteins,16-19 whereas among the various germline TCRβ mRNAs, several may potentially encode proteins.14,15 Furthermore, nonproductive rearrangement gives rise to aberrant TCR mRNAs that could encode proteins. These processes, as well as random TCR degradation, may result in the accumulation of incomplete TCR forms, endangering the integrity and functionality of newly formed TCR complexes.
We show herein that incomplete single chain TCRβ proteins are not massively degraded, as reported for complete forms of this molecule,20-25 but rather traffic to the outer mitochondrial membrane. This mitochondrial targeting causes apoptosis, implying a role for incomplete TCRβ molecules in the control of cell survival in the thymus.
Methods
Mice
TCRβ-deficient mice (C57BL/6J-TCRβtm1Mom; The Jackson Laboratory, Bar Harbor, ME) and C57BL/6J (Harlan, Indianapolis, IN) were used. Studies reported herein were approved by the Institutional Animal Care and Use Committee at the Weizmann Institute of Science (Rehovot, Israel).
Plasmid construction, cell culture, and transfections
Plasmid construction is described in detail in “Plasmid construction” in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). COS-7 monkey kidney, 293, 293T, S49.1 murine T-cell lymphoma, NSO murine myeloma (plasmacytoma) cell lines and Bax/Bak−/− mouse embryo fibroblasts were maintained in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FCS (Biological Industries, Kibbutz Beit Haemek, Israel) in an atmosphere of 10% CO2 at 37°C. Immature thymoma cells derived from severe combined immunodeficient (SCID) mice, SCID.adh, kindly provided by Dr D. Wiest (Fox Chase Cancer Center, Philadelphia, PA), and Jurkat human T-cell line were maintained in RPMI 1640 (GIBCO Cell Culture System; Invitrogen) as described above in this section. DNA transfection of COS-7 was performed using LipofectAMINE Reagent (Invitrogen). DNA transfection of 293 and 293T cells was performed by the calcium-phosphate/DNA precipitation method.26 The following transfections were preformed with Amaxa Biosystems (Gaithersburg, MD) solutions, S49.1, Jurkat, and SCID.adh cells using the V solution; Bax/Bak−/− and NIH3T3 cells using the MEF2 solution; and NSO cells using the T solution. Freshly isolated thymocytes (107 cells) either from C57BL/6J or from TCRβ-deficient mice were transfected using the Mouse T-cell Nucleofector Kit (Amaxa Biosystems).
Fluorescence microscopy
COS-7 cells were grown on 13-mm coverslips, and 24 hours after transfection they were either directly stained before fixation with mitochondria or lysosome-specific markers MTR CMXRos and LysoTracker Red DND-99, respectively, or were first fixed, permeabilized, and then immunostained with different antibodies. For immunostaining cells were fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.2% Triton X-100 in PBS. Endoplasmic reticulum (ER) and early endosome stainings were performed using mouse anti-KDEL (Calbiochem, San Diego, CA) and mouse anti–early endosomes antigen 1 (Transduction Laboratories, Lexington, KY), respectively, followed by Cy3-conjugated goat anti–mouse antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA). Immunofluorescence was viewed and analyzed using a Radiance 2000 confocal microscope (Bio-Rad, Hemel Hempstead, United Kingdom) based on a Nikon TE300 inverted microscope. S49.1, NSO, SCID.adh cells and thymocytes from either C57BL/6J or TCRβ-deficient mice were collected and used for cytospin preparations. Slides were air-dried, fixed with 4% PFA, and then either permeabilized with cold 0.1% Triton X-100 in 0.1% sodium citrate for 2 minutes (S49.1) or blocked and permeabilized with 1% BSA and 0.2% Triton X-100 in PBS (NSO, SCID.adh, and thymocytes). The cells were stained with anti-Hsp60 (LK-1), gift of Prof Irun R. Cohen (Weizmann Institute of Science) followed by a Cy3-conjugated goat anti–mouse. The cells were further stained with Hoechst and mounted with Elvanol. Immunofluorescence was viewed and photographed using a Nikon (Tokyo, Japan) E 1000 camera and the Openlab 4.0.1 software (Improvision, Coventry, United Kingdom).
Transmission electron microscopy
High-pressure freezing was performed as previously reported.27 Details and immunostaining for electron microscopy are described in “Transmission electron microscopy” in Document S1.
Isolation of mitochondria, proteinase K treatment, and cell fractionation
293T cells were transfected and 48 hours later were harvested and homogenized in 300 μL ice-cold MB buffer (mannitol 210 mM, sucrose 70 mM, Hepes 10 mM, EDTA 1 mM, pH 7.5). Nuclei and cellular debris were removed by centrifugation at 500g for 5 minutes. The supernatant was then centrifuged at 10 700g for 10 minutes to pellet mitochondria. Isolated mitochondria were resuspended in 200 μL MB buffer. Half of the sample was incubated with proteinase K at a final concentration of 10 μg/mL for 30 minutes on ice, and the other half was left untreated. Reaction was terminated by the addition of PMSF to a final concentration of 1 mM plus 1/100 protease inhibitor cocktail (Sigma-Aldrich, St Louis, MO). Mitochondria were washed once, pelleted, resuspended in sample buffer, and analyzed by immunoblotting. Cell fractionation was preformed with mitochondrial extraction kit (Pierce Chemical, Rockford, IL).
Western blot and immunoprecipitation
Proteins and immunoprecipitation samples were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by immunoblotting using anti-GFP (JL-8; BD Biosciences, San Jose, CA), anti-Hsp60 (LK-1) or anti-TCRβ (H-197; Santa Cruz Biotechnology, Santa Cruz, CA), followed by horseradish peroxidase–coupled antibodies and enhanced chemiluminescence reagents. TCR immunoprecipitation method is detailed in “TCR immunoprecipitation” in Document S1.
Affinity purification and mass spectrometry analysis of TCRβ
Anti-TCRβ H57-597 was coupled to a HiTrap NHS-activated column (GE Healthcare, Little Chalfont, United Kingdom). Thymocyte extract was loaded on the column and eluted with 0.14 N NH4OH. The eluted protein was precipitated by TCA-deoxycholate and analyzed by SDS-PAGE and Coomassie staining. The stained gel was analyzed for protein identification by mass spectrometry as detailed in “Mass spectrometry analysis of TCRβ” in Document S1.
Flow cytometry
After transfection, 293 cells (48 hours) and Jurkat cells (11 hours) were fixed in cold methanol (−20°C) for 30 minutes. Cells transfected with Flag-tagged constructs were then stained with a mouse anti-Flag M2 mAb (Sigma-Aldrich) followed by biotin-SP–conjugated anti–mouse antibody (Jackson ImmunoResearch Laboratories) and streptavidin Oregon Green 488 conjugate (Invitrogen). Cells transfected with EGFP-tagged or Flag-tagged constructs were then washed in PBS and resuspended in 0.4 mL PBS containing RNase A (50 μg/mL; Sigma-Aldrich) and propidium iodide (100 μg/mL; Sigma-Aldrich). The cells were then subjected to flow cytometric analysis using a FACSort flow cytometer (BD Biosciences, San Jose, CA). Analysis of cell cycle was performed using CellQuest analysis software (Becton Dickinson, Franklin Lakes, NJ).
TUNEL assay
The terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) method was used for detection of apoptotic cells, with the ApopTag Kit (Chemicon International, Temucula, CA) according to the manufacturer's instructions with some modifications as detailed in “TUNEL assay” in Document S1.
