Abstract
Chronic myelogenous leukemia (CML) is a hematopoietic disorder originating from p210BCR/ABL-transformed stem cells, which begins as indolent chronic phase (CP) but progresses into fatal blast crisis (BC). To investigate molecular mechanism(s) underlying disease evolution, CML-exhibiting p210BCR/ABL transgenic mice were crossed with BXH2 mice that transmit a replication-competent retrovirus. Whereas nontransgenic mice in the BXH2 background exclusively developed acute myeloid leukemia, p210BCR/ABL transgenic littermates developed nonmyeloid leukemias, in which inverse polymerase chain reaction detected 2 common viral integration sites (CISs). Interestingly, one CIS was transgene's own promoter, which up-regulated p210BCR/ABL expression. The other was the 5′ noncoding region of a transcription factor, Zfp423, which induced aberrant Zfp423 expression. The cooperative activities of Zfp423 and p210BCR/ABL were demonstrated as follows: (1) introduction of Zfp423 in p210BCR/ABL transgenic bone marrow (BM) cells increased colony-forming ability, (2) suppression of ZNF423 (human homologue of Zfp423) in ZNF423-expressing, p210BCR/ABL-positive hematopoietic cells retarded cell growth, (3) mice that received a transplant of BM cells transduced with Zfp423 and p210BCR/ABL developed acute leukemia, and (4) expression of ZNF423 was found in human BCR/ABL-positive cell lines and CML BC samples. These results demonstrate that enhanced expression of p210BCR/ABL and deregulated expression of Zfp423/ZNF423 contribute to CML BC.
Introduction
Chronic myelogenous leukemia (CML) is a hematopoietic disorder of multipotential stem cells, which exhibits excessive proliferation of immature and mature myeloid cells.1,2 The cytogenetic hallmark of CML is the Ph chromosome, created by t(9;22)(q34;q11),3 where the amino-terminal BCR gene on chromosome 22 is fused to most of the ABL proto-oncogene on chromosome 9, thereby creating an 8.5-kb BCR/ABL chimeric mRNA encoding a 210-kDa hybrid protein (p210BCR/ABL).4-6 p210BCR/ABL possesses a much higher kinase activity in comparison with the normal 145-kDa c-ABL,7 which is believed to play a critical role in the pathogenesis of the disease
The clinical course of CML is characterized by hematologically and temporally distinct stages.1,2 In the initial stage, called chronic phase (CP), the disease is indolent and the leukemic cells retain an ability to differentiate into mature granulocytes. After several years' duration of the chronic phase, however, the disease inevitably accelerates and ultimately progresses to the terminal fatal stage, called blast crisis (BC), which involves aggressive proliferation of immature blast cells. The frequent appearances of additional chromosomal abnormalities in the blast phase strongly suggest that superimposed genetic events would account for the disease evolution,8 but the underlying molecular mechanism(s) has remained largely unknown.
To understand the complex processes involved in the clinical course of human CML, it is necessary to develop animal models that express p210BCR/ABL and recapitulate the clinical features of the disease. Major attempts have been focused on bone marrow transplantation (BMT) experiments. Mice that have been lethally irradiated and received a transplant of bone marrow (BM) cells infected with p210BCR/ABL-expressing retroviruses exhibited a CML-like myeloproliferative disorder.9-11 On the other hand, generation of transgenic mice expressing p210BCR/ABL under various promoters also provides useful models.12-17 We generated p210BCR/ABL transgenic mice using the promoter from the mouse TEC gene, a gene encoding protein-tyrosine kinase preferentially expressed in hematopoietic progenitor cells.18,19 Although the founder mouse died of T-cell acute lymphoblastic leukemia (ALL) with a short latency, transgenic offspring reproducibly exhibited a myeloproliferative disorder after a long latency period.14 Peripheral blood smear showed remarkable myeloid hyperplasia with maturation, the BM was hypercellular with a predominance of myeloid cells at various stages of differentiation, and the spleen was enlarged with proliferation and expansion of myeloid cells.14 These pictures represent cardinal features of human CML, allowing us to consider these transgenic mice an animal model for CML.
To examine whether this transgenic model is applicable for investigating pathogenic processes from CP to BC of CML, we crossed p210BCR/ABL transgenic mice with mice heterozygous for p53, a gene frequently inactivated in CML BC, and generated mice transgenic for p210BCR/ABL and heterozygous for p53.20 Interestingly, p210BCR/ABL transgenic, p53 heterozygous mice died of acute leukemia with a short latency, and the analysis of p53 status revealed that the residual normal p53 allele was frequently and preferentially lost in the tumor tissues.20 In addition, we crossed p210BCR/ABL transgenic mice with Dok-1/Dok-2 knockout mice and showed that the absence of Dok-1 and Dok-2 accelerated the disease phenotype and caused BC, defining the role of Dok-1 and Dok-2 in tumor suppression.21 Based on these results, our transgenic mice can be regarded as a useful model for investigating molecular mechanism(s) underlying the progression from CP to BC of human CML.
In this report, to identify genes whose altered expression causes CML BC, p210BCR/ABL transgenic mice were subjected to retroviral insertional mutagenesis, by backcrossing to BXH2 mice, a recombinant inbred mouse strain that develop myeloid leukemia mainly due to a horizontally transmitted replication-competent retrovirus and intrinsic myeloid tropism induced by a mutation in the Icsbp1/Irf8 locus.22-24
Methods
Mice
p210BCR/ABL transgenic mice were generated as described.14 To allow for retroviral insertional mutagenesis, p210BCR/ABL transgenic males were backcrossed 4 generations to BXH2 females, because the ecotropic retrovirus in the BXH2 strain is transmitted to the progeny through the milk. Genotyping of the mice was carried out as described.14 All the mice used in this study were kept according to the guidelines of the Institute of Laboratory Animal Science, Hiroshima University, and all murine studies were approved by the animal care committee at the Japanese Foundation for Cancer Research.
Hematologic and pathologic analyses
Peripheral blood counts were routinely examined. Smears and stamp specimens of leukemic tissues were stained with Wright-Giemsa (WG). Tissues from dead or moribund animals were fixed in 10% buffered formaldehyde and examined by light microscopy. All organs were examined grossly and representative slices were prepared for hematoxylin-eosin staining.
Southern and Northern blot analyses
To detect gene rearrangements, genomic DNAs were digested with appropriate restriction enzymes and blotted with a genomic fragment adjacent to the integration site. For transgene promoter, a BglI-SmaI fragment in the promoter region was used as a probe, and for Zfp423, a genomic fragment generated by polymerase chain reaction (PCR) (primer sequences are 5′-GTGCGCACGTTTGTGAGGAGCTATA-3′ and 5′-CCAGCTATTCTGTCCAGGAGCAAGA-3′), which corresponds to a part of the first intron, was used as a probe. To detect RNA expression, total RNA extracted using TRIzol (Invitrogen, Carlsbad, CA) or mRNA purified using Oligo-Tex (Takara Bio, Tokyo, Japan) was blotted with p210BCR/ABL cDNA, Zfp423 cDNA, or a part of coding region of ZNF423 cDNA generated by genomic PCR (primer sequences are 5′-CAACCAGAAACACAAGTGCCCCATG-3′ and 5′-GTTGCAGTGGAAGGCAGAGATGTTG-3′).
