Abstract
Junctional adhesion molecule A (JAM-A) is a transmembrane adhesive glycoprotein that participates in the organization of endothelial tight junctions and contributes to leukocyte transendothelial migration. We demonstrate here that cultured endothelial cells not only express a cellular 43-kDa variant of JAM-A but also release considerable amounts of a 33-kDa soluble JAM-A variant. This release is enhanced by treatment with proinflammatory cytokines and is associated with the down-regulation of surface JAM-A. Inhibition experiments, loss/gain-of-function experiments, and cleavage experiments with recombinant proteases indicated that cleavage of JAM-A is mediated predominantly by the disintegrin and metalloproteinase (ADAM) 17 and, to a lesser extent, by ADAM10. Cytokine treatment of mice increased JAM-A serum level and in excised murine aortas increased ADAM10/17 activity correlated with enhanced JAM-A release. Functionally, soluble JAM-A blocked migration of cultured endothelial cells, reduced transendothelial migration of isolated neutrophils in vitro, and decreased neutrophil infiltration in a murine air pouch model by LFA-1– and JAM-A–dependent mechanisms. Therefore, shedding of JAM-A by inflamed vascular endothelium via ADAM17 and ADAM10 may not only generate a biomarker for vascular inflammation but could also be instrumental in controlling JAM-A functions in the molecular zipper guiding transendothelial diapedesis of leukocytes.
Introduction
Leukocyte adhesion and migration through the vascular wall are governed by soluble mediators and membrane-bound adhesion molecules. At the cell surface, adhesion molecules mediate capture and adhesion of leukocytes from the bloodstream to the endothelium. Adhesion molecules also contribute to leukocyte transmigration through the endothelium. Within endothelial junctions, adhesion molecules promote the establishment of interendothelial cell contacts and are also implicated in the transendothelial migration of leukocytes along the endothelial junctions.
Junctional adhesion molecule-A (JAM-A) is a transmembrane adhesive glycoprotein of the immunoglobulin superfamily (Figure 1A) expressed by endothelial and epithelial cells as well as by leukocytes and platelets.1-3 In vitro, JAM-A participates in the organization of tight junctions, where it is thought to contribute to the establishment of the vascular diffusion barrier.4,5 JAM-A is also involved in the migration and angiogenesis of cultured endothelial cells6,7 and in the transendothelial migration of isolated neutrophils or mononuclear cells,8-10 indicating a role in vascular inflammation and remodeling. Endothelial cell stimulation with interferon-γ (IFN-γ) and tumor necrosis factor-α (TNF-α) has been reported to result in an increased expression of JAM-A at the apical site of the endothelium,11 where JAM-A contributes to cell adhesion.12 However, it is not clear whether this redistribution is the only modality for regulation of JAM-A functions.
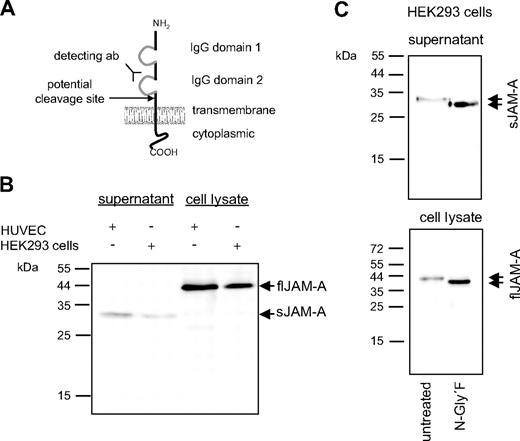
Western blot analysis of full-length and soluble JAM-A. (A) Schematic representation of the domain structure of transmembrane JAM-A and its potential cleavage site at the cell membrane. (B) HUVECs and HEK293 cells were cultured for 8 hours in the absence of fetal calf serum. Cell lysates and conditioned media were then examined for the presence of full-length (fl) and soluble (s) JAM-A by SDS-polyacrylamide gel electrophoresis under reducing conditions and subsequent Western blotting using a monoclonal antibody directed against the second IgG domain of JAM-A. (C) Cell lysates and conditioned media from HUVECs were treated with 2 U/mL N-Glycosidase F for 2 hours at 37°C, and subsequently fl and sJAM-A were analyzed by Western blotting. Data shown are representative of at least 3 independent experiments.
Western blot analysis of full-length and soluble JAM-A. (A) Schematic representation of the domain structure of transmembrane JAM-A and its potential cleavage site at the cell membrane. (B) HUVECs and HEK293 cells were cultured for 8 hours in the absence of fetal calf serum. Cell lysates and conditioned media were then examined for the presence of full-length (fl) and soluble (s) JAM-A by SDS-polyacrylamide gel electrophoresis under reducing conditions and subsequent Western blotting using a monoclonal antibody directed against the second IgG domain of JAM-A. (C) Cell lysates and conditioned media from HUVECs were treated with 2 U/mL N-Glycosidase F for 2 hours at 37°C, and subsequently fl and sJAM-A were analyzed by Western blotting. Data shown are representative of at least 3 independent experiments.
Within the vasculature, most adhesion molecules are expressed as type I single transmembrane domain proteins on the surface of either endothelial cells or leukocytes.13,14 Besides their surface expression, soluble forms have been identified for L-selectin, vascular cell adhesion molecule 1 (VCAM-1), and intercellular adhesion molecule 1, but also for the transmembrane chemokines CX3CL1 and CXCL16, and their increased appearance in blood serum has been associated with various inflammatory processes and leukocyte recruitment to sites of inflammation.15 Proteolytic cleavage at the cell surface has been identified as the underlying mechanism leading to the release of soluble variants of these adhesion molecules in a process termed shedding. The shedding activity can be up-regulated on cell activation with phorbol-12-myristyl-13-acetate (PMA), ionomycin, chemotactic mediators, apoptotic signals, or crosslinking antibodies, which, however, depends strongly on the type of shedding target.16 The disintegrin and metalloproteinase ADAM17 has been identified as the most relevant protease implicated in the PMA-induced shedding of L-selectin, VCAM, and CX3CL1.17-20 The constitutive cleavage of CX3CL1 and CXCL16, however, involves the activity of the closely related ADAM10.21-23 Whereas L-selectin shedding is thought to influence rolling velocity and capture of leukocytes, shedding of CX3CL1 promotes detachment of bound leukocytes.24 Therefore, the activity of sheddases on the endothelium or bound leukocytes probably influences leukocyte recruitment during inflammation. However, the activity of the sheddases is probably not limited to molecules expressed at the luminal side of vascular cells but also affects those expressed in intraendothelial junctions.
Here, we demonstrate that JAM-A is constitutively released from endothelial and epithelial cells via proteolytic shedding and that this shedding is enhanced by cell activation with IFN-γ and TNF-α or platelet activating factor (PAF). Multiple evidence is provided that ADAM10 and ADAM17 are involved in this shedding process. As potential consequences of JAM-A shedding, we observed increased endothelial migration and inhibition of neutrophil transmigration by released JAM-A in vitro and in vivo.
Methods
Cytokines, antibodies, expression vectors, and inhibitors
Monoclonal antibodies to human JAM-A clones BV16 and M.Ab.F11 (both IgG1) were purchased from HyCult Biotechnology (Uden, The Netherlands), and clone 43 was from BD Biosciences (San Jose, CA). Specific antibodies used for enzyme-linked immunosorbent assay (ELISA) determination of murine JAM-A were purchased from R&D Systems (Minneapolis, MN). Purified rabbit polyclonal antibodies against the c-terminus of ADAM10 and ADAM17, respectively, were from Chemicon/Millipore (Billerica, MA). Monoclonal antibodies against the ectodomains of murine ADAM10 (IgG2b, clone 163003) and ADAM17 (IgG1, clone 111633), IgG1 and IgG2b isotype controls, recombinant mouse KC, and recombinant human ADAM17 catalytic domain were from R&D Systems. Mouse monoclonal anti–β-actin antibody was from Abcam (Cambridge, United Kingdom). Peroxidase (POD)–conjugated secondary antibodies to mouse or rabbit IgG were obtained from Perbio (Bonn, Germany). Goat anti–mouse IgG conjugated to Alexa Fluor 594 and goat anti–rabbit IgG conjugated to Alexa Fluor 488 were from Invitrogen (Carlsbad, CA). Phycoerythrin/Cy5- and fluorescein isothiocyanate-labeled antibodies to monocyte markers F4/80 and Gr1 were from BioLegend (San Diego, CA) and Serotec (Oxford, United Kingdom), respectively. Recombinant human and murine IFN-γ and TNF-α were obtained from PeproTech (Rocky Hill, NJ). Recombinant human interleukin-8 (IL-8) was from ImmunoTools (Friesoythe, Germany). The expression vectors for full-length human JAM-A or the fusion protein of human JAM-A with the Fc part of human IgG (JAM-A.Fc) were described previously.25 Recombinant JAM-A. Fc was either purchased from R&D Systems or produced in CHO cells and purified by affinity chromatography with protein A and subsequent ion exchange chromatography with Mono Q as described.25 Monovalent JAM-A was generated by limited proteolysis of JAM-A.Fc using TEV protease (Invitrogen) following the manufacturer's instructions and separated from the Fc fragment using protein A and Mono Q chromatography. Fc control (irrelevant human IgG) was from Sigma Chemie (Deisenhofen, Germany), and Fc block was from Miltenyi Biotec (Auburn, CA). The metalloproteinase inhibitors GW280264X and GI254023X were synthesized and assayed for inhibition of recombinant human ADAM17 and ADAM10 as described.26 PMA was obtained from Calbiochem (Darmstadt, Germany).