Quantitative analysis of intracellular proteins
Jurkat cells were transfected with EGFP-iJC or Zap70 YFP using V solutions (Amaxa Biosystems). EGFP-iJC–positive cells were sorted. Total proteins were extracted from both transfectants, and an immunoblot analysis was performed using anti-Zap70 mAb (2F3.2; Upstate Biotechnology, Charlottesville, VA). The number of endogenous Zap70 molecules in Jurkat cells has previously been determined to be 1.2 × 106 molecules/cell.28 According to this number, and the densitometry ratio between the bands of the endogenous and recombinant Zap70 (Zap70 YFP), we were able to calculate the number of recombinant Zap70 YFP molecules/cell. The blot was than stripped and analyzed using the anti-GFP antibody (JL-8; BD Biosciences). According to the densitometry ratio between bands of Zap70 YFP and EGFP-iJC in the anti-GFP immunoblot, and using the calculated number of Zap70 YFP molecules/cell, we were able to calculate the number of EGFP-iJC molecules/cell.
Further details of plasmid construction, primers (Table S1), TCR immunoprecipitation, mass spectrometry analysis of TCRβ, TUNEL assay, and transmission electron microscopy, are provided in Document S1.
Image acquisition
Immunofluorescence was viewed and analyzed using a Radiance 2000 confocal microscope (Bio-Rad) based on a Nikon TE300 inverted microscope and LaserSharp 5.2 software (W. M. Keck Laboratory for Biological Imaging, University of Wisconsin, Madison, WI). The lens used was a Nikon Plan APO 60×/1.4 oil, 00/0.17 WD 0.21.
Immunofluorescence was viewed and photographed using a Nikon (Tokyo, Japan) E1000 microscope and a Hamamatsu ORCA-ER camera (Hamamatsu City, Japan) and Openlab 4.0.1 software (Improvision, Coventry, United Kingdom). The lens used was a Nikon Plan Fluor 100×/1.3 oil, 00/0.17 WD 0.2. All pictures were taken using immersion oil type DF (Cargille Laboratories, Cedar Grove, NJ).
Results
A germline species of TCRβ mRNA encodes a protein that localizes to mitochondria
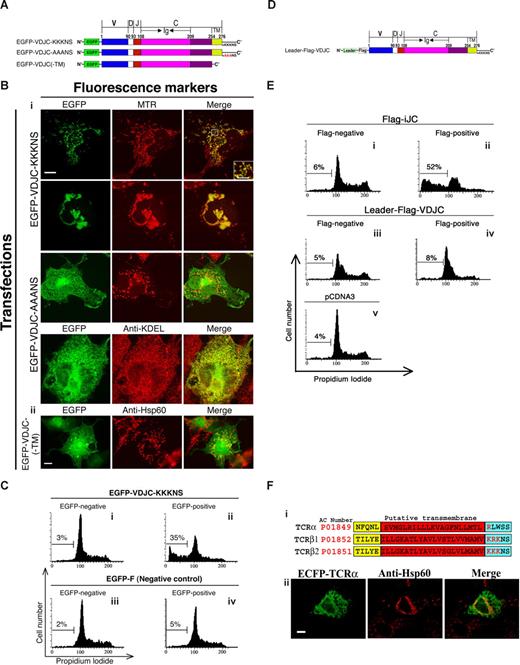
Germline transcripts of TCR have been reported to be expressed in primary thymocytes,1,3,4 as well as in primary mesenchyme and in mesenchymal cell lines.13 We cloned a few of these mRNAs and determined the complete sequence of several species. One murine TCRβ transcript, found in the thymus and in mesenchyme, is studied here in detail. It initiates within the corresponding intronic region upstream of Jβ2.6, spans the Jβ2.6 stretch, as well as the complete Cβ2 region, and is therefore designated iJC-TCRβ (Figure S1). The entire sequence is in frame, from an ATG and a partial kozak sequence, within the sequence derived from the intron, and including the J and the C regions. This sequence encodes a unique protein, different from TCRβ; such a protein would lack the V region, which is replaced by a novel peptide sequence encoded from the intronic noncoding sequence. To examine the cellular localization of this protein, we prepared a fusion construct with enhanced green fluorescent protein (EGFP) at the 5′ end of the iJC-TCRβ cDNA (EGFP-iJC; Figure 1A). After expression in the T-lymphocyte cell line S49.1, EGFP-iJC partially colocalized with the mitochondrial heat shock chaperonin, Hsp60 (Figure 1Bi). Similar results were obtained using a SCID mouse–derived T-cell line (Figure 1Bii), which is TCR deficient. Expression of iJC-TCRβ in primary wild-type thymocytes, as well as in thymocytes from TCRβ-deficient mice, resulted in similar mitochondrial localization (Figure 1Biii and iv, respectively). Thus, iJC-TCRβ encodes a protein that is found associated with the mitochondrion, in both lymphoid cell lines and in primary T lymphocytes, under conditions of overexpression as well as when being expressed on the TCR knockout background.
iJC-TCRβ localizes to mitochondria in both T and non-T cells. (A) Schematic representation of EGFP-iJC and different modified and truncated versions of this molecule. (B) Different T cells [S49.1 (i), SCID.adh (ii)], and thymocytes from either wild-type (iii) or TCRβ-deficient mice (iv) were transfected with EGFP-iJC (green) and 5 hours later were stained with anti-Hsp60 antibody followed by a Cy3-conjugated secondary antibody (red) and with Hoechst for nuclear staining (blue). The marked area is magnified in the inset. Bar, 1 μM. (C) COS-7 cells were transfected with EGFP-iJC (green) and 24 hours later were incubated with MTR (red) for 30 minutes (i). COS-7 cells were transfected with Flag-iJC, 24 hours later the cells were incubated with MTR (red) for 30 minutes and stained by either anti-Flag (ii) or anti-TCR (iii) antibodies followed by Alexa 488–conjugated secondary antibody (green). COS-7 cells were transfected with EGFP-iJC (green) and 24 hours later stained with anti-Hsp60 antibody followed by a Cy3-conjugated secondary antibody (red) (iv). (D) The cell line 293T was transfected with EGFP or EGFP-iJC-TCRβ. Forty-eight hours after transfection cell fractionation was preformed, followed by Western analysis using anti-GFP antibody. Fluorescent images (B,Civ) were taken by fluorescence microscopy (100× oil-immersion objectives) and fluorescent images (Ci-iii) were taken by confocal microscopy. TCR mitochondrial localization was found in all of a total of 100 cells examined in each experimental group, within this figure and the figures below. Constructs are described in Figure 1A. Bars, 5 μM (B), 10 μM (C).
iJC-TCRβ localizes to mitochondria in both T and non-T cells. (A) Schematic representation of EGFP-iJC and different modified and truncated versions of this molecule. (B) Different T cells [S49.1 (i), SCID.adh (ii)], and thymocytes from either wild-type (iii) or TCRβ-deficient mice (iv) were transfected with EGFP-iJC (green) and 5 hours later were stained with anti-Hsp60 antibody followed by a Cy3-conjugated secondary antibody (red) and with Hoechst for nuclear staining (blue). The marked area is magnified in the inset. Bar, 1 μM. (C) COS-7 cells were transfected with EGFP-iJC (green) and 24 hours later were incubated with MTR (red) for 30 minutes (i). COS-7 cells were transfected with Flag-iJC, 24 hours later the cells were incubated with MTR (red) for 30 minutes and stained by either anti-Flag (ii) or anti-TCR (iii) antibodies followed by Alexa 488–conjugated secondary antibody (green). COS-7 cells were transfected with EGFP-iJC (green) and 24 hours later stained with anti-Hsp60 antibody followed by a Cy3-conjugated secondary antibody (red) (iv). (D) The cell line 293T was transfected with EGFP or EGFP-iJC-TCRβ. Forty-eight hours after transfection cell fractionation was preformed, followed by Western analysis using anti-GFP antibody. Fluorescent images (B,Civ) were taken by fluorescence microscopy (100× oil-immersion objectives) and fluorescent images (Ci-iii) were taken by confocal microscopy. TCR mitochondrial localization was found in all of a total of 100 cells examined in each experimental group, within this figure and the figures below. Constructs are described in Figure 1A. Bars, 5 μM (B), 10 μM (C).