RT-PCR
RNA was extracted using TRI zol. Reverse-transcription (RT)–PCR was performed as described (primer sequences are 5′-GAATGTCATCGTCCACTCAGCC-3′ and 5′-GGCCACAAAATCATACAGTGCA-3′ for p210BCR/ABL, 5′-GAGGATACCCCTACGACGTG-3′ and 5′-GACTTGTCACGCTGTTCCTGTC-3′ for Zfp423, and 5′-GGCATCAACCACGAGTGTAAGC-3′ and 5′-CTTCTGCGGAGAGGTGTCCTGT-3′ for ZNF423).25
Western blot analysis
Proteins extraction and Western blot were performed as described.14
Flow cytometric analysis
Cells were stained with monoclonal antibodies and second reagents. FITC-, PE-, and biotin-labeled monoclonal antibodies were purchased from BD PharMingen (San Diego, CA; Thy-1.2, CD19, CD45R/B220, Mac-1, Gr-1, and CD3) or from eBioscience (San Diego, CA; CD43, IgM, BP-1, and CD20). Biotinylated antibodies were revealed with streptavidin-APC (BD PharMingen). Clone 2.4G2 anti-CD32:CD16 was used to block Fc receptors. Fluorescence-activated cell sorter (FACS) analysis was performed on a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ), and the data were analyzed with FlowJo software (TreeStar, Ashland, OR).
Identification of retroviral integration sites
Genomic DNAs were digested with restriction enzymes, self-ligated, and subjected to inverse PCR as described,22 except that the CUA and CAU repeats were deleted from the secondary PCR primers. The position mapping on the mouse chromosome was done by BLAST searching using the University of Colombo School of Computing (UCSC) Genome Bioinformatics database (http://genome.ucsc.edu/)26 and the definition of a common integration site (CIS) was the same as in the mouse retrovirus tagged cancer gene database (RTCGD; http://rtcgd.abcc.ncifcrf.gov/).23,27
Retrovirus-mediated gene transfer, colony formation assay, and bone marrow transplantation
Retroviral preparation and retrovirus-mediated gene transfer were performed as described.28 For colony assay, BM cells of 5-fluorouracil (5FU)–treated transgenic or nontransgenic littermates were cultured in αMEM plus 20% FCS supplemented with 10 ng/mL IL-6, 10 ng/mL IL-3, and 100 ng/mL SCF (R&D Systems, Minneapolis, MN). Retrovirus was generated using plat-E cells29 and added into the medium containing BM cells with 6 mg/mL polybrene (Sigma-Aldrich, St Louis, MO), and retrovirus-infected BM cells were subjected to B-cell colony assay using MethoCult M3630 (StemCell Technologies, Vancouver, BC) that contains rhIL7. After 7 to 12 days' incubation, green colony numbers were counted under a fluorescent microscope.
For BM transplantation, BM cells extracted from 5FU-untreated Balb/c mice were cultured for 24 hours in IMDM plus 15% FCS supplemented with 10 ng/mL IL-6, 10 ng/mL IL-3, 100 ng/mL SCF, and 10 ng/mL IL-7 (R&D Systems). Retrovirus infection and BM transplantation were performed as described.30
Retrovirus-mediated transduction of shRNA for ZNF423
Two short hairpin RNA (shRNA) target sequences for ZNF423 (5′-GACATACCAGTGCATCAAG-3′ for shRNA-1 and 5′-CTGTAAGTTCTGCAGCAAG-3′ for shRNA-2) were chosen according to the siRNA Hairpin Oligonucleotide Sequence Designer (Clontech, Mountain View, CA). Annealed double-strand oligonucleotides were subcloned into RNA-ready pSIREN-retroQ retroviral expression vector (Clontech). Human hematopoietic cells were first transduced with ecotropic retrovirus receptor (EcoRVR) to render these cells competent for ecotropic retrovirus infection.31 Plat-E cells were then transfected with a pSIREN-retroQ vector harboring shRNA and the culture supernatant was used for infecting the ecotropic retrovirus to EcoRVR-expressing cells using Viro Mag (OZ Biosciences, Marseille, France). These procedures routinely yielded a high infection efficiency (∼ 60%) as judged by GFP fluorescence (not shown). Infected cells were selected with puromycin (0.4 mg/mL) for 2 weeks, subsequently cultured in puromycin-free medium for at least another 2 weeks, and subjected to Northern blot and cell proliferation assay.
Cell proliferation assay
On day 1, 105 cells of the parental and shRNA-transduced sublines were plated in a 10-cm2 dish and cultured in RPMI plus 10% FCS. Cell numbers were counted on day 3 and day 5.
Cell lines and patient samples
Ph-positive and Ph-negative human hematopoietic cell lines were kindly provided by Drs Hiroya Aso (Hiroshima, Japan) and Toshiya Inaba (Hiroshima, Japan). Patient samples were taken after informed consent was obtained in accordance with the Declaration of Helsinki and approval from the institutional review board at Hiroshima University was granted.32 Diagnosis of CML CP or CML BC (myeloid or B-lymphoid lineage) was performed based on morphologic, cytogenetic, immunophenotypic, and molecular analyses.
Results
Acute leukemias in p210BCR/ABL transgenic mice on a BXH2 background
To identify gene(s) whose alteration by retrovirus insertion contributes to blast crisis of CML, p210BCR/ABL transgenic mice were backcrossed to BXH2 mice that contain and transmit a replication-competent retrovirus. p210BCR/ABL transgenic and wild-type (nontransgenic) littermates from the N4 BXH2 backcross generation were used for this study (designated as p210BCR/ABL/BXH2 and WT/BXH2, respectively).
WT/BXH2 mice began to develop acute leukemia at 6 months after birth (Figure 1A thin continuous line). Macroscopically, the leukemic mice exhibited hepatosplenomegaly and lymph node (LN) swelling, which were occasionally associated with thymic enlargement. Pathologic analysis showed that leukemic cells having morphology of myeloblasts proliferated in the peripheral blood and infiltrated into the liver, spleen, LNs, and other tissues (data not shown). Flow cytometric analysis of the leukemic tissues showed that the blast cells were exclusively positive for Mac-1 and Gr-1 but negative for Thy1.2 and CD19, indicating that they all were of myeloid origin (data not shown).