Cell preparation, cell culture, and transfection
The adherent bladder carcinoma cell line ECV304, the human embryonic kidney cell-line HEK293, and human umbilical vein endothelial cells (HUVECs) were cultured as described.21 Liposomal transfection of HEK293 cells has been reported previously.19 Human neutrophils from peripheral blood of healthy volunteers were isolated by sedimentation on Ficoll hypaque (GE Healthcare, Little Chalfont, United Kingdom) and subsequent incubation for 5 minutes in red blood cell lysis buffer (155 mM NH4Cl, 10 mM NaHCO3, 5 mM ethylenediaminetetraacetic acid, pH 7.4). Murine embryonic fibroblasts (MEFs) derived from mice with targeted disruption of adam17,27 adam10,28 or from wild-type mice were cultured and transfected as described.19 Transfection of HUVECs with small interfering RNA (siRNA) was carried out using a HUVEC nucleofector kit from Amaxa Biosystems (Cologne, Germany) following instructions of the manufacturer. Studies involving mice and those with HUVECs were approved by the institutional review board of Rheinisch-Westfaelische Technische Hochschule Aachen, and volunteers' informed consent was obtained in accordance with the Declaration of Helsinki.
siRNA
Down-regulation of endogenous ADAM10/ADAM17 expression was carried out as previously described.29 The following RNA oligonucleotides (Stealth RNAi, Invitrogen) were used: ADAM10-construct, 5′-UAC ACC AGU CAU CUG GUA UUU CCU C 3′; ADAM17-construct, 5′-UAC UGU ACA GGG CUU UCC UUU CCU C 3′. As negative control, unspecific stealth RNA duplexes with low GC content were used.
JAM-A cleavage assays
Cells expressing JAM-A were grown in 6-well dishes to 80% to 90% confluence in fully supplemented medium for 48 hours. Cells were washed once with sterile phosphate-buffered saline (PBS) and received 1 mL of serum-free medium with GI254023X (5 μM), GW280264X (5 μM), or dimethyl sulfoxide (DMSO). After 1 hour, the cells were stimulated with PMA (200 ng/mL), ionomycin (1 μM), PAF (50 nM), TNF-α/IFN-γ (both 10 ng/mL), or recombinant ADAM17 catalytic domain (5 ng/mL) for the indicated periods of time, and conditioned media were harvested and supplemented with a protease inhibitor mixture (Complete; Roche Diagnostics, Mannheim, Germany). Conditioned media were cleared by high-speed centrifugation and analyzed by Western blotting (see “Western blotting”). Cells were washed twice with ice-cold PBS and either lysed in 5 mM Tris HCl containing 1 mM ethyleneglycoltetraacetic acid, 250 mM saccharose 1% Triton X-100, and Complete protease inhibitor mix for analysis by Western blotting (see “Western blotting”) or scraped off in 1 mL ice-cold PBS and examined by flow cytometry (see “Flow cytometry”).
For cell-free cleavage assays, 20 ng recombinant human JAM-A.Fc fusion protein (R&D Systems) was incubated with 5 ng catalytic domain of human ADAM17 (R&D Systems) for 3 hours in 200 μL of 5 mM Tris, pH 9.0, 2.5 μM ZnCl2, 0.005% Brij 35 at 37°C and subsequently analyzed by Western blotting for JAM-A cleavage fragments.
Preparation of Triton-soluble and -insoluble cell extracts
HUVECs were washed twice with PBS and then lysed for 30 minutes with lysis buffer containing 0.5% Triton X-100, 150 mM NaCl, 50 mM Tris-HCl, and protease inhibitors (Roche Diagnostics). The lysate was then centrifuged at 10 000g for 15 minutes to collect the supernatant (Triton-soluble fraction). The pellet was incubated for 30 minutes with lysis buffer supplemented with 0.02% sodium dodecyl sulfate (SDS) and cleared by centrifugation at 10 000g for 15 minutes (Triton-insoluble fraction).
Enzymatic deglycosylation
HUVECs were cultured in 6-well dishes for 16 hours in serum-free medium, and subsequently conditioned media were harvested and cells were lysed in incubation buffer containing 25 mM ethylenediaminetetraacetic acid 1% Triton X-100, 0.1% SDS, and 1% mercaptoethanol and cleared by high-speed centrifugation. Fifty-fold concentrated media were diluted 5-fold in incubation buffer, and subsequently 2 U/mL N-glycosidase F (Roche Diagnostics) was added to the conditioned media and cell lysates. After 1 hour of incubation at 37°C, conditioned media and cell lysates were analyzed by Western blotting.
Western blotting
Western blot analysis was carried out as described previously.19,21 Samples were then subjected to SDS-polyacrylamide gel electrophoresis under reducing conditions using 10% Tris-glycine gels. Proteins were transferred onto polyvinylidene difluoride membranes (Hybond-P, GE Healthcare) that were probed with dilutions of rabbit antibodies against ADAM10 (1:1000) or ADAM17 (1:1000) or mouse antibody against JAM-A (clone 43; 1:1000) followed by incubation with POD-coupled goat anti–rabbit Ig or POD-coupled goat anti–mouse (both diluted 1:10 000 in PBS-T). After addition of chemiluminescence substrate (enhanced chemiluminescence advanced, GE Healthcare), signals were recorded and quantified using a luminescent image analyzer LAS3000 and Multi Gauge 3.0 software (Fujifilm, Tokyo, Japan). Equal loading and transfer of proteins to the membrane were verified by subsequent detection of β-actin using a specific monoclonal antibody.
In-gel digestion of JAM-A
JAM-A.Fc (90 μg) was deglycosylated, desalted, and subsequently incubated with recombinant ADAM17 ectodomain (5 μg) for 48 hours at 37°C (“JAM-A cleavage assays”). The incubation mixture was resolved by 10% SDS-polyacrylamide gel electrophoresis, and proteins were stained by colloidal Coomassie staining (Fermentas, St Leon-Rot, Germany). A protein band that migrated at the expected molecular weight of sJAM-A was excised, and the resulting gel fragment was destained through washing with acetonitrile/sodium carbonate buffer. Proteins in the gel fragment were reduced and alkylated with dithiothreitol and iodoacetamide, and the gel band was incubated with proteomics-grade trypsin (Sigma Chemie) for 3 hours at 37°C. After quenching the digestion with trifluoroacetic acid, tryptic fragments were extracted from the gel and analyzed using an Applied Biosystems/MDS Sciex (Foster City, CA) 4800 steel matrix-assisted laser desorption/ionization (MALDI) tandem time-of-flight mass spectrometry (MS/MS) system.
Mass spectrometric analysis and peptide mass fingerprinting
Solutions containing tryptic fragments were mixed 1:1 with a saturated matrix solution of α-cyano-4-hydroxycinnamic acid (5 mg/mL) in 50% acetonitrile/water/0.1% trifluoracetic acid, spotted in duplicate on a MALDI–time of flight (MALDI-TOF) target, and a total of 500 spectra were accumulated over an m/z range of 500 to 3000. The recorded tryptic cleavage peptide mass profiles (from m/z 900-3000) were determined by protein identification algorithms through Mascot database scoring (Matrix Science, Boston, MA; http://www.matrixscience.com/). Using a peptide mass fingerprint confirmation mode, JAM-A peptides matching top protein hits in MS mode were automatically selected for MS/MS analysis to confirm protein identification.