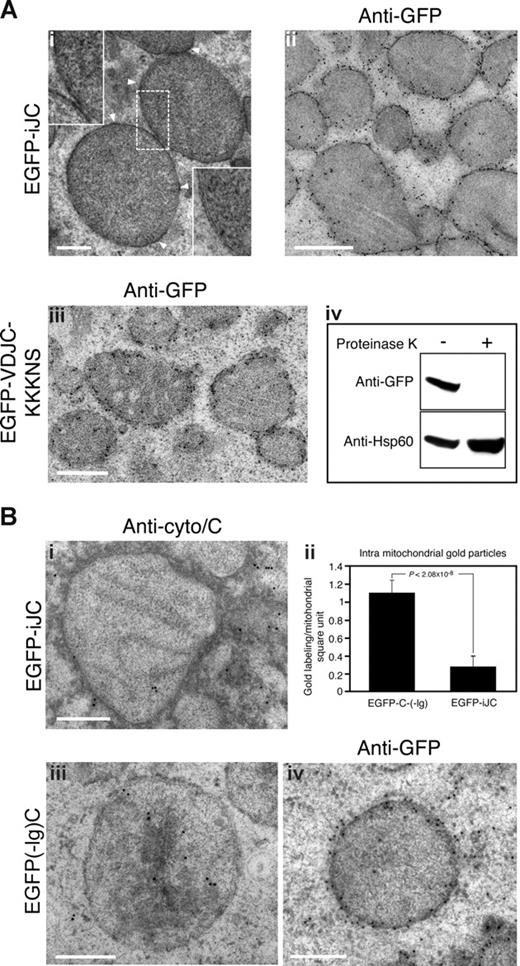
To further analyze this phenomenon we chose COS-7 cells. In these cells the EGFP-iJC signal surrounded the mitochondrion as confirmed by MitoTracker (MTR; Figure 1Ci) and Hsp60 (Figure 1Civ) in what appeared to be membrane staining. We observed that in a significant number of the cells expressing EGFP-iJC, the mitochondria lost their normal distribution throughout the cytoplasm (Figure 1Ci first row merge) and, instead, were clustered in a perinuclear position (Figure 1Ci second row merge). Flag-tagged iJC-TCRβ (Flag-iJC) (Figure 1A), colocalized with the MTR signal, indicating that the unexpected mitochondrial localization of TCR was not related to the GFP fusion (Figure 1Cii). Antibodies directed against the C region of TCR detected iJC-TCRβ in an identical association with the mitochondrion, confirming the presence of a nondegraded fusion protein at this site (Figure 1Ciii). No colocalization of EGFP-iJC with lysosomes, Golgi, ER, or early endosomes was observed (not shown). Subcellular fractionation verified that EGFP-iJC is preferentially found in mitochondria rather than in the cytosol (Figure 1D). Transmission electron microscope (TEM) examination, of germline TCR-transfected cells, showed thickening of the mitochondrial membrane with electron dense material (Figure 2Ai), blurring the mitochondrial double membrane structure. This suggests the presence of the protein in close contact with the mitochondrial membrane. Immuno-TEM analysis confirmed that EGFP-iJC localizes to the mitochondrial membrane, probably to the mitochondrial outer membrane (MOM; Figure 2Aii). To confirm the MOM localization of EGFP-iJC, isolated mitochondria were either treated with proteinase K or left untreated. Immunoblotting of extracted proteins indicated that the band representing EGFP-iJC was destroyed after the proteinase K treatment, whereas Hsp60, which resides inside the mitochondrion, remained intact (Figure 2Aiv). These results indicate that the mitochondria were undamaged and that EGFP-iJC localizes to the MOM. The membrane insertion of EGFP-iJC is further implicated because the protein can be effectively extracted from cells only in lysis buffer containing at least 0.5% SDS and not by gentle detergents (Figure S2A). Because such mitochondrial targeting of TCR molecules has not been previously described, we next set to determine the molecular mechanism underlying this phenomenon.
iJC-TCRβ localizes to the mitochondrial outer membrane and induces cytochrome c release from mitochondria. (A) COS-7 cells were transfected with either EGFP-iJC (i,ii) or EGFP-VDJC-KKKNS (iii), and 24 hours later they were fixed using the high-powered fields (HPF) method for TEM. The cells were either left unstained (i) or stained by anti-GFP (ii,iii) followed by goat anti–rabbit antibodies coupled to 10-nm gold particles. Arrowheads indicate the thickening of the mitochondrial membrane in transfected cells, which is shown at higher magnification in the top left insert. The insert at the bottom right corner of Ai shows a normal double mitochondrial membrane negatively stained. (iv) 293T cells were transfected with EGFP-iJC, and 24 hours later mitochondria were isolated. Mitochondria were then either treated with proteinase K or left untreated. Proteins were extracted, separated by SDS-PAGE, and blotted with anti-GFP stripped and reblotted with anti-Hsp60. (B) COS-7 cells were transfected with either EGFP-iJC (i) or EGFP(-Ig)C (iii,iv) and 24 hours later were fixed using the HPF method. The cells were than stained by either anti–cytochrome c (i,iii) or anti-GFP antibodies (iv) followed by goat anti–rabbit IgG coupled to 10-nm gold particles. Each of the images in subpanels i, iii, and iv shows a representative mitochondria. The amount of intramitochondrial gold particles in a square U of mitochondria was compared between 13 cells that were transfected with either EGFP-iJC or EGFP(-Ig)C, and the results are represented as a graph (Biib). Bar, 0.5 μM.
iJC-TCRβ localizes to the mitochondrial outer membrane and induces cytochrome c release from mitochondria. (A) COS-7 cells were transfected with either EGFP-iJC (i,ii) or EGFP-VDJC-KKKNS (iii), and 24 hours later they were fixed using the high-powered fields (HPF) method for TEM. The cells were either left unstained (i) or stained by anti-GFP (ii,iii) followed by goat anti–rabbit antibodies coupled to 10-nm gold particles. Arrowheads indicate the thickening of the mitochondrial membrane in transfected cells, which is shown at higher magnification in the top left insert. The insert at the bottom right corner of Ai shows a normal double mitochondrial membrane negatively stained. (iv) 293T cells were transfected with EGFP-iJC, and 24 hours later mitochondria were isolated. Mitochondria were then either treated with proteinase K or left untreated. Proteins were extracted, separated by SDS-PAGE, and blotted with anti-GFP stripped and reblotted with anti-Hsp60. (B) COS-7 cells were transfected with either EGFP-iJC (i) or EGFP(-Ig)C (iii,iv) and 24 hours later were fixed using the HPF method. The cells were than stained by either anti–cytochrome c (i,iii) or anti-GFP antibodies (iv) followed by goat anti–rabbit IgG coupled to 10-nm gold particles. Each of the images in subpanels i, iii, and iv shows a representative mitochondria. The amount of intramitochondrial gold particles in a square U of mitochondria was compared between 13 cells that were transfected with either EGFP-iJC or EGFP(-Ig)C, and the results are represented as a graph (Biib). Bar, 0.5 μM.