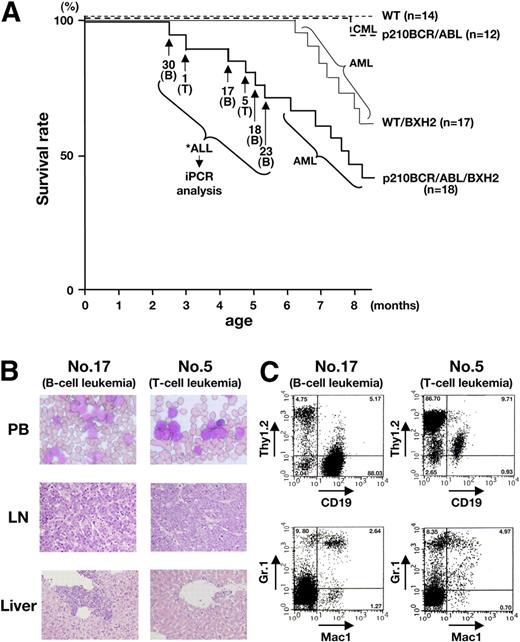
Survival curves and pathologic and flow cytometric analyses of leukemic mice. (A) Survival curves of the mice. The survival curves of WT/BXH2 and p210BCR/ABL/BXH2 are shown by thin and thick continuous lines, respectively, whereas those of BXH2-nonbackcrossed WT and p210BCR/ABL are shown by thin and thick dotted lines, respectively. As for the 6 p210BCR/ABL/BXH2 animals that died in a short latency and exhibited nonmyeloid phenotypes (nos. 30, 1, 17, 5, 18, and 23), the death points are indicated by → and the immunophenotypes of the disease are shown in the parentheses. T indicates T-cell leukemia; and B, B-cell leukemia. (B) Pathologic analysis of the leukemic mice. WG-stained peripheral blood smears (PB) and HE-stained lymph node (LN) and liver slices of a representative mouse for B-cell leukemia (no. 17) or T-cell leukemia (no. 5) are shown. PB smears show proliferation of blast cells and LN specimen shows the destruction of the basal structure by blast cell infiltration. In the liver, blast cells are observed around the vessel and in the sinusoids. (C) Flow cytometric analysis of mice that developed B-cell or T-cell leukemia. Blast cells of no. 17 were positive for CD19 but negative for Thy1.2, Mac-1, and Gr-1 and those of no. 5 were positive for Thy1.2 but negative for CD19, Mac-1, and Gr-1, indicating that they were of B- and T-lymphoid origins, respectively. The percentages of positive cells in each quadrant are shown.
Survival curves and pathologic and flow cytometric analyses of leukemic mice. (A) Survival curves of the mice. The survival curves of WT/BXH2 and p210BCR/ABL/BXH2 are shown by thin and thick continuous lines, respectively, whereas those of BXH2-nonbackcrossed WT and p210BCR/ABL are shown by thin and thick dotted lines, respectively. As for the 6 p210BCR/ABL/BXH2 animals that died in a short latency and exhibited nonmyeloid phenotypes (nos. 30, 1, 17, 5, 18, and 23), the death points are indicated by → and the immunophenotypes of the disease are shown in the parentheses. T indicates T-cell leukemia; and B, B-cell leukemia. (B) Pathologic analysis of the leukemic mice. WG-stained peripheral blood smears (PB) and HE-stained lymph node (LN) and liver slices of a representative mouse for B-cell leukemia (no. 17) or T-cell leukemia (no. 5) are shown. PB smears show proliferation of blast cells and LN specimen shows the destruction of the basal structure by blast cell infiltration. In the liver, blast cells are observed around the vessel and in the sinusoids. (C) Flow cytometric analysis of mice that developed B-cell or T-cell leukemia. Blast cells of no. 17 were positive for CD19 but negative for Thy1.2, Mac-1, and Gr-1 and those of no. 5 were positive for Thy1.2 but negative for CD19, Mac-1, and Gr-1, indicating that they were of B- and T-lymphoid origins, respectively. The percentages of positive cells in each quadrant are shown.
In contrast to the WT/BXH2 mice, several p210BCR/ABL/BXH2 mice (named as nos. 30, 1, 17, 5, 18, and 23; Figure 1A thick continuous line) developed nonmyeloid leukemias with a shorter latency. Among them, 4 mice (nos. 30, 17, 18, and 23) displayed splenomegaly and LN swelling but did not show apparent hepatomegaly. The other 2 mice (nos. 1 and 5) exhibited massive thymic enlargement with pleural effusion and splenomegaly. Pathologic analysis showed that leukemic cells having morphology of lymphoblasts were evident in the peripheral blood and infiltration of the blast cells was observed in the LNs, liver, and other tissues examined (Figure 1B and not shown). Flow cytometric analysis revealed that the leukemic cells of the former 4 mice (nos. 30, 17, 18, and 23) were positive for CD19 but negative for Thy1.2, Mac-1, and Gr-1, and those of the latter 2 mice (nos. 1 and 5) were positive for Thy1.2 but negative for CD19, Mac-1, and Gr-1, indicating that they were of B-lymphoid and T-lymphoid origins, respectively (Figure 1C and not shown). Three CD19+ samples (nos. 17, 18, and 30) were further analyzed with antibodies against CD20, B220, BP-1, CD43, and IgM to investigate the differentiation stages (pro-B, pre-B, or mature B). As shown in Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article), all the samples were positive for CD20, B220, BP-1, and CD43 but negative for IgM, indicating that they were pre–B-cell leukemias.
The characteristics of the 6 p210BCR/ABL/BXH2 leukemic mice with an early disease onset are summarized in Table 1. As for the leukemias developed in the remaining p210BCR/ABL/BXH2 mice after 6 months of age, macroscopic appearances and the results of flow cytometric analysis were indistinguishable from those of the WT/BXH2 mice (data not shown). During the observation period, no mice developed hematologic disease in BXH2-nonbackcrossed WT mice and one mouse died of CML in BXH2-nonbackcrossed p210BCR/ABL transgenic mice (Figure 1A thin and thick dotted lines, respectively).
Enhanced expression of p210BCR/ABL and aberrant expression of Zfp423 in the leukemic tissues with B-cell phenotype
We focused on the 6 p210BCR/ABL/BXH2 mice that developed nonmyeloid leukemias in a shortened period, because diseases in these mice would not be only due to the BXH2 background-derived intrinsic mechanism but caused by cooperation of p210BCR/ABL with retrovirus-inserted altered gene expression. To identify virus-affected genes in these mice, inverse PCR (iPCR) was performed and sequences of PCR fragments were subjected to BLAST searching using the UCSC Genome Bioinformatics database. Among candidate genes (listed in Table S1), we found 2 common integration sites (CISs) in B-lineage leukemias (shown by asterisks and in boldface in Table S1).
The first one was the promoter region of the mouse TEC gene. This CIS was found in nos. 17 and 30 and the viral integration sites were approximately 1.5-kb and approximately 200-bp upstream of the transcription initiation site,19 respectively. Interestingly, in both cases, sequencing of the entire PCR fragment revealed that the mouse TEC promoter sequences were interrupted at +22 from the transcription initiation site and followed by human BCR/ABL cDNA. This result indicated that the integration sites were not in the endogenous mouse TEC gene but in the promoter region of the transgene itself. Another CIS observed in nos. 18 and 23 was in the noncoding region of the first exon of mouse Zfp423 (Zinc finger protein 423, also known as Early B-cell factor-associated zinc-finger protein, Ebfaz) gene.33 In these cases, the retroviruses were integrated almost in the same position, approximately 100-bp upstream of the translational initiation ATG.33 The schematic models of the integration sites are shown in Figure 2A.