Flow cytometry
Cells (3 × 106 cells/mL) were incubated with monoclonal mouse anti–human JAM-A IgG1 antibody (M.Ab.F11; 0.5 μg/mL) or monoclonal antibodies against the ectodomains of either ADAM10 or ADAM17 (1 μg/mL and 2.5 μg/mL, respectively) or respective IgG1 and IgG2b controls in PBS containing 1% bovine serum albumin (BSA) and 0.01% NaN3 for 1 hour on ice followed by detection with fluorescein-conjugated anti–mouse F(ab′)2 fragment (Jackson, Newmarket, United Kingdom) and subsequent analysis by flow cytometry (FACSCalibur, BD Biosciences). CellQuest software (BD Biosciences) was used to generate histograms and to calculate mean fluorescence intensities.
Immunocytochemistry
For JAM-A indirect immunofluorescence staining, endothelial cells were grown onto collagen-coated glass coverslips and subsequently fixed with 4% paraformaldehyde. Unspecific antibody binding was blocked with 3% BSA (PAA Laboratories, Linz, Austria) in PBS for 30 minutes. Cells were incubated with monoclonal mouse anti–human JAM-A IgG1 antibody (BV16; 2 μg/mL) for 1 hour at room temperature. After washing, Alexa 488–conjugated goat anti–mouse IgG secondary antibody (Invitrogen) was added for 1 hour, and slides were analyzed by fluorescence microscopy using a Leica DM IRB inverted microscope with a Leica 20×/0.4 NPlan air objective, a Leica DFC 310 FX camera, and Leica application suite software LAS 3.2.0 (Leica, Wetzlar, Germany).
Endothelial cell migration assays
A wound-induced endothelial migration assay (scratch assay) was performed by destroying a confluent HUVEC layer by a defined scratch of approximately 1-mm width as described.30 After removal of detached cells, indicated concentrations of sJAM-A.Fc or human IgG control were added, and migration of endothelial cells into the scratch area was monitored for 24 hours. The experiment was performed in triplicate.
RT-PCR
Total messenger RNA from HUVECs was isolated (RNeasy Mini Kit, QIAGEN, Hilden, Germany) and reverse-transcribed with oligo-dT primers (M-MLV reverse transcriptase, Promega, Madison, WI). A real-time polymerase chain reaction with up to 100 ng of cDNA was performed with the QuantiTect Kit with SYBRGreen (SYBR Advantage, Clontech, Mountain View, CA) and specific primers for JAM-A (5′-TCGAGAGGAAACTGTTGTGC-3′, 5′-ACCAGTTGGCAAGAAGGTCA-3) and GAPDH, (5′-GCCTCAAGATCATCAGC-3′, 5′-ACCACTGACACGTTGGC-3′), respectively, following the manufacturer's protocol. Amplification (40 cycles, with annealing at 55°C) was performed via the MJ Research Opticon 2 (Biozym, Oldendorf, Germany). Determinations were performed in triplicates.
Transmigration assay
Isolated neutrophils were preincubated with sJAM-A.Fc, hIgG control, or Fc-block for 30 minutes, washed, and investigated for transmigration across a HUVEC monolayer grown on collagen-coated 24-well transwell tissue culture inserts (5-μm pore size; Corning Life Sciences, Acton, MA). In some experiments, HUVECs were pretreated with the recombinant catalytic domain of ADAM17 (10 ng/mL) for 1 hour at 37°C, and the monolayer was either washed or engaged directly in the transmigration assay. As a chemotactic stimulus, IL-8 (10 nM in RPMI-1640 with 0.2% BSA) was added to the lower compartment, and the upper compartment was filled with neutrophil suspension (150 000 cells in 100 μL RPMI-1640 with 0.2% BSA). After 45 minutes of incubation, transmigrated cells in the lower compartment were counted using a hematocytometer.
Cytokine treatment of excised murine aortas
Aortas (∼ 20 μg) were prepared from male C57BL/6 mice (15 weeks old) stimulated with murine TNF-α and murine IFN-γ (both 20 ng/mL) in 1 mL Dulbecco modified Eagle medium in the absence or presence of metalloproteinase inhibitor GW280264 (5 μM) for 16 hours. Subsequently, released JAM-A in the conditioned media was determined by ELISA specific for murine JAM-A (R&D Systems). Tissue extracts were analyzed for ADAM10/17 activity using a fluorogenic peptide-based assay kit from R&D Systems following the manufacturer's instructions. The enzymatic activity in the cell lysates was expressed in relation to that of a serially diluted recombinant ADAM17 standard run in parallel.
Cytokine treatment of mice
Male C57BL/6 mice (22 weeks old) were treated intravenously with a combination of murine TNF-α (50 ng/g) and murine IFN-γ (40 ng/g) in 150 μL of sterile 0.9% NaCl solution or received sterile vehicle solution (n = 3 per group). After 2.5 hours, animals were killed, and blood was drawn from the heart and centrifuged for 1 hour at 500g. Serum samples were then analyzed for the presence of JAM-A by ELISA specific for murine JAM-A (R&D Systems).
Leukocyte accumulation in the mouse air pouch
For assessment of leukocyte extravasation in vivo, wild-type and jam-a−/− mice (kindly provided by Elisabetta Dejana, University of Milan, Italy) were used.31 Air pouches were generated in female C57BL/6 mice as described.32 In brief, 5 mL of sterile air was injected subcutaneously into the back of anesthetized mice and 3 days later the air pouch was reinflated with 2.5 mL sterile air. After further 4 days, 60 μg sJAM-A.Fc or human IgG control or 40 μg monovalent sJAM-A was injected intravenously. One hour later, 1 mL PBS containing murine KC (0.5 μg/mL) or vehicle control was injected into the air pouch. Four hours after KC injection, the animals were killed and the air pouches lavaged with 1 mL of 5 mM ethylenediaminetetraacetic acid in PBS. After centrifugation of the lavage, cells were resuspended in PBS with 0.1% BSA, manually counted, and differentiated by flow cytometry. Neutrophils were identified as CD45+/Gr1+/F480−/CD115−. All animal experiments were approved by an institutional review board of the Rheinisch-Westfaelische Technische Hochschule Aachen for animal experimentation.
Statistical analysis and presentation of data
Data are shown either as one representative of at least 3 independent experiments or as mean plus or minus SD of 3 independent experiments. Data were statistically analyzed by one-way analysis of variance. Two populations of data were considered significantly different at P less than .05.
Results
Constitutive release of soluble JAM-A
HUVECs and HEK293 cells were investigated for the expression of JAM-A in the cell lysates and its release into the culture media by Western blotting using an antibody against the second Ig domain of JAM-A (Figure 1A). The majority of JAM-A from the cell lysates migrated at 43 kDa. This variant was termed full-length JAM-A (flJAM). Interestingly, a smaller JAM-A variant that migrated at 33 kDa was also detected in the culture supernatant of HUVECs and HEK293 cells (Figure 1B). This variant was termed soluble JAM-A (sJAM-A).
JAM-A carries a single site for potential glycosylation at Asn 185 within the second extracellular domain. To investigate whether sJAM-A and flJAM-A represent differentially glycosylated variants of the JAM-A protein, deglycosylation experiments with N-Glycosidase F were carried out. The apparent molecular weight of deglycosylated flJAM-A was 38 kDa, whereas deglycosylated sJAM-A migrated at 28 kDa (Figure 1C). Thus, both flJAM-A and sJAM-A carry a very similar degree of N-linked glycosylation of approximately 5 kDa. The difference in molecular weight between deglycosylated flJAM-A and sJAM-A is therefore rather the result of a variation of the protein backbone. Our observation that antibodies against the second extracellular loop but not antibodies against the C-terminus of JAM-A were able to detect sJAM-A (data not shown) led us to speculate that the released molecule was C-terminally truncated (Figure 1A).
Induced release of soluble JAM-A
We next asked whether the release of endogenous JAM-A would be modulated by activation of endothelial cells or HEK293 cells. Treatment of HUVECs with the phorbol ester PMA rapidly induced release of sJAM-A within 2 hours (Figure 2A). Stimulation with PMA led to an approximately 3-fold increase of sJAM (JAM-A content in 1 mL of conditioned media from 2 × 105 cells increased from 5 ng/mL to 15 ng/mL). This PMA-induced release was associated with a reduction of flJAM-A in the cell lysates. Proinflammatory cytokines (a combination of IFN-γ and TNF-α) or PAF also enhanced release of sJAM-A but required longer time periods (16 hours) compared with PMA (2 hours; Figure 2A,B). This stimulated release of sJAM-A was associated with the down-regulation of surface-expressed JAM-A as demonstrated by fluorescence-activated cell sorter analysis (Figure 2C). The latter observation suggests that the induced release of sJAM-A is not the result of an induction of jam-a gene transcription, which was confirmed by quantitative real time RT-PCR analysis showing no differences in JAM-A mRNA expression in unstimulated and cytokine-stimulated HUVECs (Figure 2D). We then asked whether sJAM-A would also be released in vivo. To investigate this issue, we analyzed the JAM-A content in blood sera of healthy mice and mice with systemic inflammation (n = 3 per group) using an ELISA for murine JAM-A. Basal levels of sJAM-A were present in untreated wild-type mice (3.5 ng/mL) but not detectable in mice with targeted disruption of the JAM-A gene (jam-a−/−), indicating the specificity of the method for detection of serum JAM-A. Systemic injection of TNF-α and IFN-γ significantly (P < .05) increased the level of sJAM-A approximately 2-fold (from 4 ng/mL in mice receiving vehicle to approximately 8 ng/mL in cytokine-treated mice) within 2.5 hours of treatment (Figure 2E).