iJC-TCRβ is a tail-anchored–like protein
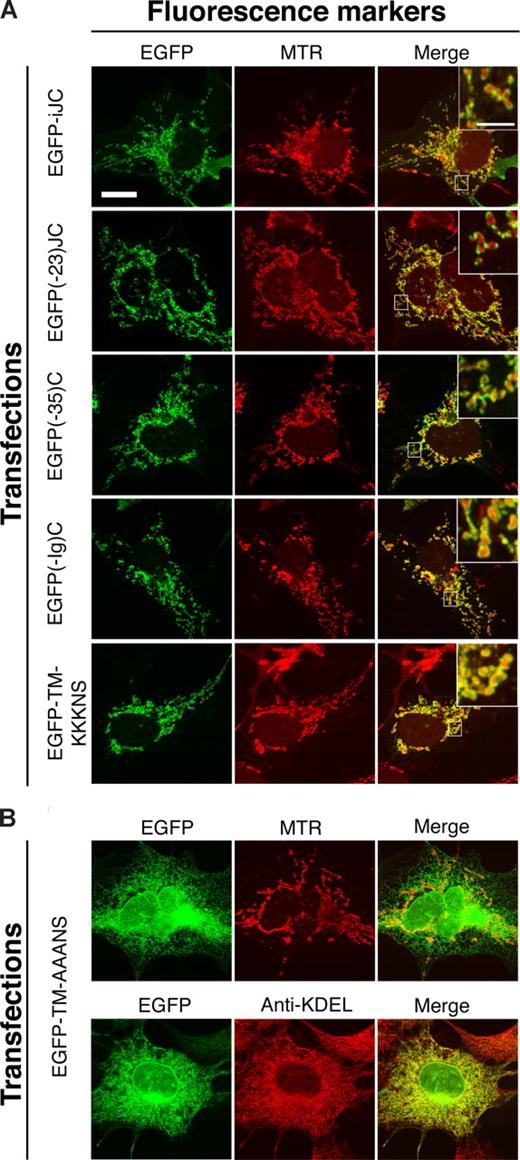
To identify the mitochondrial-targeting signal of germline TCRβ, stepwise deletions of the cDNA were prepared; the most extensive one leaving only the transmembrane domain (TMD) and downstream sequences intact. All of these deleted molecules were fused to the COOH-terminal end of EGFP (Figure 1A) and were expressed in COS-7 cells. The molecular sizes of the proteins encoded by the deleted molecules were confirmed by immunoblotting, to assure that they are not degraded on expression (Figure S2B). The merged images of the EGFP and MTR fluorescence, for the various deleted molecules, are shown in Figure 3A. All 4 variants were able to localize to mitochondria, indicating that the mitochondrial-targeting sequence of TCRβ protein resides within the most COOH-terminal part of the protein, which contains the TMD and the residues that follow it. A construct in which the EGFP was fused to the COOH-terminus of iJC-TCRβ was unable to target mitochondria (data not shown), implying that the proximity of GFP to the COOH-terminus sequences interfered with traffic of the fusion protein.
The TMD of iJC-TCRβ and the residues downstream to it are sufficient to target EGFP to mitochondria. COS-7 cells were transfected with the indicated constructs, and 24 hours later they were either incubated with MTR for 30 minutes or stained with anti-KDEL followed by anti–mouse Cy3 antibodies. The left column shows EGFP images, whereas the middle column shows the MTR or KDEL signal. The right column is the merge of the EGFP and MTR or KDEL signal. Fluorescent images were taken by confocal microscopy. All the constructs shown in panel A encode peptides that target the mitochondrion whereas panel B shows lack of such localization by a mutant lacking charged amino acids. The different constructs are described in Figure 1A. Bar, 10 μM.
The TMD of iJC-TCRβ and the residues downstream to it are sufficient to target EGFP to mitochondria. COS-7 cells were transfected with the indicated constructs, and 24 hours later they were either incubated with MTR for 30 minutes or stained with anti-KDEL followed by anti–mouse Cy3 antibodies. The left column shows EGFP images, whereas the middle column shows the MTR or KDEL signal. The right column is the merge of the EGFP and MTR or KDEL signal. Fluorescent images were taken by confocal microscopy. All the constructs shown in panel A encode peptides that target the mitochondrion whereas panel B shows lack of such localization by a mutant lacking charged amino acids. The different constructs are described in Figure 1A. Bar, 10 μM.
The mitochondrial targeting of a variety of tail-anchored (TA) proteins depends on a rather short presumed TMD (∼20 residues), flanked by basic amino acids (aa).29,30 Similarly to TA proteins, iJC-TCRβ contains 3 lysine residues proximal to the COOH-terminus of the TMD at positions 204, 205, and 206. The construct EGFP-TM-AAANS, in which the 3 lysines in the most COOH-terminus were exchanged by alanines (Figure 1A), showed a reduced mitochondrial localization in most of the cells examined (Figure 3B); instead, the molecule formed a netlike structure resembling ER and was also observed in the proximity of the nuclear envelope. However, some mitochondrial localization was retained. To confirm the ER localization of EGFP-TM-AAANS, a colocalization experiment with an ER marker (using the anti-KDEL antibody) was performed. The experiment confirmed that a significant portion of EGFP-TM-AAANS indeed localized to the ER (Figure 3B). Thus, iJC-TCRβ behaves as a TA-like protein, because its mitochondrial localization is guided by sequences at the molecule's COOH-terminus. Such MOM localization of proteins is characteristic of molecules involved in the regulation of cell survival such as the Bcl-2 family proteins that are either proapoptotic or antiapoptotic.31
Expression of iJC-TCRβ induces apoptotic cell death in both T and non-T cells
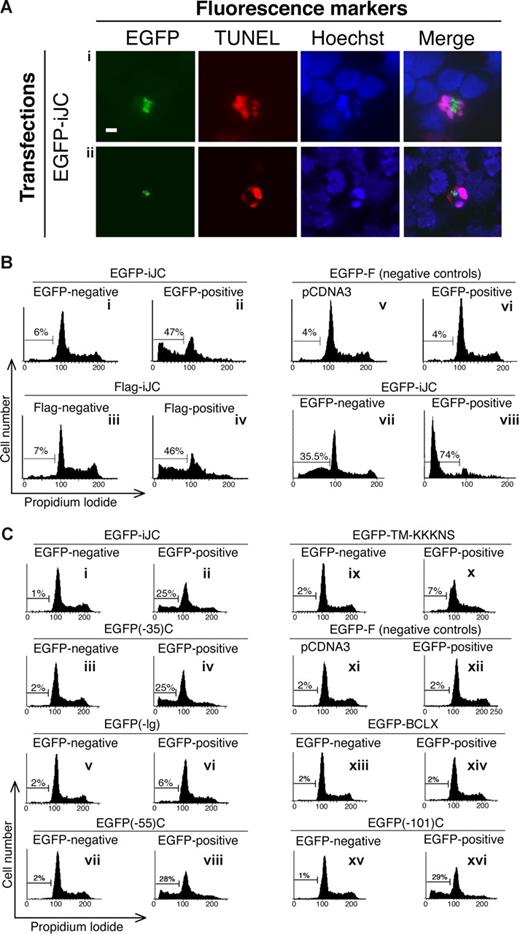
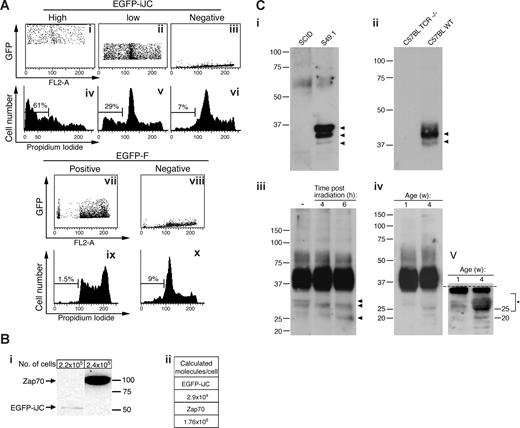
Many of the EGFP-positive cells transfected with iJC-TCRβ had reduced size and distorted morphology. We therefore set to examine whether these cells were undergoing a process of apoptosis. Approximately 60% of the S49.1 T lymphocytes transfected with EGFP-iJC were TUNEL positive 5 hours after transfection (Figure 4Ai). To confirm the specificity of this effect, we examined a control EGFP-farnesylated (F) construct that is directed to the cell membrane, affects the cell cycle, but is not known to cause apoptosis. This construct caused only 7% of the transfectants to become TUNEL positive. After 12 hours, most of the cells expressing iJC-TCRβ completely lost their nucleus. NSO cells transfected with EGFP-iJC showed similar results (data not shown). After 7 hours of expression of iJC-TCRβ in primary T cells from TCRβ−/− mice, 32% were TUNEL-positive (Figure 4Aii) and 33% of the cells lost their nucleus. The expression of EGFP-F in primary T cells from TCRβ−/− mice resulted in only 6% TUNEL-positive cells after 7 hours. To complement the TUNEL data, we analyzed the cell-cycle distribution of 293 cells transfected with either the EGFP (Figure 4Bii) or Flag (Figure 4Biv) tagged iJC-TCRβ. A marked increase in sub-G1 cell population specifically occurred after expression of iJC-TCRβ, indicating apoptosis, as further confirmed in the same cells and in COS-7 by TUNEL assay at 48 hours after transfection (Figure S3). Furthermore, the Jurkat T-cell line transfected with EGFP-iJC (Figure 4Bviii) showed a dramatic shift to sub-G1 and a total loss of normal cell-cycle distribution.