Retrovirus integration sites, genomic rearrangements, and altered gene expressions in mice with B-cell leukemia. (A) Schematic models of retrovirus integration sites. The retrovirus integration sites are indicated by vertical arrows. The left panel illustrates the transgene structure, where the mouse TEC promoter, p210BCR/ABL cDNA, and polyA and splicing signals are shown by dotted, filled, and shaded boxes, respectively. In mice nos. 17 and 30, retroviruses were integrated approximately 1.5-kb and approximately 200-bp upstream of the transcriptional initiation site, respectively. In the right panel, the noncoding and coding regions of Zfp423 exon 1 are shown by blank and filled boxes, respectively. In mice nos. 18 and 23, the viral integration occurred almost in the same site, approximately 100-bp upstream of the translational initiation site. The positions of probes used for Southern blots are also shown. (B) Southern blots to confirm the CISs as major integration sites. Genomic DNAs extracted from the spleen of a control transgenic mouse (C) and tumor tissues of the diseased mice (nos. 17, 30, 18, and 23) were digested with BamHI and blotted with a DNA fragment adjacent to the integration site. Probe A (A) was used for transgene rearrangement (nos. 17 and 30, left panel) and probe B was used for Zfp423 gene rearrangement (nos. 18 and 23, right panel). The positions of germline (G) and rearranged bands are indicated by → and ◀, respectively. Molecular markers are shown on the left. (C) Enhanced expression of p210BCR/ABL in mice nos. 17 and 30 and up-regulated expression of Zfp423 in mice nos. 18 and 23. For detecting p210BCR/ABL message, 20 μg total RNAs extracted from the spleen of a control p210BCR/ABL transgenic mouse (C) and tumor tissues of the diseased mice (nos. 17, 30, 18, and 23) were blotted with p210BCR/ABL cDNA (top left panel) and for detecting Zfp423 message, 3 μg mRNAs from the same tissues were blotted with a part of Zfp423 cDNA (top right panel). The result of β-actin hybridization is shown as an internal control. Molecular markers are shown on the left and the positions of p210BCR/ABL and Zfp423 messages are indicated by →. Enhanced expression of p210BCR/ABL protein in mice nos. 17 and 30 was detected by blotting the proteins extracted from the same tissues with an anti-ABL antibody (bottom left panel). Protein markers are shown on the left and the positions of p210BCR/ABL and c-ABL (145 kDa) are indicated by →.
Retrovirus integration sites, genomic rearrangements, and altered gene expressions in mice with B-cell leukemia. (A) Schematic models of retrovirus integration sites. The retrovirus integration sites are indicated by vertical arrows. The left panel illustrates the transgene structure, where the mouse TEC promoter, p210BCR/ABL cDNA, and polyA and splicing signals are shown by dotted, filled, and shaded boxes, respectively. In mice nos. 17 and 30, retroviruses were integrated approximately 1.5-kb and approximately 200-bp upstream of the transcriptional initiation site, respectively. In the right panel, the noncoding and coding regions of Zfp423 exon 1 are shown by blank and filled boxes, respectively. In mice nos. 18 and 23, the viral integration occurred almost in the same site, approximately 100-bp upstream of the translational initiation site. The positions of probes used for Southern blots are also shown. (B) Southern blots to confirm the CISs as major integration sites. Genomic DNAs extracted from the spleen of a control transgenic mouse (C) and tumor tissues of the diseased mice (nos. 17, 30, 18, and 23) were digested with BamHI and blotted with a DNA fragment adjacent to the integration site. Probe A (A) was used for transgene rearrangement (nos. 17 and 30, left panel) and probe B was used for Zfp423 gene rearrangement (nos. 18 and 23, right panel). The positions of germline (G) and rearranged bands are indicated by → and ◀, respectively. Molecular markers are shown on the left. (C) Enhanced expression of p210BCR/ABL in mice nos. 17 and 30 and up-regulated expression of Zfp423 in mice nos. 18 and 23. For detecting p210BCR/ABL message, 20 μg total RNAs extracted from the spleen of a control p210BCR/ABL transgenic mouse (C) and tumor tissues of the diseased mice (nos. 17, 30, 18, and 23) were blotted with p210BCR/ABL cDNA (top left panel) and for detecting Zfp423 message, 3 μg mRNAs from the same tissues were blotted with a part of Zfp423 cDNA (top right panel). The result of β-actin hybridization is shown as an internal control. Molecular markers are shown on the left and the positions of p210BCR/ABL and Zfp423 messages are indicated by →. Enhanced expression of p210BCR/ABL protein in mice nos. 17 and 30 was detected by blotting the proteins extracted from the same tissues with an anti-ABL antibody (bottom left panel). Protein markers are shown on the left and the positions of p210BCR/ABL and c-ABL (145 kDa) are indicated by →.
To confirm that these CISs were major integration sites in the leukemic samples, Southern blot was performed using a genomic DNA fragment adjacent to the integration site. DNA extracted from a spleen of a BXH2-nonbackcrossed p210BCR/ABL transgenic mouse was used as a negative control. As shown in Figure 2B, a rearranged band is evident in each sample (indicated by an arrowhead in Figure 2B), indicating that tumor cells with the CISs were predominant in the related tumors and were clonal in origin. The clonality and B-cell commitment of the leukemic cells in these mice (nos. 17, 30, 18, and 23) were further demonstrated by Southern blot using a mouse JH probe (Figure S2).
To investigate the alteration in gene expression of p210BCR/ABL and Zfp423 by virus integration, RNAs extracted from tumor tissues of the 4 leukemic mice were blotted with p210BCR/ABL cDNA or mouse Zfp423 cDNA. RNA extracted from a spleen of a BXH2-nonbackcrossed p210BCR/ABL transgenic mouse was used as control. The results are shown in upper panels of Figure 2C.
As for p210BCR/ABL, it is not surprising that the p210BCR/ABL message was not detected in the control transgenic mouse spleen (Figure 2C top left panel, “C”), because our previous data showed that the basal transgene expression was quite low, probably due to the nature of the promoter used (Honda et al14 and data not shown). In contrast, a clear p210BCR/ABL message was evident in the tumors of nos. 17 and 30 (∼ 7 kb, Figure 2C top left top panel, arrow). The quantitative p210BCR/ABL mRNA expression in these samples is shown in Figure S3. As expected from the result of the Northern blot (Figure 2C top left panel), the p210BCR/ABL mRNA in the control transgenic spleen was quite low and was significantly enhanced by the transgene integration (nos. 17 and 30). The enhanced expression of p210BCR/ABL at the protein level was confirmed by Western blot using an anti-ABL antibody (Figure 2C bottom left panel).
As for Zfp423, no clear message was observed in the control transgenic spleen (Figure 2C top right panel, “C”), which is in accordance with our previous report showing that Zfp423 message was barely detectable in the spleen when using polyA+ RNA.34 In contrast, in nos. 18 and 23, an enhanced expression of the Zfp423 message was observed (∼ 6 kb, Figure 2C top right panel, arrow). These results indicated that the retrovirus integrations up-regulated p210BCR/ABL expression and induced aberrant Zfp423 expression.
Expression of Zfp423 in transgenic BM cells enhanced B-cell colony-forming ability, and suppression of ZNF423 in ZNF423-expressing, p210BCR/ABL-positive CML BC cells retarded cell growth
We next investigated the effect of up-regulation and down-regulation of Zfp423 on the proliferative ability of p210BCR/ABL-positive cells. We first examined whether introduction of Zfp423 confers a growth advantage to transgenic BM cells by a colony formation assay. BM cells purified from wild-type (WT) or p210BCR/ABL transgenic mice were infected with control pMysIG or Flag-HA–tagged Zfp423 (FHZfp423)–expressing pMysIG (pMysIG/FHZfp423) retrovirus, and the infected cells were cultured in methylcellulose-based media (Figure 3A). Because mice with Zfp423 activation (nos. 18 and 23) developed B-lineage leukemia, the virus-infected cells were subjected to a B-cell colony assay, and as the retrovirus vector contains GFP as a detection marker, colonies with green fluorescence were counted.