Enhanced JAM-A shedding in response to PMA and proinflammatory mediators. (A) HUVECs were stimulated with PMA (200 ng/mL) or vehicle control (DMSO) for 2 hours, and subsequently conditioned media and cell lysates were subjected to Western blot analysis for sJAM-A and flJAM-A, respectively. A standard of serially diluted recombinant sJAM-A was run in parallel. (B) HUVECs were stimulated with PAF (100 nM) or TNF-α/IFN-γ (10 ng/mL) or treated with vehicle control (DMSO) for 16 hours, and conditioned media were analyzed by Western blotting using an anti–hJAM-A monoclonal antibody. Data are shown as representative Western blots of least 3 independent experiments. (C) HUVECs or HEK293 cells were left unstimulated or stimulated as described in panels A and B, and subsequently JAM-A surface expression was assessed by flow cytometry. Data were shown as representative histograms (top panel) and as mean plus or minus SD of the fluorescence signals obtained in 3 independent experiments (bottom panel). (D) Effect of IFN-γ and TNF-α on JAM-A mRNA expression. HUVECs were stimulated with IFN-γ and TNF-α or vehicle control (PBS) for 16 hours and analyzed for JAM-A mRNA expression by quantitative real-time RT-PCR. Expression of JAM-A mRNA was measured in 3 independent experiments and expressed as percentage of GAPDH mRNA expression. (E) Serum levels of murine JAM-A. Wt mice and jam-a−/− mice were intravenously treated with IFN-γ and TNF-α (40 and 50 ng/g, respectively) or vehicle control (0.9% NaCl) or left untreated (n = 3 per group), and after 2.5 hours blood serum was investigated for released JAM-A by ELISA. *Significant increase in the JAM-A level induced by the cytokine treatment compared with the vehicle-treated control (P < .05).
Enhanced JAM-A shedding in response to PMA and proinflammatory mediators. (A) HUVECs were stimulated with PMA (200 ng/mL) or vehicle control (DMSO) for 2 hours, and subsequently conditioned media and cell lysates were subjected to Western blot analysis for sJAM-A and flJAM-A, respectively. A standard of serially diluted recombinant sJAM-A was run in parallel. (B) HUVECs were stimulated with PAF (100 nM) or TNF-α/IFN-γ (10 ng/mL) or treated with vehicle control (DMSO) for 16 hours, and conditioned media were analyzed by Western blotting using an anti–hJAM-A monoclonal antibody. Data are shown as representative Western blots of least 3 independent experiments. (C) HUVECs or HEK293 cells were left unstimulated or stimulated as described in panels A and B, and subsequently JAM-A surface expression was assessed by flow cytometry. Data were shown as representative histograms (top panel) and as mean plus or minus SD of the fluorescence signals obtained in 3 independent experiments (bottom panel). (D) Effect of IFN-γ and TNF-α on JAM-A mRNA expression. HUVECs were stimulated with IFN-γ and TNF-α or vehicle control (PBS) for 16 hours and analyzed for JAM-A mRNA expression by quantitative real-time RT-PCR. Expression of JAM-A mRNA was measured in 3 independent experiments and expressed as percentage of GAPDH mRNA expression. (E) Serum levels of murine JAM-A. Wt mice and jam-a−/− mice were intravenously treated with IFN-γ and TNF-α (40 and 50 ng/g, respectively) or vehicle control (0.9% NaCl) or left untreated (n = 3 per group), and after 2.5 hours blood serum was investigated for released JAM-A by ELISA. *Significant increase in the JAM-A level induced by the cytokine treatment compared with the vehicle-treated control (P < .05).
Inhibition of JAM-A release by metalloproteinase inhibitors
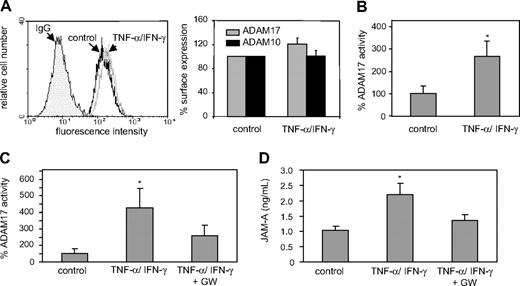
Soluble JAM-A could be generated by limited proteolysis (shedding) of transmembrane JAM-A at the cell surface. The metalloproteinases ADAM10 and ADAM17 have been implicated in various shedding events on endothelial cells, including the cleavage of VCAM-1, VE-cadherin, and transmembrane chemokines CX3CL1 and CXCL16.17-19,21,23,33 Both ADAM10 and ADAM17 are constitutively expressed by HUVECs, and stimulation with proinflammatory cytokines (IFN-γ and TNF-α) does not significantly affect ADAM10 or ADAM17 expression (Figure 3A) but increases their activity by more than 3-fold as demonstrated by a fluorogenic assay using a synthetic substrate (Figure 3B). A profound increase in ADAM10/ADAM17 activity was also observed when murine aortas were stimulated with IFN-γ and TNF-α (Figure 3C), and this was associated with enhanced release of JAM-A from the aortas into the culture medium (Figure 3D). A combined inhibitor of ADAM10 and ADAM17 (GW280264X) not only reduced the sheddase activity but also reduced JAM-A release from activated mouse aortas (Figure 3C,D).
Up-regulation of ADAM10/17 activity by proinflammatory mediators. (A) HUVECs were stimulated with IFN-γ and TNF-α (each 10 ng/mL) or vehicle control (PBS) for 16 hours and analyzed for ADAM10 and ADAM17 surface expression by flow cytometry. Data are shown as representative histogram (left) or as mean plus or minus SD of the fluorescence signals determined for ADAM10 and ADAM17 determined in 3 independent experiments (right). Surface expression was expressed in relation to that determined for the PBS-treated control. (B) HUVECs were stimulated with IFN-γ and TNF-α (each 10 ng/mL) or vehicle control (PBS) for 16 hours, and analyzed for cell-associated ADAM10/17 activity using a fluorogenic peptide-based assay. The shedding activity (mean plus or minus SD) was determined in 3 independent experiments using a recombinant ADAM17 standard run in parallel and expressed in relation to that of the vehicle-treated control. (C,D) Isolated murine aortas were treated with vehicle control (PBS, n = 4) or IFN-γ and TNF-α (each 20 ng/mL) in the absence (n = 4) or presence (n = 3) of the combined ADAM10/17 inhibitor GW280264 (GW, 5 μM) for 16 hours. Subsequently, cell lysates were analyzed for ADAM10/17 activity (C), and conditioned media were assayed for release of JAM-A into the culture medium (D). *Significant increase compared with the vehicle-treated controls (P < .05).
Up-regulation of ADAM10/17 activity by proinflammatory mediators. (A) HUVECs were stimulated with IFN-γ and TNF-α (each 10 ng/mL) or vehicle control (PBS) for 16 hours and analyzed for ADAM10 and ADAM17 surface expression by flow cytometry. Data are shown as representative histogram (left) or as mean plus or minus SD of the fluorescence signals determined for ADAM10 and ADAM17 determined in 3 independent experiments (right). Surface expression was expressed in relation to that determined for the PBS-treated control. (B) HUVECs were stimulated with IFN-γ and TNF-α (each 10 ng/mL) or vehicle control (PBS) for 16 hours, and analyzed for cell-associated ADAM10/17 activity using a fluorogenic peptide-based assay. The shedding activity (mean plus or minus SD) was determined in 3 independent experiments using a recombinant ADAM17 standard run in parallel and expressed in relation to that of the vehicle-treated control. (C,D) Isolated murine aortas were treated with vehicle control (PBS, n = 4) or IFN-γ and TNF-α (each 20 ng/mL) in the absence (n = 4) or presence (n = 3) of the combined ADAM10/17 inhibitor GW280264 (GW, 5 μM) for 16 hours. Subsequently, cell lysates were analyzed for ADAM10/17 activity (C), and conditioned media were assayed for release of JAM-A into the culture medium (D). *Significant increase compared with the vehicle-treated controls (P < .05).