iJC-TCRβ induces apoptotic cell death in both non-T and T cells that is regulated by its Ig domain. (A) S49.1 T cells (i) or thymocytes from TCRβ-deficient mice (ii) were transfected with EGFP-iJC (green) and 5 hours or 7 hours later, respectively, were labeled by the TUNEL method (rhodamine staining) and counterstained with Hoechst for nuclear staining. The merge panels show colocalization of the Hoechst and the TUNEL signals. All pictures were taken by fluorescence microscopy (100× oil-immersion objectives). (B,C) 293 cells (Bi-vi,Ci-xvi) or Jurkat cells (Bvii,viii) were transfected with the indicated constructs. At 48 hours (293) or 11 hours (Jurkat) after transfection, the cells were fixed, and those transfected with Flag-iJC were stained with anti-Flag antibody, a biotin-conjugated anti–mouse antibody, and a streptavidin Oregon Green 488 conjugate. All cells were stained with propidium iodide and analyzed for their cell-cycle distribution. EGFP-negative or Flag-negative cells are the cell populations that remained untransfected in each experiment; EGFP-positive or Flag-positive cells are the transfected cell populations; cells transfected with empty vectors are pCDNA3 (Bv,Cxi) and pEGFP-F (Bvi,Cxii). Bar, 5 μM.
iJC-TCRβ induces apoptotic cell death in both non-T and T cells that is regulated by its Ig domain. (A) S49.1 T cells (i) or thymocytes from TCRβ-deficient mice (ii) were transfected with EGFP-iJC (green) and 5 hours or 7 hours later, respectively, were labeled by the TUNEL method (rhodamine staining) and counterstained with Hoechst for nuclear staining. The merge panels show colocalization of the Hoechst and the TUNEL signals. All pictures were taken by fluorescence microscopy (100× oil-immersion objectives). (B,C) 293 cells (Bi-vi,Ci-xvi) or Jurkat cells (Bvii,viii) were transfected with the indicated constructs. At 48 hours (293) or 11 hours (Jurkat) after transfection, the cells were fixed, and those transfected with Flag-iJC were stained with anti-Flag antibody, a biotin-conjugated anti–mouse antibody, and a streptavidin Oregon Green 488 conjugate. All cells were stained with propidium iodide and analyzed for their cell-cycle distribution. EGFP-negative or Flag-negative cells are the cell populations that remained untransfected in each experiment; EGFP-positive or Flag-positive cells are the transfected cell populations; cells transfected with empty vectors are pCDNA3 (Bv,Cxi) and pEGFP-F (Bvi,Cxii). Bar, 5 μM.
iJC-TCRβ–induced apoptosis is regulated by a portion of 35 residues within the immunoglobulin domain of the protein
To investigate the sequences within iJC-TCRβ that are involved in mediation of the apoptotic process, we used the above-discussed deletion constructs that localized to mitochondria. As can be seen in Figure 4C, transfection of cells with the TCRβ constructs containing the entire C region [EGFP-iJC and EGFP(-35)C], resulted in accumulation of 25% of the cells in sub-G1 phase of the cell cycle (Figure 4Cii and iv, respectively). Cells expressing the TCRβ constructs that lack the immunoglobulin (Ig) domain [EGFP(-Ig)C and EGFP-TM-KKKNS], lost the capacity to cause apoptosis almost entirely (Figure 4Cvi and x, respectively). The construct that lack only 20 aa from the NH2-terminal end of the Ig domain [EGFP(-55)C] (Figure 4Cviii) and an additional one in which most of the Ig domain was deleted, leaving only 35 residues of it [EGFP(-101)C] (Figure 4Cxvi), induced apoptosis in a similar manner to iJC-TCRβ. The mere localization of a control protein (Bcl-xL) in the mitochondrion, under similar experimental conditions, did not cause a comparable accumulation of cells in sub-G1 phase (Figure 4Cxiii,xiv). Thus, the 35 residues within the Ig domain seem to control the apoptosis-inducing activity of iJC-TCRβ.
Apoptotic stimuli cause the release of mitochondrial cytochrome c into the cytosol where it activates caspases. Immunogold TEM analysis of cells, with antibodies to cytochrome c, showed (Figure 2Bi,ii) that, in iJC-TCRβ– transfected cells, cytochrome c was detectable mostly outside of the mitochondria. A mutant lacking the Ig domain did not show this phenomenon when transfected into cells (Figure 2Bii,iii), although it retained the ability to target the MOM (Figure 2Biv). iJC-TCRβ–induced apoptosis was reduced by the caspase inhibitors p35 (Figure S4Bb) and by zVAD (not shown), indicating the requirement of these enzymes in iJC-TCRβ–mediated cell death. The Bcl-2–associated protein Bax and Bcl-2–antagonist/killer (Bak) are essential regulators of lymphocyte apoptosis. A recent study indicated that the involvement of Bax and Bak in T-cell antigen-specific proliferation.32 The apoptotic process mediated by iJC-TCRβ was partially dependent on Bax and Bak signaling; expression of this truncated TCR in cells deficient for both Bax and Bak quantitatively reduced cell death compared with control cells (Figure S5).
Rearranged TCRβ, harboring a complete V region and lacking the leader sequence, localizes to the mitochondrion and induces apoptosis
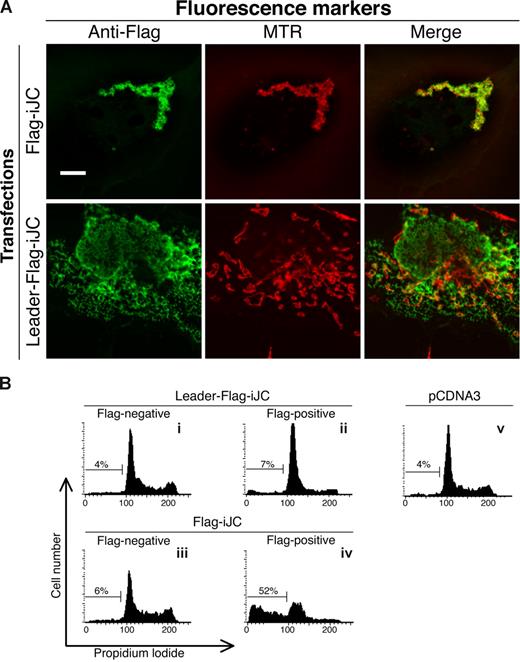
In contrast to full-length, productively rearranged TCRβ, the truncated germline form lacks the NH2-terminal leader sequence. We therefore examined whether artificial addition of such a sequence at the 5′ end of iJC-TCRβ would affect the subsequent cellular localization of the protein. Indeed, iJC-TCRβ fused to a leader peptide sequence in its most NH2-terminus (Figure 1A) displayed no specific mitochondrial localization (Figure 5A), nor did this construct induce apoptosis (Figure 5Bii). Thus, the leader sequence overrides the sorting signal of the COOH-terminus, and the apoptotic effect of iJC-TCRβ strictly depends on its mitochondrial localization.