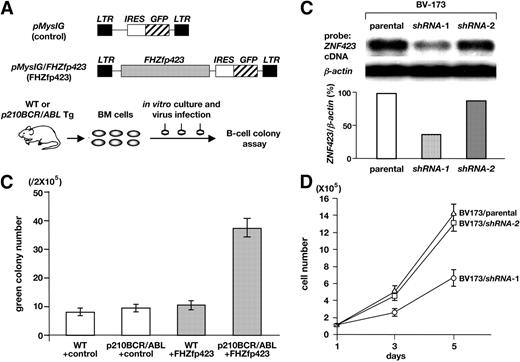
Effects of Zfp423 expression on the colony formation and proliferation of p210BCR/ABL-expressing cells. (A) Schematic structures of the retroviruses and the illustration of the experimental procedure. BM cells were extracted from WT or p210BCR/ABL transgenic mice, infected with empty retrovirus (pMyIG, control) or Flag-HA–tagged Zfp423 (FHZfp423)–expressing retrovirus (pMyIG/FHZfp423, FHZfp423), and subjected to the B-cell colony assay. (B) Results of B-cell colony assay. The mean green colony number of 3 independent experiments for each group (WT+conrol, p210BCR/ABL+control, WT+FHZfp423, and p210BCR/ABL+FHZfp423) is shown with error bars. (C) Suppression of ZNF423 expression by shRNAs. mRNA (5 μg) extracted from the parental BV-173 line and 2 shRNA-introduced sublines (shRNA-1 and shRNA-2) were blotted with a part of the human ZNF423 coding region. β-Actin hybridization was performed as an internal control and the relative expression ratio of ZNF423 to β-actin in each cell line is shown as a vertical column. (D) Results of cell proliferation assay. Cells of the parental BV-173 line and 2 shRNA-introduced sublines (BV-173/shRNA-1 and BV-173/shRNA-2) were plated at a density of 105/10 cm2 on day 1 and cell numbers were counted on day 3 and day 5. The mean cell number of 3 independent experiments of each line is plotted with error bars.
Effects of Zfp423 expression on the colony formation and proliferation of p210BCR/ABL-expressing cells. (A) Schematic structures of the retroviruses and the illustration of the experimental procedure. BM cells were extracted from WT or p210BCR/ABL transgenic mice, infected with empty retrovirus (pMyIG, control) or Flag-HA–tagged Zfp423 (FHZfp423)–expressing retrovirus (pMyIG/FHZfp423, FHZfp423), and subjected to the B-cell colony assay. (B) Results of B-cell colony assay. The mean green colony number of 3 independent experiments for each group (WT+conrol, p210BCR/ABL+control, WT+FHZfp423, and p210BCR/ABL+FHZfp423) is shown with error bars. (C) Suppression of ZNF423 expression by shRNAs. mRNA (5 μg) extracted from the parental BV-173 line and 2 shRNA-introduced sublines (shRNA-1 and shRNA-2) were blotted with a part of the human ZNF423 coding region. β-Actin hybridization was performed as an internal control and the relative expression ratio of ZNF423 to β-actin in each cell line is shown as a vertical column. (D) Results of cell proliferation assay. Cells of the parental BV-173 line and 2 shRNA-introduced sublines (BV-173/shRNA-1 and BV-173/shRNA-2) were plated at a density of 105/10 cm2 on day 1 and cell numbers were counted on day 3 and day 5. The mean cell number of 3 independent experiments of each line is plotted with error bars.
The results are shown in Figure 3B. No obvious difference in the colony numbers was found between control pMysIG virus-infected WT (WT+control) and p210BCR/ABL transgenic (p210BCR/ABL+control) BM cells. This result indicates that the basal p210BCR/ABL expression in this transgenic system does not affect the proliferative ability of B cells, probably due to the nature of the promoter used, and is in accordance with our observation that the p210BCR/ABL transgenic mice have not developed B-cell disease so far.14 pMysIG/FHZfp423-infected WT BM cells (WT+FHZfp423) showed a slight increase in the colony number in this system. In contrast, pMysIG/FHZfp423-infected p210BCR/ABL transgenic BM cells (p210BCR/ABL+FHZfp423) generated a significantly increased number of colonies.
We next tried to down-regulate endogenous ZNF423 (the human homologue of Zfp423) by RNA interference and examined its effect on the growth rate. We designed 2 short hairpin RNAs targeted to ZNF423 mRNA (shRNA-1 and shRNA-2) and introduced them into BV-173, a p210BCR/ABL-positive and ZNF423-expressing human hematopoietic cell line (see Figure 5A). As shown in Figure 3C, introduction of shRNA-1 effectively decreased ZNF423 mRNA to approximately 40% of that in the parental cells, whereas shRNA-2 was less effective. Concurrently, as shown Figure 3D, BV-173 cells transduced with shRNA-1 displayed a significantly reduced growth rate, whereas cells expressing shRNA-2 showed only marginal growth retardation. To confirm that the shRNAs did not affect the growth of cells without ZNF423 expression, the same shRNAs were introduced into KOPN67, a p210BCR/ABL-positive but ZNF423-nonexpressing cell line (see Figure 5A). As expected, no difference in cell growth was observed in the parental line and shRNAs-transduced sublines (Figure S4), confirming the specificity of the shRNAs on the ZNF423-dependent cell growth. These results indicated that Zfp423/ZNF423 cooperated with p210BCR/ABL and enhanced proliferation of p210BCR/ABL-expressing hematopoietic cells.
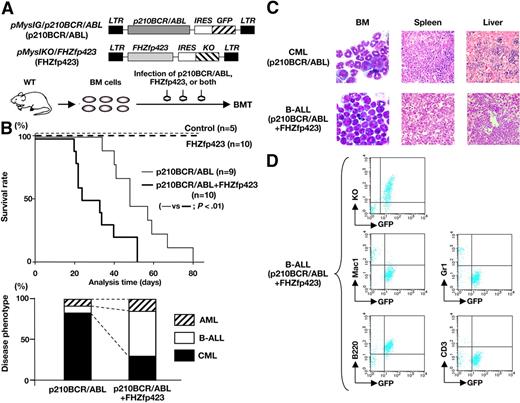
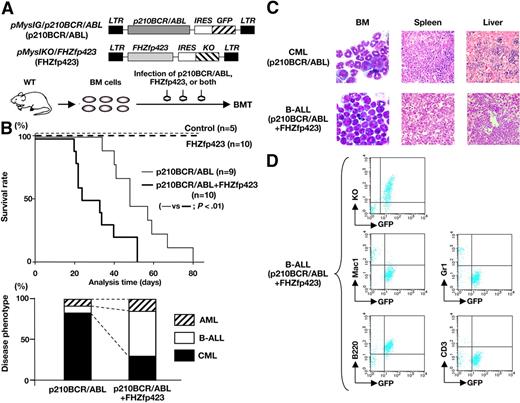
Survival and disease phenotype in mice that received a transplant of p210BCR/ABL- and/or FHZfp423-expressing BM cells. (A) Schematic structures of the retroviruses and the illustration of the experimental procedure. BM cells infected with p210BCR/ABL-expressing retrovirus (pMyIG/p210BCR/ABL), FHZfp423-expressing retrovirus (pMyIKO/FHZfp423), or both types of viruses were subjected to the BMT assay. KO indicates Kusabira Orange. (B) Acceleration of disease onset and altered disease phenotype by cotransduction of Zfp423 and p210BCR/ABL. In the top panel, survival curves of mice reconstituted with BM cells transduced with control retrovirus (control, n = 5), pMyIKO/FHZfp423 (FHZfp423, n = 10), pMyIG/p210BCR/ABL (p210BCR/ABL, n = 9), and both viruses (p210BCR/ABL+FHZfp423, n = 10) are shown as thin dotted, thick dotted, thin continuous, and thick continuous lines, respectively. In the bottom panel, the percentages of samples diagnosed as CML, B-ALL, and AML are shown by black, white, and shaded boxes, respectively. (C) Representative results of pathologic analysis of CML and B-ALL developed in mice transduced with p210BCR/ABL and p210BCR/ABL+FHZfp423, respectively. In the BM smears, proliferation of differentiated myeloid cells is observed in the CML case (top left panel), whereas monotonous proliferation of immature lymphoid tumor cells is apparent in the B-ALL case (bottom left panel). Massive infiltration of leukemic cells is shown in the spleen and liver (middle and right panels). (D) Representative results of flow cytometric analysis of B-ALL developed in mice transduced with p210BCR/ABL and FHZfp423. Leukemic cells are positive for both GFP and KO (top panel), confirming that they were originated from hematopoietic progenitor cells infected with both p210BCR/ABL and FHZfp423. GFP-positive leukemic cells showed positive staining for B220, but are negative for Mac-1, Gr-1, and CD3 (middle and bottom panels).