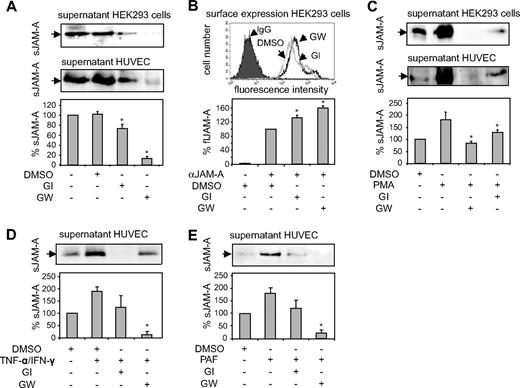
Cultured HUVECs were then used to further analyze the involvement of ADAM10 and ADAM17 in the release of sJAM-A by specific inhibition of these proteases. The inhibition experiments revealed that the constitutive release of JAM-A (Figure 4A) by HEK293 cells as well as by HUVECs was only slightly reduced by the inhibitor GI254023X, which blocks ADAM10 with high and ADAM17 with 100-fold lower potency.19,26 JAM-A release was almost completely suppressed by GW280264X, which blocks both proteases with high potency. Inhibition of JAM-A shedding was associated with an increased surface expression of JAM-A (Figure 4B). The JAM-A release by HEK293 cells or HUVECs stimulated with PMA, cytokines, or PAF (Figure 4C-E) was also only partially suppressed by GI254023X but almost completely abolished by GW280264X. The identical inhibition profiles obtained for HEK293 cells and HUVECs indicated that there are no differences between the 2 cell types with respect to the ADAMs involved. Moreover, the difference in inhibition between the 2 inhibitors points toward a predominant contribution of ADAM17 to the cleavage of JAM-A, but an involvement of ADAM10 is not excluded.
Inhibition of constitutive and inducible JAM-A shedding. (A) HEK293 cells and HUVECs were incubated with the preferential ADAM10 inhibitor GI254023X (GI, 5 μM), the combined ADAM10/17 inhibitor GW280264 (GW, 5 μM), or vehicle control (DMSO) for 16 hours. Conditioned media were analyzed by Western blotting using an anti–hJAM-A monoclonal antibody. (B) Surface expression of JAM-A on HEK293 cells treated with DMSO or inhibitor as described in panel A was analyzed by flow cytometry. (C) Cells were pretreated with the ADAM10 inhibitor GI254023X (5 μM) or ADAM10/17 inhibitor GW280264X (5 μM) or DMSO. Subsequently, cells were incubated in the presence or absence of PMA (200 ng/mL) for 2 hours. Supernatants were harvested, and soluble JAM-A was determined by Western blotting using an anti–hJAM-A monoclonal antibody. (D,E) Cells were stimulated with TNF-α/IFN-γ (10 ng/mL) or PAF (50 nM) or treated with vehicle control (DMSO) for 16 hours in the absence or presence of GI254023X or GW280264 (both 5 μM), and released JAM-A was determined by Western blotting. Quantified signals in panels A through E were normalized and shown as mean plus or minus SD for 3 independent experiments. *Statistically significant reduction in JAM-A surface expression caused by cell stimulation (P < .05).
Inhibition of constitutive and inducible JAM-A shedding. (A) HEK293 cells and HUVECs were incubated with the preferential ADAM10 inhibitor GI254023X (GI, 5 μM), the combined ADAM10/17 inhibitor GW280264 (GW, 5 μM), or vehicle control (DMSO) for 16 hours. Conditioned media were analyzed by Western blotting using an anti–hJAM-A monoclonal antibody. (B) Surface expression of JAM-A on HEK293 cells treated with DMSO or inhibitor as described in panel A was analyzed by flow cytometry. (C) Cells were pretreated with the ADAM10 inhibitor GI254023X (5 μM) or ADAM10/17 inhibitor GW280264X (5 μM) or DMSO. Subsequently, cells were incubated in the presence or absence of PMA (200 ng/mL) for 2 hours. Supernatants were harvested, and soluble JAM-A was determined by Western blotting using an anti–hJAM-A monoclonal antibody. (D,E) Cells were stimulated with TNF-α/IFN-γ (10 ng/mL) or PAF (50 nM) or treated with vehicle control (DMSO) for 16 hours in the absence or presence of GI254023X or GW280264 (both 5 μM), and released JAM-A was determined by Western blotting. Quantified signals in panels A through E were normalized and shown as mean plus or minus SD for 3 independent experiments. *Statistically significant reduction in JAM-A surface expression caused by cell stimulation (P < .05).
Role of ADAM17 and ADAM10 in the release of JAM-A
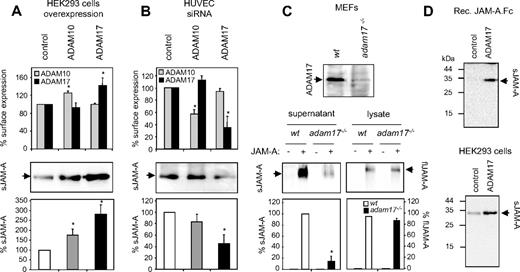
To further investigate whether ADAM17 and ADAM10 are potential JAM-A sheddases, both proteases were separately overexpressed in HEK293 cells. The successful overexpression of the proteases was demonstrated by flow cytometry (Figure 5A) and by Western blotting (Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article). Overexpression of ADAM17 profoundly increased the release of endogenous sJAM-A, whereas overexpression of ADAM10 had a less pronounced effect (Figure 5A). To examine the role of endogenous ADAM10 and ADAM17 in JAM-A cleavage, we used siRNA to decrease ADAM10 or ADAM17 expression in HUVECs. The successful down-regulation of the proteases was demonstrated by flow cytometry (Figure 5B) and Western blotting (Figure S1B). The release of endogenous JAM-A was considerably reduced when ADAM17 was suppressed, and only slightly affected when ADAM10 was down-regulated (Figure 5A). Results similar to these were obtained for constitutive and PMA-induced JAM-A shedding in HEK293 cells treated with ADAM10- or ADAM17-siRNA (Figure S1C). Notably, simultaneous down-regulation of ADAM17 and ADAM10 resulted in most pronounced inhibition of JAM-A shedding in HUVECs and HEK293 cells (Figure S1D). Although ADAM17 appears to be the predominant JAM-A sheddase, the latter observation indicates that ADAM10, nevertheless, contributes to the residual JAM-A shedding in the absence of ADAM17.
Loss- and gain-of-function experiments indicate a role for ADAM17 in JAM-A shedding. (A) Overexpression of ADAM10 and 17 in human HEK293 cells. Cells were transiently transfected with ADAM10 and ADAM17 or the empty expression vector as a control. Overexpression was controlled by flow cytometric analysis of ADAM10/17 surface expression (top panel). Conditioned media (middle panel) were analyzed for the presence of JAM-A by Western blotting, and signals were quantified by densitometry and calculated as percentage of the mock-transfected control (bottom panel). (B) Down-regulation of endogenous ADAM10/17 using siRNA. HUVECs were transiently transfected with ADAM10- ADAM17- or control-siRNA. Down-regulation of ADAM10 and ADAM17 surface expression was analyzed by flow cytometry (top panel). Soluble JAM-A released into the conditioned media was detected Western blotting using an anti–hJAM-A monoclonal antibody and quantified by densitometry (bottom panels). (C) JAM-A shedding in adam17−/− fibroblasts. Total cell extracts from adam17−/− and wild-typet MEFs were controlled for the absence and presence of ADAM17 by Western blotting (top panel). Adam17−/− and wild-type MEFs were transiently transfected with JAM-A. After 48 hours, conditioned media and cell lysates were harvested and analyzed by Western blotting for the presence of sJAM-A and flJAM-A (bottom panels, left and right, respectively). (D) Recombinant JAM-A.Fc was incubated with the recombinant catalytic domain of ADAM17 for 3 hours, and subsequently cleavage products were analyzed by Western blotting using a monoclonal antibody against JAM-A. (Top panel) HEK293 cells were incubated with the recombinant catalytic domain of ADAM17 for 3 hours, and conditioned media were analyzed for the presence of JAM-A by Western blotting. (Bottom panel) All Western blots were shown as representative experiments, and quantified signals were calculated as mean plus or minus SD for 3 independent experiments. *Statistically significant changes in JAM-A expression compared with the control (P < .05).