Mitochondrial localization of iJC-TCRβ is essential for its ability to induce apoptosis. (A) COS-7 cells were transfected with the indicated constructs and 24 hours after transfection were stained with mouse anti-Flag antibodies followed by Alexa 488–conjugated anti–mouse antibodies. All cells were counterstained with MTR for mitochondrial detection, as indicated. Fluorescent images were taken by confocal microscopy. (B) 293 cells were transfected with the indicated constructs. At 48 hours after transfection cells were subjected to a flow cytometry cell-cycle analysis. Cells transfected with an empty pCDNA3 vector are shown (v) as indicated. Constructs are described in Figure 1A. Bar, 10 μM.
Mitochondrial localization of iJC-TCRβ is essential for its ability to induce apoptosis. (A) COS-7 cells were transfected with the indicated constructs and 24 hours after transfection were stained with mouse anti-Flag antibodies followed by Alexa 488–conjugated anti–mouse antibodies. All cells were counterstained with MTR for mitochondrial detection, as indicated. Fluorescent images were taken by confocal microscopy. (B) 293 cells were transfected with the indicated constructs. At 48 hours after transfection cells were subjected to a flow cytometry cell-cycle analysis. Cells transfected with an empty pCDNA3 vector are shown (v) as indicated. Constructs are described in Figure 1A. Bar, 10 μM.
The data presented above show that a germline form of TCRβ, which is unrearranged and lacks V regions, invariably sort to the mitochondrion. The question was raised as to whether the presence of the V region would impinge on this trafficking. The cDNA of a rearranged TCRβ chain from the mouse T-cell clone 2B4,33 containing the complete Vβ3, was tagged like the other TCR molecules described earlier, by fusion with EGFP at the NH2-terminus, and termed EGFP-VDJC-KKKNS (Figure 6A). This molecule localized effectively to the mitochondrion. The 2 different phenotypes of mitochondria, those dispersed throughout the cytoplasm or, alternatively, clustered in a perinuclear position, occurred after expression of the rearranged TCRβ, to the same extent as with the germline forms (Figure 6Bi). Flow cytometric analysis of 293 cells and immunofluorescence TUNEL assay of both 293 and COS-7 cells transfected with EGFP-VDJC-KKKNS showed an apoptotic response (Figure 6C and Figure S3, respectively) similar to that induced by germline TCR, which was associated with release of cytochrome c and involved caspase activity (Figure S4). By contrast, a construct of the rearranged TCRβ, in which the positive lysine residues after the TMD were changed to alanines (EGFP-VDJC-AAANS; Figure 6A), lost the mitochondrial localization (Figure 6Bi). Similarly, deletion of the entire TMD abolished the capacity of TCR to target the mitochondrion (Figure 6Bii). It is thus evident that the V region of TCRβ has no bearing on the mitochondrial localization of the encoded protein. Addition of a leader sequence, upstream at the NH2-terminus, of the rearranged (Figure 6D) as well as the germline forms of TCR abolished the capacity of these molecules to induce apoptosis (Figures 6E and 5B, respectively). TCRα lacks the sequences required for mitochondrial localization (Figure 6Fi). In cells transfected with TCRα, no localization of the recombinant molecule to the mitochondria was observed (Figure 6Fii). It can be therefore generalized that all of the incomplete TCRβ single chain proteins tested were sorted to the mitochondrion and induced apoptosis. These processes depended on an intact TMD and associated downstream positively charged residues, along with specific residues within the Ig domain upstream of the TMD.
Rearranged TCRβ, harboring a complete V region and lacking the leader sequence, localizes to the mitochondrion and induces apoptosis. (A) Schematic presentation of the different constructs of a rearranged TCRβ (B) COS-7 cells were transfected with the indicated VDJC-TCRβ constructs and 24 hours later were either incubated with MTR for 30 minutes or stained with either anti-KDEL or anti-Hsp60 followed by anti–mouse Cy3 antibodies. The left column shows EGFP images, whereas the middle column shows the MTR or KDEL signal (i) or anti-Hsp60 (ii). The right column is the merge of the EGFP and MTR, KDEL or Hsp60 signal. Fluorescent images were taken by confocal microscopy (i) or fluorescence microscopy (100× oil-immersion objectives) (ii). Bar, 10 μM. (C) 293 cells were transfected with the indicated constructs and 48 hours after transfection were subjected to a flow cytometry cell-cycle analysis. Cells transfected with empty vector are pEGFP-F (Civ). (D) Schematic presentation of Leader-Flag-VDJC. (E) 293 cells were transfected with the indicated constructs. At 48 hours after transfection cells were subjected to a flow cytometric cell-cycle analysis. Cells transfected with an empty pCDNA3 vector are shown (Ev) as indicated. (F) The most COOH-terminal residues of mouse TCRα, β1, and β2 are shown. The positively charged residues adjacent to the putative TMD are marked in red (i). COS-7 cells were transfected with ECFP-TCRα and 24 hours later were stained with anti-Hsp60 antibody followed by a Cy3-conjugated secondary antibody (red) (ii). Fluorescent images were taken by fluorescence microscopy (100× oil-immersion objectives). Bar, 10 μM.
Rearranged TCRβ, harboring a complete V region and lacking the leader sequence, localizes to the mitochondrion and induces apoptosis. (A) Schematic presentation of the different constructs of a rearranged TCRβ (B) COS-7 cells were transfected with the indicated VDJC-TCRβ constructs and 24 hours later were either incubated with MTR for 30 minutes or stained with either anti-KDEL or anti-Hsp60 followed by anti–mouse Cy3 antibodies. The left column shows EGFP images, whereas the middle column shows the MTR or KDEL signal (i) or anti-Hsp60 (ii). The right column is the merge of the EGFP and MTR, KDEL or Hsp60 signal. Fluorescent images were taken by confocal microscopy (i) or fluorescence microscopy (100× oil-immersion objectives) (ii). Bar, 10 μM. (C) 293 cells were transfected with the indicated constructs and 48 hours after transfection were subjected to a flow cytometry cell-cycle analysis. Cells transfected with empty vector are pEGFP-F (Civ). (D) Schematic presentation of Leader-Flag-VDJC. (E) 293 cells were transfected with the indicated constructs. At 48 hours after transfection cells were subjected to a flow cytometric cell-cycle analysis. Cells transfected with an empty pCDNA3 vector are shown (Ev) as indicated. (F) The most COOH-terminal residues of mouse TCRα, β1, and β2 are shown. The positively charged residues adjacent to the putative TMD are marked in red (i). COS-7 cells were transfected with ECFP-TCRα and 24 hours later were stained with anti-Hsp60 antibody followed by a Cy3-conjugated secondary antibody (red) (ii). Fluorescent images were taken by fluorescence microscopy (100× oil-immersion objectives). Bar, 10 μM.
Degraded forms of TCRβ consisting of COOH-terminal end peptides naturally occur in thymocytes
The above experiments raised the question as to whether iJC-TCRβ–induced cell death is due to nonphysiologic overexpression of this molecule. To examine this issue we transfected 293 cells with EGFP-iJC and examined the fluorescence pattern by flow cytometry. As shown in Figure 7A, analysis of low fluorescing cells indicated that they nevertheless were undergoing apoptosis. Apparently, low titer of EGFP-iJC is therefore sufficient to induce apoptosis. To critically examine this point we determined the precise number of TCR molecules per cell, based on the known number of native TCR and Zap-70 molecules in T cells.28 In EGFP-iJC–transfected Jurkat cells the number of TCR molecules per cell was estimated to be 2.9 × 104. This value is in the range found in normal thymocytes (Figure 7Bi,ii). The ability of iJC-TCRβ to induce cell death, when present at relatively low peptide copies per cell, implies a possible similar role of truncated TCRβ forms in T lymphocytes.