Survival and disease phenotype in mice that received a transplant of p210BCR/ABL- and/or FHZfp423-expressing BM cells. (A) Schematic structures of the retroviruses and the illustration of the experimental procedure. BM cells infected with p210BCR/ABL-expressing retrovirus (pMyIG/p210BCR/ABL), FHZfp423-expressing retrovirus (pMyIKO/FHZfp423), or both types of viruses were subjected to the BMT assay. KO indicates Kusabira Orange. (B) Acceleration of disease onset and altered disease phenotype by cotransduction of Zfp423 and p210BCR/ABL. In the top panel, survival curves of mice reconstituted with BM cells transduced with control retrovirus (control, n = 5), pMyIKO/FHZfp423 (FHZfp423, n = 10), pMyIG/p210BCR/ABL (p210BCR/ABL, n = 9), and both viruses (p210BCR/ABL+FHZfp423, n = 10) are shown as thin dotted, thick dotted, thin continuous, and thick continuous lines, respectively. In the bottom panel, the percentages of samples diagnosed as CML, B-ALL, and AML are shown by black, white, and shaded boxes, respectively. (C) Representative results of pathologic analysis of CML and B-ALL developed in mice transduced with p210BCR/ABL and p210BCR/ABL+FHZfp423, respectively. In the BM smears, proliferation of differentiated myeloid cells is observed in the CML case (top left panel), whereas monotonous proliferation of immature lymphoid tumor cells is apparent in the B-ALL case (bottom left panel). Massive infiltration of leukemic cells is shown in the spleen and liver (middle and right panels). (D) Representative results of flow cytometric analysis of B-ALL developed in mice transduced with p210BCR/ABL and FHZfp423. Leukemic cells are positive for both GFP and KO (top panel), confirming that they were originated from hematopoietic progenitor cells infected with both p210BCR/ABL and FHZfp423. GFP-positive leukemic cells showed positive staining for B220, but are negative for Mac-1, Gr-1, and CD3 (middle and bottom panels).
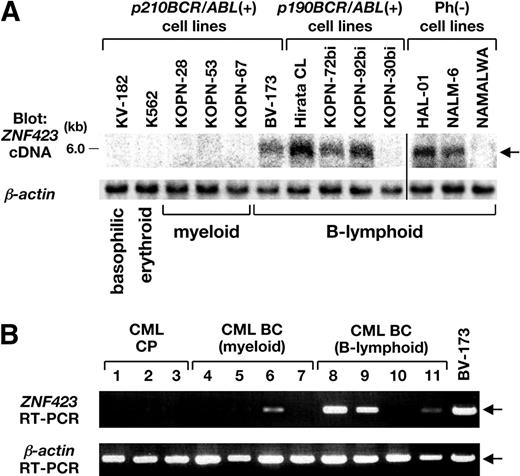
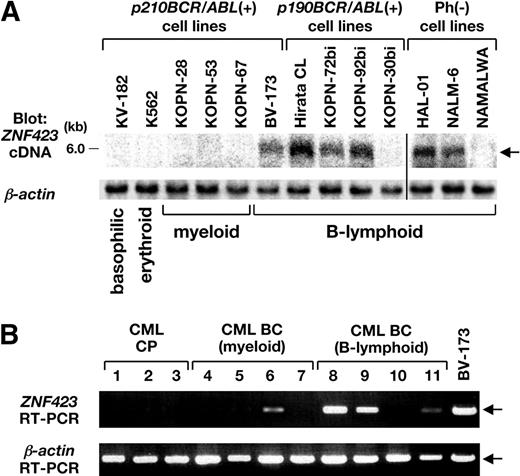
Expression of ZNF423 in BCR/ABL-positive cell lines and in CML BC samples. (A) mRNA (3 μg) extracted from 6 p210BCR/ABL-positive, 4 p190BCR/ABL-positive, and 3 Ph-negative cell lines was blotted with a part of the human ZNF423 coding region. β-Actin hybridization was performed as an internal control. The position of the ZNF423 message is indicated by → and the immunophenotypes of the cell lines are shown at the bottom. A vertical line has been inserted to indicate a repositioned gel lane. (B) Total RNAs extracted BM samples from 3 CML CP and 8 CML BC cases (4 myeloid and 4 B-lymphoid) were subjected to RT-PCR for ZNF423 expression. β-Actin RT-PCR was performed as an internal control.
Expression of ZNF423 in BCR/ABL-positive cell lines and in CML BC samples. (A) mRNA (3 μg) extracted from 6 p210BCR/ABL-positive, 4 p190BCR/ABL-positive, and 3 Ph-negative cell lines was blotted with a part of the human ZNF423 coding region. β-Actin hybridization was performed as an internal control. The position of the ZNF423 message is indicated by → and the immunophenotypes of the cell lines are shown at the bottom. A vertical line has been inserted to indicate a repositioned gel lane. (B) Total RNAs extracted BM samples from 3 CML CP and 8 CML BC cases (4 myeloid and 4 B-lymphoid) were subjected to RT-PCR for ZNF423 expression. β-Actin RT-PCR was performed as an internal control.
Zfp423 accelerated disease onset of p210BCR/ABL-induced leukemia and increased incidence of B-ALL
We then investigated the in vivo cooperative activity of Zfp423 and p210BCR/ABL by a BMT approach. Because p210BCR/ABL transgenic mice were not congenic enough for BMT, we generated p210BCR/ABL-expressing retrovirus and FHZfp423-expressing retrovirus separately, and infected them with BM cells of Balb/c mice, the strain that has been successfully used to develop CML in the BMT experiments.10,11 And because Zfp423-positive leukemias (nos. 18 and 23) were of B-cell phenotype, BM cells were not pretreated with 5FU and in vitro cultures were performed using a cytokine cocktail with IL-6, IL-3, SCF, and IL-7, as previously described.11
BM cells infected with control retrovirus, p210BCR/ABL-expressing retrovirus, FHZfp423-expressing retrovirus, or both types of viruses were transplanted into sublethally irradiated syngeneic mice (Figure 4A). To detect p210BCR/ABL- and FHZfp423-positive cells by flow cytometry, cells expressing p210BCR/ABL and FHZfp423 were labeled with GFP and KO (Kusabira Orange),35 respectively (Figure 4A). The protein expression of the inserted cDNA by retrovirus infection was confirmed by Western blot (Figure S5).