Loss- and gain-of-function experiments indicate a role for ADAM17 in JAM-A shedding. (A) Overexpression of ADAM10 and 17 in human HEK293 cells. Cells were transiently transfected with ADAM10 and ADAM17 or the empty expression vector as a control. Overexpression was controlled by flow cytometric analysis of ADAM10/17 surface expression (top panel). Conditioned media (middle panel) were analyzed for the presence of JAM-A by Western blotting, and signals were quantified by densitometry and calculated as percentage of the mock-transfected control (bottom panel). (B) Down-regulation of endogenous ADAM10/17 using siRNA. HUVECs were transiently transfected with ADAM10- ADAM17- or control-siRNA. Down-regulation of ADAM10 and ADAM17 surface expression was analyzed by flow cytometry (top panel). Soluble JAM-A released into the conditioned media was detected Western blotting using an anti–hJAM-A monoclonal antibody and quantified by densitometry (bottom panels). (C) JAM-A shedding in adam17−/− fibroblasts. Total cell extracts from adam17−/− and wild-typet MEFs were controlled for the absence and presence of ADAM17 by Western blotting (top panel). Adam17−/− and wild-type MEFs were transiently transfected with JAM-A. After 48 hours, conditioned media and cell lysates were harvested and analyzed by Western blotting for the presence of sJAM-A and flJAM-A (bottom panels, left and right, respectively). (D) Recombinant JAM-A.Fc was incubated with the recombinant catalytic domain of ADAM17 for 3 hours, and subsequently cleavage products were analyzed by Western blotting using a monoclonal antibody against JAM-A. (Top panel) HEK293 cells were incubated with the recombinant catalytic domain of ADAM17 for 3 hours, and conditioned media were analyzed for the presence of JAM-A by Western blotting. (Bottom panel) All Western blots were shown as representative experiments, and quantified signals were calculated as mean plus or minus SD for 3 independent experiments. *Statistically significant changes in JAM-A expression compared with the control (P < .05).
We next used adam17−/− MEFs to confirm the importance of ADAM17 for JAM-A release. JAM-A was transfected into wild type and adam17−/− MEFs to yield approximately equal expression of the molecule in the cell lysates. However, considerably less sJAM-A was released in the absence of ADAM17 (Figure 5C). No such effect was observed when JAM-A–transfected adam10−/− MEFs were analyzed for JAM-A shedding (Figure S1E). Although the release of JAM-A was not completely suppressed by either down-regulation or genetic knockout of ADAM17, these data nevertheless confirm that ADAM17 is critically involved in the mechanism of JAM-A shedding in the different cell types tested. To provide evidence for the ability of ADAM17 to directly cleave JAM-A recombinant soluble fusion protein of JAM-A (amino acids 28-242, comprising the ectodomain plus part of the transmembrane domain) linked to the Fc part of human IgG (JAM-A.Fc)25 was incubated with the recombinant catalytic domain of ADAM17, leading to the generation of a 32-kDa JAM-A fragment as demonstrated by Western blotting. A JAM-A fragment of comparable size was also obtained when HEK293 cells were treated with the recombinant catalytic domain of ADAM17 (Figure 5D). To narrow down the cleavage site, recombinant JAM-A.Fc was treated with ADAM17 and subsequently deglycosylated using N-Glycosidase F. After in-gel trypsin digestion, cleavage fragments were then analyzed by mass spectrometry and assigned according to the sequence of JAM-A (Figure S2). The most C-terminal assigned fragment corresponded to the sequence MEAVER (positions 229-234 of JAM-A), which is separated from the predicted transmembrane domain (positions 239-259 of flJAM-A) by 5 intervening amino acids. These data suggest that JAM-A is shed at a site very close to the cell membrane.
Regulation of JAM-A at interendothelial junctions
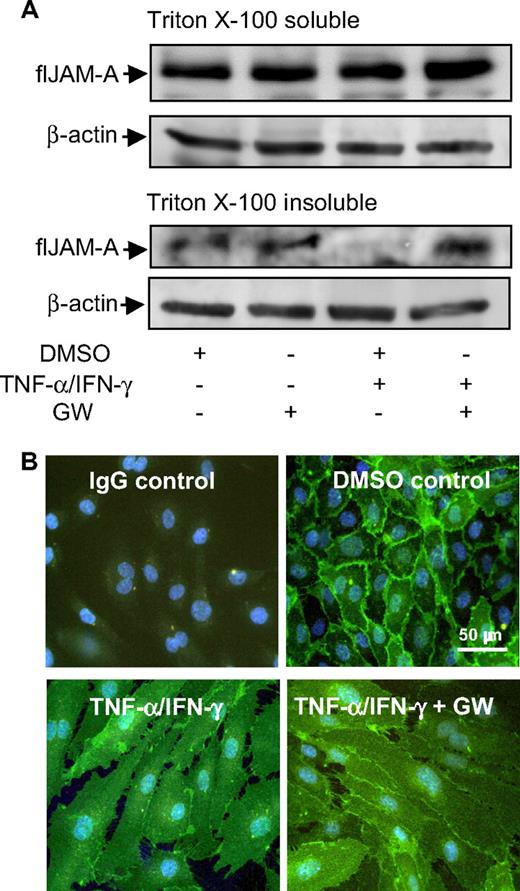
IFN-γ and TNF-α have been demonstrated to stimulate endothelial cells to redistribute JAM-A from the basal to the apical site,11,12 which was associated with a shift of JAM-A from Triton-insoluble to Triton-soluble fractions of endothelial cell extracts.34 We therefore questioned where JAM-A shedding would occur in endothelial cells. Western blot analysis revealed that JAM-A shedding was restricted to the Triton-insoluble fraction, as indicated by the absence of JAM-A in the respective extracts after cytokine stimulation and complete restoration of JAM-A in the presence of the ADAM17/10 inhibitor GW280264X (Figure 6A). Immunocytochemistry confirmed that JAM-A disappeared from lateral contacts on cytokine stimulation, which was rescued to a large extent by GW280264X (Figure 6B). These data suggest that ADAM-mediated shedding is a relevant process controlling the subcellular localization of the JAM-A pool, organized in Triton-insoluble microdomains, and preferentially expressed at lateral sites.
Effect of TNF-α/IFN-γ on the subcellular localization of JAM-A. (A) HUVECs were incubated for 16 hours in the absence or presence of TNF-α/IFN-γ and GW254023X. Triton X-100–soluble and –insoluble JAM-A was analyzed by Western blotting using an anti–hJAM-A monoclonal antibody. Detection of β-actin in the lysates confirmed equal loading. (B) Confluent HUVECs were incubated for 16 hours with TNF-α/IFN-γ in the absence or presence of GW254023X (GW) or vehicle control (DMSO). Samples were fixed and stained with anti–hJAM-A monoclonal antibody, followed by Alexa Fluor 488–conjugated goat anti–mouse IgG secondary antibody (green) and counterstaining of nuclei (blue).
Effect of TNF-α/IFN-γ on the subcellular localization of JAM-A. (A) HUVECs were incubated for 16 hours in the absence or presence of TNF-α/IFN-γ and GW254023X. Triton X-100–soluble and –insoluble JAM-A was analyzed by Western blotting using an anti–hJAM-A monoclonal antibody. Detection of β-actin in the lysates confirmed equal loading. (B) Confluent HUVECs were incubated for 16 hours with TNF-α/IFN-γ in the absence or presence of GW254023X (GW) or vehicle control (DMSO). Samples were fixed and stained with anti–hJAM-A monoclonal antibody, followed by Alexa Fluor 488–conjugated goat anti–mouse IgG secondary antibody (green) and counterstaining of nuclei (blue).
In vitro activity of soluble JAM-A
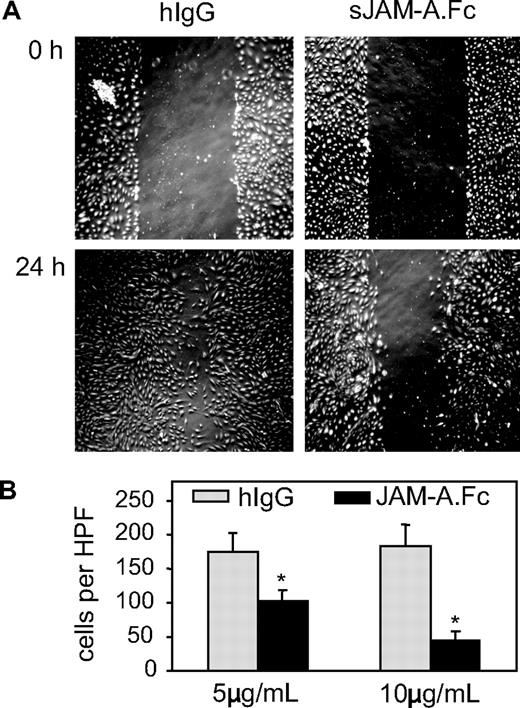
We next explored potential antagonistic effects of soluble JAM-A on endothelial cell functions. JAM-A is involved in the migration of endothelial cells after wounding of the endothelial cell layer.7 We therefore analyzed endothelial cell migration in the absence or presence of sJAM-A using an in vitro scratch assay, in which endothelial cell migration into a defined scratch area was monitored for 24 hours. For these experiments, the soluble fusion protein JAM-A.Fc and an irrelevant human IgG1 control were used. JAM-A.Fc but not IgG control dose-dependently blocked endothelial wound closure, suggesting that JAM-A.Fc suppresses endothelial cell migration (Figure 7).