Evidence for the physiologic role of incomplete forms of TCRβ in thymocytes. (A) 293 cells were transfected with the indicated constructs. At 48 hours after transfection the cells were fixed and stained with propidium iodide. Cells were than separated by analysis to different populations according to their GFP expression (i-iii,vii,viii). Each population was then further analyzed for its cell-cycle distribution. (B) Twenty-four hours after transfection with Zap70-YFP or EGFP-iJC, the protein extracts of Jurkat cells were loaded onto 10% SDS-PAGE gels. Immunoblot analyses were preformed with anti-Zap70 antibody and after stripping with anti-GFP (i). The 2 lanes shown were cut from the same blot. The calculated molecule/cell numbers of the rZap70-YFP or EGFP-iJC proteins are displayed in the table (ii). (C) The cell lines (S49.1 or SCID, 5 × 107 cells/sample) (i) or thymocytes (WT and TCR−/−, 2 × 108 cells/sample) (ii) were lysed, and TCRβ was immunoprecipitated by beads cross-linked to anti-TCRβ antibodies. Samples were separated on SDS-PAGE and probed with anti-TCRβ antibodies. The lack of corresponding bands in lymphocytes from SCID and TCR−/− cells show the specificity of the antibodies. Analysis of extracts from thymocytes from either untreated mice, cells from mice irradiated at 6 Gy (iii) or thymocytes at 1 or 4 weeks of age (iv) shows the presence of low-molecular-weight forms reactive with the antibody. The blot was cut (dotted line) and redeveloped to allow intensification of the lower molecular-weight bands (v). Peptides were isolated.
Evidence for the physiologic role of incomplete forms of TCRβ in thymocytes. (A) 293 cells were transfected with the indicated constructs. At 48 hours after transfection the cells were fixed and stained with propidium iodide. Cells were than separated by analysis to different populations according to their GFP expression (i-iii,vii,viii). Each population was then further analyzed for its cell-cycle distribution. (B) Twenty-four hours after transfection with Zap70-YFP or EGFP-iJC, the protein extracts of Jurkat cells were loaded onto 10% SDS-PAGE gels. Immunoblot analyses were preformed with anti-Zap70 antibody and after stripping with anti-GFP (i). The 2 lanes shown were cut from the same blot. The calculated molecule/cell numbers of the rZap70-YFP or EGFP-iJC proteins are displayed in the table (ii). (C) The cell lines (S49.1 or SCID, 5 × 107 cells/sample) (i) or thymocytes (WT and TCR−/−, 2 × 108 cells/sample) (ii) were lysed, and TCRβ was immunoprecipitated by beads cross-linked to anti-TCRβ antibodies. Samples were separated on SDS-PAGE and probed with anti-TCRβ antibodies. The lack of corresponding bands in lymphocytes from SCID and TCR−/− cells show the specificity of the antibodies. Analysis of extracts from thymocytes from either untreated mice, cells from mice irradiated at 6 Gy (iii) or thymocytes at 1 or 4 weeks of age (iv) shows the presence of low-molecular-weight forms reactive with the antibody. The blot was cut (dotted line) and redeveloped to allow intensification of the lower molecular-weight bands (v). Peptides were isolated.
We next set to examine whether T cells harbor short versions of TCRβ. To enable identification of both extracellular and intracellular TCR species, we used immunoprecipitation of TCRβ with monoclonal H57-597 anti-TCRβ C region antibodies, followed by immunoblotting with an anti-TCR antibody (H-197) rabbit polyclonal, raised against aa 122 to 312 of human TCRβ. This procedure detected 3 protein bands in the S49.1 T-cell line extract, that were absent from a T-cell line derived from SCID mice, showing the specificity of the polyclonal H-197 antibody to TCRβ gene-encoded peptides (Figure 7Ci). We further analyzed primary thymocytes. The specificity of the antibody was confirmed by comparison of thymocytes from C57BL/6 mice to those from TCR−/− mice (Figure 7Cii). Apart from an expected wide spread of molecules in the range of 37 to 45 kDa, lower bands were observed at weaker intensity down to approximately 28 kDa (Figure 7Ciii). The size of these lower molecular bands further reduced after exposure of mice to ionizing irradiation (Figure 7Ciii). These low molecular weight bands were found more abundantly in mouse thymus at 4 compared with 1 week of age (Figure 7Civ,v). Thymocyte protein extracts were then enriched for TCRβ using affinity column purification, followed by SDS-PAGE separation. Strips were cut from the blots at approximately 25 to 29 and 29 to 32 kDa, to avoid the major rearranged TCR band, and were examined by mass spectrometry. The sequencing data confirmed the presence of peptides derived from the Cβ (ATLVCLAR) and from 1 of the 2 optional Jβs that exist in the mouse, Jβ2.1 (VSLFEPSKAEIANK, and an overlapping sequence, VSLFEPSK). By contrast, no V region fragments could be detected. When slices were obtained from just below the major TCR band, both V (FLIGQEGQK) and Cβ (ATLVCLAR) region peptides were identified. These findings, along with the apparent molecular size of the molecules in the part of the gel examined, indicate the abundance of short peptides derived form TCRβ COOH-terminus.
Discussion
Our study is the first description of TCR molecules within the mitochondrion and specifically in the MOM. The ER localization is a default for membrane deposition of complete TCR molecules, whereas the mitochondrial localization seems to be a primary route for truncated-incomplete TCR forms. Biochemical analysis of normal and irradiated thymus cell extracts, confirmed the presence of short forms of TCRβ, containing peptide sequences from the COOH-terminal end of the molecule. These low molecular forms of TCRβ naturally occur at a far lower concentration, compared with membrane-associated TCR complex. This, however, is clearly expected because of the capacity of truncated TCR forms to mediate apoptosis.
Analysis of the molecular requirements for TCR localization in the mitochondrion showed that even a minimal truncation, involving removal of the leader sequence in the most NH2-terminal end of the molecule, was sufficient to direct the protein to the mitochondrion. However, most of the TCR chain is dispensable, as far as mitochondrial trafficking is concerned, because the TMD, which is located at its most COOH-terminus and the 5 residues that follow it, were sufficient for mitochondrial targeting. Proteins that are targeted to the membrane by a COOH-terminal hydrophobic tail are classified into a growing group, termed TA proteins34 (recently reviewed by Borgese et al35 ). The orientation of the insertion is such that their NH2-terminus, constituting most of the protein, is exposed to the cytosol, whereas the remaining residues, which are located at the most COOH-terminus of the protein, following the hydrophobic TMD, probably protrude into the intermembrane space. TA proteins are found in most cellular membranes; however, they are directly inserted in vivo only to the mitochondria, ER, and peroxisome membranes. The mechanisms of targeting and insertion of TA proteins to the different membranes are largely unsolved.36-44 A recent study uncovered a new complex, termed the TMD recognition complex, that is involved in the targeting of TA proteins to the ER.45 All the information that is required for the efficient targeting and insertion of TA proteins is concentrated in the most COOH-terminal portion, including the TMD, and is less than 20 aa, including the residues directly upstream or downstream to it.46,47 In addition, it was found that specific residues within the TMD are important for efficient mitochondrial targeting.48 However, mitochondrial targeting of TA proteins seems to occur through a targeting signal, containing strong positive charge both upstream and downstream to the hydrophobic portion, whereas a weak or complete lack of positive charge and a TMD of more than 20 residues targets proteins to ER.29,30,49,50 Because the 5 residues downstream to the TMD of TCRβ include 3 positively charged lysine residues, we assumed that they were responsible for its specific mitochondrial targeting. Indeed, constructs in which the 3 lysine residues were replaced with alanines lost most of the mitochondrial targeting and, instead, localized mostly to ER. Thus, although the mitochondrial targeting signal of incomplete TCRβ does not include a positive charge upstream to the TMD, it behaves as a genuine TA protein. This finding is in line with the new definition of the MOM targeting signal,51 suggesting that the net positive charge of all the residues downstream to the last uncharged residue of the TMD, of MOM-targeted TA, should be positive. In the case of TCRβ it is +2.