The mice that underwent transplantation were continuously observed and peripheral blood parameters were routinely examined for morphologic changes by Wight-Giemsa staining and for GFP and/or KO positivities by flow cytometry. The survival rate of each group evaluated using the Kaplan-Meier test is shown in the upper panel of Figure 4B. No disease developed in the control virus-transduced mice. In addition, no hematologic abnormalities were observed in FHZfp423-transduced mice, indicating that overexpression of Zfp423 does not possess a transforming ability on primary hematopoietic cells, which is in accordance with the result that introduction of Zfp423 in BM cells did not apparently increase colony numbers (Figure 3B). As expected from the results of previous studies, most of the p210BCR/ABL-transduced mice developed CML, except 2 cases that developed acute myeloid leukemia (AML) and B-cell ALL (B-ALL, Figure 4B bottom panel, left bar). In contrast, mice transduced with both types of viruses died in a shortened period and exhibited different phenotypes. The mean survival periods of mice reconstituted with p210BCR/ABL+FHZfp423 and those reconstituted with p210BCR/ABL alone were 29.5 and 48 days, respectively (Figure 4B top panel, thick and thin continuous lines), and the difference was statistically significant (P < .01). In addition, compared with mice reconstituted with p210BCR/ABL, those reconstituted with p210BCR/ABL+FHZfp423 exhibited an increased incidence of B-ALL (Figure 5B bottom panel, right bar), although the difference was not statistically significant (P = .119), probably due to the limited sample numbers. These results demonstrated that Zfp423 possesses a cooperative oncogenecity with p210BCR/ABL in vivo, which accelerated disease onset and induced a more aggressive phenotype mainly of B-cell lineage. The representative results of pathologic analyses of mice that developed CML by p210BCR/ABL and that developed B-ALL by p210BCR/ABL plus FHZfp423 are shown in Figure 4C and the results of flow cytometry of the latter are shown in Figure 4D. The expression of p210BCR/ABL and Zfp423 mRNAs in tumors developed in p210BCR/ABL plus FHZfp423-transduced mice was confirmed by RT-PCR (Figure S6).
Expression of ZNF423 in human BCR/ABL-positive hematopoietic cell lines and CML BC samples
We finally investigated the clinical relevance of ZNF423 expression in the progression from CML CP to BC using human Ph-positive hematopoietic cell lines and clinical samples. For the cell line experiment, cells expressing p190BCR/ABL (an alternative form of the BCR/ABL fusion gene) were also examined, because the expression of p190BCR/ABL is exclusively associated with B-ALL,36 the same phenotype as the leukemias developed in the mice with Zfp423 integration (nos. 18 and 23). In addition, Ph-negative B-ALL lines were included in this study to investigate the role of ZNF423 in the development of B-cell malignancy without BCR/ABL.
mRNAs extracted from 6 p210BCR/ABL-positive cell lines (1 B-lymphoid, 3 myeloid, 1 erythroid, and 1 basophilic), 4 p190BCR/ABL-positive B-lymphoid cell lines, and 3 Ph-negative B-lymphoid cell lines were blotted with human ZNF423 cDNA. As shown in Figure 5A, ZNF423 mRNA expression was detected in 1 of 6 p210BCR/ABL-positive (BV-173), 3 of 4 p190BCR/ABL-positive (Hirata CL, KOPN-72bi, and KOPN-92bi), and 2 of 3 Ph-negative cell lines (HAL-01 and NALM-6), all of which were of B-cell phenotype.
We then examined ZNF423 expression in human clinical samples diagnosed as CML CP or BC. BM samples of 3 CML CP and 4 CML BC patients (4 myeloid and 4 B-lymphoid lineages) with informed consent were subjected to RT-PCR using ZNF423-specific primers. BV-173 cells that express ZNF423 (Figure 5A) were used as a control. As shown in Figure 5B, whereas no ZNF423 expression was detected in CML CP sample (nos. 1-3), 4 of 8 CML BC samples were found to express ZNF423, where 1 was of myeloid (no. 6) and the other 3 were of B-lymphoid (nos. 8, 9, and 11) phenotypes. These results strongly indicated that aberrant expression of ZNF423 clinically contributes to the malignant transformation of BCR/ABL-positive cells and to the progression to CML BC, mainly of B-cell lineage.
Discussion
CML provides an appropriate disease model for multistep carcinogenesis in which generation of p210BCR/ABL initiates CML CP and an additional genetic event(s) contributes to the evolution to CML BC.8 We developed a transgenic mouse model for human CML, which expresses p210BCR/ABL in hematopoietic progenitor cells and reproducibly exhibits a CML-like myeloproliferative disorder.14 To investigate molecular mechanism(s) responsible for disease progression, the p210BCR/ABL transgenic mice were subjected to retroviral insertional mutagenesis. BXH2 mice that harbor a horizontally transmittable replication-competent retrovirus22 were used as a virus donor strain, because it has been successfully used to detect second hit genes in previous studies.37-39
The inbred BXH2 mice were reported to develop acute myeloid leukemia at 7 to 12 months of age due to ecotropic virus integration and intrinsic myeloid tropism.22-24 In line with this report, all the WT/BXH2 mice died of myeloid leukemia (Figure 1A). In contrast, 6 p210BCR/ABL/BXH2 mice developed nonmyeloid leukemias with a shorter latency. The early disease onset and different phenotypes in these mice indicated that the diseases were caused by the cooperation of p210BCR/ABL with altered expression of virus-affected gene(s). Therefore, in this study, we intended to identify virus-integrated genes in the tumors of the 6 mice and iPCR analysis detected 2 CISs in B-cell leukemia samples. These CISs were considered to be strong candidates for CML BC, because leukemia with B-cell phenotype has not been detected in p210BCR/ABL transgenic mice14 and very rarely reported in BXH2 mice (http://rtcgd.abcc.ncifcrf.gov/).22,23,27
It is to be noted that one CIS was in the promoter region of the transgene. Interestingly, our previous retrovirus insertional mutagenesis study, in which newborn p210BCR/ABL transgenic mice were directly injected with retroviruses, also identified the transgene as a CIS in B-cell BC cases, where retrovirus integration resulted in overexpression and/or enhanced kinase activity of the transgene product.40 The up-regulation of p210BCR/ABL in the blast phase is especially interesting, because it corresponds to double Ph, which is one of the most frequently observed chromosomal abnormalities found in CML BC.8 Therefore, our observation provides in vivo experimental evidence that acquired enhancement of p210BCR/ABL expression accelerates the disease and causes BC. The reason why mice with transgene integration exhibited B-cell leukemia is not clear. It could be possible that p210BCR/ABL originally expressed by the TEC promoter and then up-regulated by retrovirus insertion might predispose the infected mice to develop B-ALL by an unknown mechanism. In human CML BC, double Ph was reported to be occasionally associated with B-cell BC samples.41,42
Another CIS was the 5′ noncoding region of Zfp423, which encodes a transcription factor with multiple zinc-finger repeats.33 Zfp423 was originally identified as a binding partner of Ebf (early B-cell factor, also denoted as Olf1), a protein essential for B-cell and olfactory nervous system development,43,44 and was subsequently shown to interact with SMADs in response to bone morphogenic protein 2 (BMP2) in the Xenopus laevis.45 Zfp423 was also cloned as a target in B-cell lymphoma in AKXD27 mice by retroviral insertional mutagenesis.33 In that study, retrovirus integration occurred upstream of the translation initiation codon resulting in a high level of expression, as observed in our cases (Figure 2).33 A recent study demonstrated that Zfp423 knockout mice exhibited abnormal cerebellum development but appeared to have a normal hematopoiesis,46 suggesting that its ectopic expression would be involved in leukemogenesis.