Antagonistic effects of sJAM-A on endothelial cells. (A,B) A confluent HUVEC layer was wounded by a defined scratch; and after of 24 hours of incubation in the presence or absence of dimeric sJAM-A.Fc or human IgG (both 5 and 10 μg/mL), endothelial cells that had migrated into the scratch area were counted. (A) A representative experiment. (B) Mean plus or minus SD values were calculated for 3 independent experiments. *Statistically significant changes in endothelial migration compared with the control receiving hIgG (P < .05).
Antagonistic effects of sJAM-A on endothelial cells. (A,B) A confluent HUVEC layer was wounded by a defined scratch; and after of 24 hours of incubation in the presence or absence of dimeric sJAM-A.Fc or human IgG (both 5 and 10 μg/mL), endothelial cells that had migrated into the scratch area were counted. (A) A representative experiment. (B) Mean plus or minus SD values were calculated for 3 independent experiments. *Statistically significant changes in endothelial migration compared with the control receiving hIgG (P < .05).
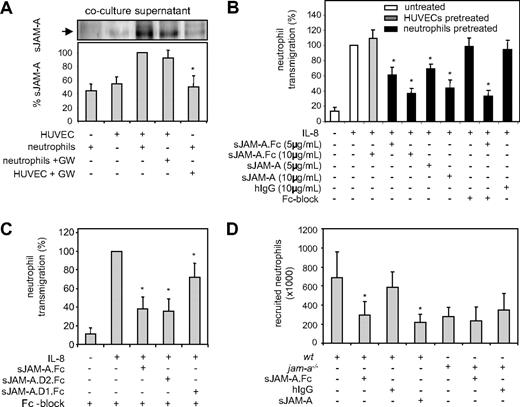
Besides endothelial cell migration, JAM-A also affects leukocyte recruitment by promoting their migration through endothelial cells.8 Western blot analysis indicated that only very small amounts of JAM-A were released from HUVECs alone or isolated neutrophils over a period of 2 hours. However, when HUVECs were cocultured with neutrophils for the same time period, considerable amounts of JAM-A were released (Figure 8A). Pretreatment of HUVECs with GW280264 for 2 hours before coculture markedly blocked JAM-A release, whereas similar pretreatment of neutrophils had no effect. The increased JAM-A release was associated with a reduction of JAM-A surface expression on HUVECs by 42% but not on neutrophils (Figure S3A). These findings indicate that shedding of endothelial JAM-A is enhanced in the course of neutrophil interaction with endothelial cells.
Antagonistic effects of sJAM-A on leukocyte recruitment. (A) HUVECs and neutrophils were pretreated for 1 hour with GW280264X (GW) or left untreated. After removal of the inhibitor by washing, both cell types were either coincubated or incubated separately for 2 hours, and subsequently conditioned media were investigated for the release of sJAM-A by Western blotting. Western blot signals were quantified and shown as mean plus or minus SD for 3 independent experiments. *Significant inhibition of JAM-A from cocultured cells after pretreatment of neutrophils with GW280264X. (B) HUVECs or isolated neutrophils were pretreated with the indicated concentrations of sJAM-A.Fc, monovalent sJAM, or irrelevant hIgG in the presence and absence of Fc block (50 μg/mL) for 30 minutes and subsequently washed. Neutrophils were then assayed for transendothelial migration toward IL-8. *Statistically significant changes in IL-8–induced neutrophil transmigration compared with cells receiving IgG control. (C) Isolated neutrophils were pretreated with sJAM-A.Fc, sJAM-A.D1.Fc, or sJAM-A.D2.Fc (all 10 μg/mL) in the presence of Fc block (50 μg/mL) for 30 minutes and subsequently assayed for transendothelial migration as described in panel B. *Statistically significant changes in IL-8–induced neutrophil transmigration compared with the control treated with Fc block only. (D) JAM-A.Fc (60 μg), monovalent sJAM (40 μg), or human IgG (60 μg) were intravenously administered to wild-type and jam-a−/− mice (n = 4 per group). One hour later, the neutrophil attracting chemokine KC or vehicle control was injected into experimentally induced air pouches. Four hours after KC injection, neutrophil recruitment into the air pouches was determined by flow cytometric analysis of the differentially labeled leukocyte populations. *Statistically significant changes in neutrophil infiltration compared with the IgG-treated control (P < .05).
Antagonistic effects of sJAM-A on leukocyte recruitment. (A) HUVECs and neutrophils were pretreated for 1 hour with GW280264X (GW) or left untreated. After removal of the inhibitor by washing, both cell types were either coincubated or incubated separately for 2 hours, and subsequently conditioned media were investigated for the release of sJAM-A by Western blotting. Western blot signals were quantified and shown as mean plus or minus SD for 3 independent experiments. *Significant inhibition of JAM-A from cocultured cells after pretreatment of neutrophils with GW280264X. (B) HUVECs or isolated neutrophils were pretreated with the indicated concentrations of sJAM-A.Fc, monovalent sJAM, or irrelevant hIgG in the presence and absence of Fc block (50 μg/mL) for 30 minutes and subsequently washed. Neutrophils were then assayed for transendothelial migration toward IL-8. *Statistically significant changes in IL-8–induced neutrophil transmigration compared with cells receiving IgG control. (C) Isolated neutrophils were pretreated with sJAM-A.Fc, sJAM-A.D1.Fc, or sJAM-A.D2.Fc (all 10 μg/mL) in the presence of Fc block (50 μg/mL) for 30 minutes and subsequently assayed for transendothelial migration as described in panel B. *Statistically significant changes in IL-8–induced neutrophil transmigration compared with the control treated with Fc block only. (D) JAM-A.Fc (60 μg), monovalent sJAM (40 μg), or human IgG (60 μg) were intravenously administered to wild-type and jam-a−/− mice (n = 4 per group). One hour later, the neutrophil attracting chemokine KC or vehicle control was injected into experimentally induced air pouches. Four hours after KC injection, neutrophil recruitment into the air pouches was determined by flow cytometric analysis of the differentially labeled leukocyte populations. *Statistically significant changes in neutrophil infiltration compared with the IgG-treated control (P < .05).
Next, we assayed the effect of JAM-A.Fc on IL-8–induced transendothelial migration of isolated human neutrophils. Pretreatment with JAM-A–Fc of neutrophils but not of endothelial cells led to a considerably reduced transendothelial migration (Figure 8B). This indicates that leukocytes are the primary target of soluble JAM-A, probably related to the fact that endothelial JAM-A is less accessible because of its position at the intercellular junction. The blocking effect of JAM-A was not mediated by binding via Fc-receptors because monovalent JAM-A also blocked neutrophil transmigration. In addition, the irrelevant human control IgG did not have an inhibitory effect, nor was the effect of JAM-A.Fc modulated by blocking of Fc receptors on neutrophils. Moreover, a JAM-A.Fc variant containing only the second Ig domain was capable of efficiently blocking transmigration, whereas the variant containing only the first IgG domain had a minor yet significant effect (Figure 8C). These data suggest that the inhibitory activity of sJAM-A on neutrophils is predominantly mediated by the second membrane-proximal Ig domain through blocking JAM-A interactions with LFA-1. However, the first IgG domain of JAM-A may also contribute to this inhibitory activity, albeit to a lesser extent. In support of the notion that neutrophils interact with the D2 domain of JAM-A, we also found that pretreatment of endothelial cells with an antibody against the D2 domain reversed transendothelial migration of neutrophils (Figure S3B), whereas pretreatment of neutrophils with this antibody had no effect. Because recombinant ADAM17 is capable of cleaving JAM-A at the cell surface, we also investigated its effect on neutrophil transmigration. Transmigration experiments demonstrated that pretreatment of HUVECs with soluble ADAM17 catalytic domain slightly decreases transmigration only when shedding products are present during the transmigration process, supporting the view that JAM-A molecules that are shed on-site may indeed function to attenuate transmigration (Figure S3C).