The mitochondrion is a central organ in the apoptotic pathway.52 Like several members of the TA group of proteins, incomplete forms of TCR that harbor mitochondrial localization sequences induce apoptosis, which is associated with cytochrome c release, may involve caspase activity and Bax and Bak signaling. The addition of a leader sequence at the NH2-terminus imitates the structure of rearranged forms of TCRβ and is expected to destine it to the ER. Indeed, this modification by itself was sufficient to override the mitochondrial targeting signal of the COOH-terminus and abolished apoptosis. Apparently, the leader sequence at the NH2-terminal end of these proteins is a sufficient and dominant signal, directing the molecules to the ER. Thus, the expression of complete rearranged forms of TCR, as single chains, would not promote apoptosis, as verified by our experiments (Figure 6E) and by previous observations reported by others.20,21,53
Incomplete TCR proteins in T cells could occur “intentionally” by induction of translation of germline transcripts such as iJC-TCRβ described in detail herein. From all the various Jβ2-Cβ2 transcripts that were identified, one that harbors Jβ2.6 (also termed Jβ2.7) contains an upstream in-frame intronic ATG and can potentially encode a 23-kDa protein. An additional transcript that contains of Jβ2.1 has a similar structure as evidenced by cloning and sequencing (results not shown) and by database genomic sequence (KO2802; X00934). However, an additional reported sequence (AE000665) does not corroborate the above. Thus, in the mouse there are 2 possible TCRβ chains (one containing Jβ2.1 and the other Jβ2.6 sequences). In addition, there are 14 TCRα variants that may exist as proteins. However, the mouse TCRα mRNA species lack the complete mitochondrial localization signal and do not target this organelle (Figure 6C). Incomplete TCR forms could also occur “accidentally” because of nonproductive rearrangement, giving rise to mRNA forms encoding truncated proteins. The D-J-C transcripts that are formed by such partial rearrangement include, sometimes, an upstream in-frame ATG codon and may encode proteins.54 TCR activation results in internalization of this receptor complex and subsequent degradation either by lysosomes or proteasome.55 Some degradation products could be truncated proteins that still harbor the COOH-terminus. We therefore propose that the trafficking of TCR molecules to the mitochondrion is part of the mechanism that protects the cells from overaccumulation of incomplete TCR molecules. Accumulation of the latter may introduce competition with complete TCR forms and slow down TCR complex formation and surface expression. In the more extreme scenario the truncated forms may function as dominant-negative moieties and block T-cell functions. A study of CD5 expression in B cells indicates that mRNA encoding a truncated form of this molecule causes reduced surface expression of the complete form of this protein.56
There are several known mechanisms for degradation of mammalian proteins. Proteasomes degrade short-lived proteins, as well as misfolded proteins in the ER, after their ubiquitination. Lysosomes hydrolyze protein complexes and organelles through autophagy. An additional pathway for cytosolic protein degradation by lysosomes is chaperone-mediated autophagy (CMA). In contrast to autophagy that entails formation of vacuoles, which fuse with lysosomes, in CMA proteins bind to specific receptors on lysosome membranes and are then moved to the lysosome lumen for degradation. We suggest an alternative pathway for defective TCRβ molecules, in which the mitochondrion is a sequestration site that accumulates partially formed or degraded TCR molecules. In this bound form the molecules are withheld from interfering with TCR complex formation. The apoptosis resulting from expression of incomplete TCR forms, taken together with the abundance of germline TCR mRNA in the thymus, suggests that the accumulation of such incomplete transcripts and the encoded proteins is one mode of execution of negative selection and the reason for extensive cell death in the thymus. This assumption is supported by our ability to demonstrate in the thymus short peptide forms of TCR, including sequences derived from the C region of TCRβ.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Ms Varda Segal for excellent assistance, Mr Guy Landau for help in the analysis of TCR expression level, and the staff of the Smoler Proteomics Center at the Technion, Haifa, Israel.
We thank the Charles and David Wolfson Charitable Trust for the support of Stem Cell Research at the Weizmann Institute of Science. This work was supported by the Gabrielle Rich Center for Transplantation Biology and the Helen and Martin Kimmel Institute for Stem Cell Research. D.Z. is an incumbent of the Joe and Celia Weinstein Professorial Chair at the Weizmann Institute of Science.
Authorship
Contribution: N.S., H.R.-L., and Y.P.-C. performed most of the experiments and contributed to writing the paper; K.S., M.C.-S., Y.S-T., and M.B.-S. performed part of the experiments; V.S. performed the electron microscopic study; and D.Z. was the group head, coordinated the study, and contributed to the writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dov Zipori, Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, 76100, Israel; e-mail: dov.zipori@weizmann.ac.il.

![Figure 1. iJC-TCRβ localizes to mitochondria in both T and non-T cells. (A) Schematic representation of EGFP-iJC and different modified and truncated versions of this molecule. (B) Different T cells [S49.1 (i), SCID.adh (ii)], and thymocytes from either wild-type (iii) or TCRβ-deficient mice (iv) were transfected with EGFP-iJC (green) and 5 hours later were stained with anti-Hsp60 antibody followed by a Cy3-conjugated secondary antibody (red) and with Hoechst for nuclear staining (blue). The marked area is magnified in the inset. Bar, 1 μM. (C) COS-7 cells were transfected with EGFP-iJC (green) and 24 hours later were incubated with MTR (red) for 30 minutes (i). COS-7 cells were transfected with Flag-iJC, 24 hours later the cells were incubated with MTR (red) for 30 minutes and stained by either anti-Flag (ii) or anti-TCR (iii) antibodies followed by Alexa 488–conjugated secondary antibody (green). COS-7 cells were transfected with EGFP-iJC (green) and 24 hours later stained with anti-Hsp60 antibody followed by a Cy3-conjugated secondary antibody (red) (iv). (D) The cell line 293T was transfected with EGFP or EGFP-iJC-TCRβ. Forty-eight hours after transfection cell fractionation was preformed, followed by Western analysis using anti-GFP antibody. Fluorescent images (B,Civ) were taken by fluorescence microscopy (100× oil-immersion objectives) and fluorescent images (Ci-iii) were taken by confocal microscopy. TCR mitochondrial localization was found in all of a total of 100 cells examined in each experimental group, within this figure and the figures below. Constructs are described in Figure 1A. Bars, 5 μM (B), 10 μM (C).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/15/10.1182_blood-2008-07-171405/7/m_zh80020929190001.jpeg?Expires=1769287334&Signature=iQa6phYJk7q0-FpR-XXvV-voGPa~rVYJGTsPtHVqsIXgCtu7rGp-WvlwyWc5j5Vj~sw33bXBiiu1r~h7uyMuOZzIurqW9oVvPNq5k-batZ5C2uXT3VdX32UxU56eABTL1Mw~23CFUL79MOjkBHhUD1E2CajDGA2-M~tC1sONQJHUOLjeo5l2P2Nq57zOVfAWgLeNVv8Ju6Vt-k9elfYY9FL7Qmrs4E3WvWTSWvS6lOhDU8Gg7ZNCm1CCobxhazhlxfN64lT86ruBARSfdJYF~rPv3FV9MaeJYk7p8Bt9IUmpd5rQbgq9urPJ7Au-tV5e3GziLq5qoMcYZfAO4Rq55g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)