It remains to be clarified how overexpression of Zfp423 contributes to B-cell malignancy. Interestingly, although Zfp423 was originally identified as an Ebf-binding partner, it is not expressed in hematopoietic tissues including B cells.33,34 In the olfactory nervous system, Zfp423 was shown to negatively regulate Ebf function; Zfp423 forms a heterodimer with Ebf, which inhibits Ebf homodimer formation that has an ability to transactivate downstream target genes.43 Thus, it would be possible that the aberrant expression of Zfp423 in the hematopoietic system induces B-cell leukemia, at least in part by impairing Ebf-mediated signaling. Alternatively, because a recent study demonstrated that a highly conserved 12–amino acid peptide located in the extreme N-terminus of Zfp423 recruits the nucleosome remodeling and deacetylase corepressor complex (NuRD),47 it could be postulated that Zfp423 functions as a transcription repressor and contributes to leukemogenesis by suppressing downstream target genes.
In this study, Zfp423 was identified as a gene whose deregulated expression cooperates with p210BCR/ABL and induces CML BC. The cooperative activities of Zfp423 and p210BCR/ABL were demonstrated by in vitro and in vivo mouse experiments and also by human samples. Enforced expression of Zfp423 in hematopoietic cells derived from p210BCR/ABL transgenic mice enhanced B-cell colony formation, and suppression of ZNF423 expression in ZNF423-expressing, p210BCR/ABL-positive CML BC cells reduced cell growth (Figure 3). In addition, expression of Zfp423 with p210BCR/ABL in hematopoietic progenitor cells accelerated p210BCR/ABL-mediated leukemia and induced a more aggressive phenotype mainly of B-cell lineage (Figure 4). Furthermore, ZNF423 is expressed in a subset of BCR/ABL-positive hematopoietic cell lines and several CML BC samples mostly with B-cell phenotype (Figure 5). These results demonstrated that Zfp423/ZNF423 cooperates with p210BCR/ABL, confers a proliferative advantage to p210BCR/ABL-expressing hematopoietic cells, and consequently develops CML B-cell BC. It is to be noted that ZNF423 expression was detected in several Ph-negative B-ALL lines (Figure 5), which indicates that ZNF423 contributes not only to CML B-cell BC or Ph-positive B-ALL but also to de novo B-ALL without BCR/ABL.
It is intriguing that while the recipient mice transduced with both p210BCR/ABL and Zfp423 developed mainly B-ALL, 2 cases developed AML (Figure 4B). Although Zfp423 has been exclusively associated with B-ALL in retroviral insertional mutagenesis studies,33,34 this result strongly suggests that Zfp423 might contain a potency to develop AML as well as B-ALL. This idea is in accordance with the finding that one myeloid BC case expressed ZNF423 in clinical analysis (Figure 5B no. 6). Recently, Zfp521/ZNF521 (also known as EHZF [early hematopoietic zinc finger protein] and Evi3), which is homologous to Zfp423/ZNF423 and was also identified as a target of B-ALLs by mouse retrovirus insertional mutagenesis studies,34,48 was reported to be frequently involved in AML samples in human leukemias.49 Thus, a set of zinc finger–containing transcription factors that has been isolated as a target in B-ALL in mice might contribute to leukemias with different phenotypes in humans.
As for the 2 mice that developed T-ALL, although we could not identify any CIS, candidate genes, such as Hcst (hematopoietic cell signal transducer, also called DAP10/KAP10), an adaptor protein involved in T-cell signaling,50,51 and Il21r (IL21 receptor), a cytokine receptor mediating T-cell activation52,53 were detected (Table S1). In addition, several genes isolated by iPCR have been reported in cancer gene studies. For example, Avpil1, Ddx6, Runx1, Mef2d, Jak1, Cbfa2t3h, and Sox4 have already been identified as retrovirus integration sites in the mouse retrovirus tagged cancer gene database (RTCGD; http://rtcgd.abcc.ncifcrf.gov/),23,27 and IL21R, DDx6, Runx1, and Cbfa2t3h have been denoted as chromosomal translocation-associated genes in human cancer (http://www.sanger.ac.uk/genetics/CGP/Census/).54 Furthermore, MEF2D was shown to create a fusion gene in t(1;19)(q23;p13),55 and Sox4 was demonstrated to be a powerful tool to identify cooperative genes when transplanted by a replication-defective retrovirus.56 Further studies will be required to investigate whether virus insertion in these genes might affect the BC phenotype and/or lineage commitment observed in p210BCR/ABL/BXH2 mice.
In this study, we demonstrated that enhanced expression of p210BCR/ABL and aberrant expression of Zfp423/ZNF423 contribute to blastic transformation of p210BCR/ABL-expressing hematopoietic cells. Our results provide insights into the molecular mechanism(s) for disease progression of human CML and prove this transgenic system is a valuable tool in identifying genes whose altered expression cooperates with p210BCR/ABL to induce CML BC.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Yuki Sakai, Kayoko Hashimoto, and Yuko Tsukawaki for the mouse care and technical assistance; Tomoko Takahara and Yukari Yamazaki for BMT studies; and Hirotaka Matsui for the statistical analysis. We also thank Motomi Osato for helpful discussion and Hiroya Aso and Toshiya Inaba for providing us with Ph-positive human hematopoietic cell lines.
This work was supported by a Grant-in-Aid from the Ministry of Education, Science and Culture of Japan (Tokyo, Japan), a Grant-in-Aid for Cancer Research from the Ministry of Health, Labor and Welfare of Japan (13-2; Tokyo, Japan) Research Grant of the Princess Takamatsu Cancer Research Fund (Tokyo, Japan), Mitsubishi Pharma Research Foundation (Osaka, Japan), YASUDA Medical Research Foundation (Osaka, Japan), a Grant-in-Aid of The Japan Medical Association (Tokyo, Japan), and Japan Leukemia Research Fund (Tokyo, Japan).
Authorship
Contribution: K.M., N.Y., M.M., T.N., and H.H. designed and performed the research and wrote the paper; H.O. centralized the pathologic analysis; Y. Komeno, J.K., Z.-i.H., and T. Kitamura, performed the retrovirus and shRNA studies; S.W., N.A.J., and N.G.C. participated in the Zfp423 studies and wrote the paper; T. Kuwata, Y. Kanno, and T.N. contributed to the BMT analysis; and all the authors checked and agreed on the final version of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hiroaki Honda, Department of Developmental Biology, Research Institute of Radiation Biology and Medicine, Hiroshima University, 1-2-3 Kasumi, Minami-ku, Hiroshima 734-8553, Japan; e-mail: hhonda@hiroshima-u.ac.jp.