In vivo activity of soluble JAM-A
Similar as in humans, the close murine homolog of JAM-A is also found within endothelial cell contacts of mice and appears to fulfill similar functions in both species.1 Having demonstrated that human sJAM-A.Fc not only binds to human but also to murine neutrophils (Figure S3D), we then tested whether an excess of human sJAM-A can be used to antagonize murine neutrophil migration in vivo. Chemokine-induced recruitment of leukocytes was investigated in an experimentally induced air pouch model (n = 4 per group). Using wild-type and jam-a−/− mice, we first established that efficient neutrophil recruitment into the air pouch in response to the murine CXC-chemokine KC is dependent on expression of endogenous JAM-A (Figure 8D). Intravenous injection of sJAM-A.Fc or monovalent sJAM-A into wild-type mice efficiently blocked the infiltration of neutrophils (Figure 8D). This in vivo activity of exogenous sJAM-A was confirmed by a different set of experiments (n = 5 per group) using human IL-8 as a chemoattractant (Figure S3E). In jam-a−/− mice, however, KC-induced residual neutrophil infiltration was not altered by soluble JAM-A.Fc (Figure 8D), suggesting that the effect of soluble JAM-A was also dependent on the presence of endogenous JAM-A. These data show that sJAM-A can limit neutrophil migration in vivo.
Discussion
This is the first report describing the proteolytic release of JAM-A by cultured endothelial cells. Our inhibition experiments as well as loss- and gain-of-function studies provide multiple lines of evidence for an important role of disintegrin-like metalloproteinases as JAM-A sheddases. ADAM17 emerged as the major protease contributing to JAM-A shedding, whereas ADAM10 plays a minor role. By showing that recombinant ADAM17 cleaves purified recombinant JAM-A.Fc, we also provide evidence that ADAM17 is capable of cleaving JAM-A proximal to the cell membrane. These data strongly suggest that JAM-A represents a novel ADAM17 substrate, although it cannot be completely excluded that ADAM17 activates an intermediate protease, which could also participate in the cleavage of JAM-A in vivo. JAM-A shedding in vivo and in vitro is up-regulated by proinflammatory stimuli, including PAF and cytokines (IFN-γ/TNF-α) inducing increased ADAM17 activity, or by contact to neutrophils finally leading to the enhanced release of soluble JAM-A and to reduced surface expression of the membrane-expressed JAM-A variant on endothelial cells. As potential consequences of JAM-A shedding, we observed inhibition of neutrophil transmigration by released JAM-A in vitro and in vivo. We therefore propose that released JAM-A may not only serve as a biomarker for vascular inflammation, but cleavage of JAM-A may also be instrumental in controlling vascular permeability and neutrophil transmigration.
ADAM10 and ADAM17 are constitutively expressed in various tissues, including vascular cells. Proteolytic cleavage by ADAM10 and ADAM17 activity can be up-regulated by proinflammatory stimuli, such as lipopolysaccharide, pore-forming toxins, ligands of G protein–coupled receptors, and inducers of apoptosis.14,15 This implies a posttranslational regulation in proteolytic activity rather than an up-regulation in protein synthesis or surface expression of the proteases. We here provide evidence that JAM-A shedding by endothelial cells is up-regulated by the proinflammatory cytokines IFN-γ and TNF-α. Neither protein synthesis of JAM-A nor surface expression of ADAM10 and ADAM17 was altered by stimulation with both cytokines, whereas cleavage of a synthetic substrate for ADAM10 and ADAM17 was clearly up-regulated. These data suggest that the increased release of JAM-A was mediated by an up-regulation of sheddase activity and were complemented by experiments with mice undergoing systemic inflammation on injection of IFN-γ and TNF-α, which rapidly up-regulates serum levels of sJAM-A within 2.5 hours. Recently, IFN-γ and TNF-α were reported to induce redistribution of JAM-A from the basal to the apical site.11,12 Cytokine stimulation was also associated with a translocation of JAM-A from Triton-insoluble to Triton-soluble microdomains.34 However, as we demonstrate here, shedding by ADAM17 and ADAM10 is another important modality of JAM-A regulation. Increased JAM-A shedding leads to the down-regulation of the molecule from vascular junctions and to its disappearance from Triton-insoluble microdomains, and inhibition of shedding prevents these effects. These findings suggest that the decrease of JAM-A in endothelial junctions in response to proinflammatory stimuli is predominantly the result of proteolytic shedding rather than redistribution of JAM-A on the cell surface. Our coculture and inhibition experiments reveal that increased JAM-A shedding on endothelial cells is also induced by the presence of neutrophils and that the increased activity of metalloproteinases acting in cis on the endothelial surface can mediate this effect. This up-regulation of shedding activity was seen already after 2 hours of coincubation with neutrophils, suggesting that it is predominantly mediated by an increase of proteolytic activity rather than protein expression of ADAMs or JAM-A. In the onset of leukocyte transmigration, it appears feasible that enhanced JAM-A shedding is the result of activity regulation of ADAM17, whereas protein induction of the protease could potentially become relevant at later phases of the inflammatory process.
It is conceivable that, in response to enhanced proteolytic shedding activity, increased levels of JAM-A will be generated at sites of vascular inflammation, leading to the accumulation of JAM-A in the blood. Our observations therefore reveal the underlying molecular mechanism of how soluble JAM-A could function as a biomarker of vascular inflammation, such as atherosclerosis. Indeed, recent ELISA measurements demonstrated elevated JAM-A concentrations in serum samples of patients with atherosclerotic disease, and these increased levels of JAM-A correlated with high serum levels of TNF-α, suggesting a common mechanism for the release of these molecules.35 As we demonstrate here, systemic TNF-α and IFN-γ induce release of soluble JAM-A in mice. Although the serum levels are relatively low (in the nanomolar range), JAM-A may occur at higher local concentrations within the inflamed blood vessel wall at sites of JAM-A expression and cleavage. The concentration of soluble JAM-A in interendothelial junctions is not known, and it remains an open question whether sufficient JAM-A will accumulate over time to exert inhibitory functions. Pretreatment of HUVECs with soluble ADAM17 before transmigration of neutrophils suggested that the locally released JAM-A could be concentrated enough to attenuate diapedesis of neutrophils because ADAM17 treatment per se did not influence transmigration. However, it appears probable that soluble JAM-A does not act systemically. The proposed inhibitory effects of released JAM-A on endothelial cells and transmigrating leukocytes may rather represent locally confined mechanisms of control, especially within the endothelial junction at the site of leukocyte transmigration. It can be envisaged that transmigrating leukocytes induce the shedding of JAM-A, which then occupies binding molecules on neutrophils and might finally block JAM-A–dependent reverse transmigration of extravasated neutrophils.
Expressed in tight junctions, JAM-A has been found to contribute to the establishment of the endothelial and epithelial diffusion barrier.4,5,36 Our study suggests that increased shedding of JAM-A in cytokine stimulated endothelial cells leads to proteolytic down-regulation of JAM-A at the endothelial cell contacts. Therefore, shedding of JAM-A may directly affect vascular permeability. However, other molecules, such as VE-cadherin, are also involved in the establishment of the vascular diffusion barrier and recently shedding of VE-cadherin by ADAM10 has been reported.33 By cleaving several substrate molecules expressed at the interendothelial junction, ADAM17 and ADAM10 would be highly effective in controlling vascular permeability.
The mechanism how JAM-A contributes to transendothelial migration is still not completely understood.8-10 In the course of leukocyte transmigration, JAM-A may provide a molecular zipper that opens by resolving homophilic interactions at interendothelial contacts and establishes new homophilic and heterophilic interactions with JAM-A or LFA-1 on the transmigrating leukocytes.1 Indeed, recent work has revealed that binding of LFA-1 to the membrane-proximal domain 2 of JAM-A destabilizes the homophilic JAM-A–JAM-A interaction.37 This would allow the leukocytes to further penetrate the interendothelial space. It is conceivable that soluble JAM-A blocks this process by interacting with its binding partners JAM-A and LFA-1 on the leukocyte surface. Indeed, neutrophil transmigration was specifically blocked in vitro and in vivo by administration of soluble JAM-A through a neutrophil-targeted mechanism of action. Moreover, as we demonstrate here by the use of JAM-A.Fc deletion variants, the inhibitory activity of JAM-A on neutrophils is predominantly mediated by the second Ig domain but also to some degree by the first IgG domain. Inhibition experiments with antibodies that bind to the second domain support the notion that the D2 domain on endothelial cells plays an important role in neutrophil transmigration. The second IgG domain is responsible for the interaction with LFA-1, which is critically involved in leukocyte transmigration,12 whereas the N-terminal IgG domain mediates homophilic interactions.38 In addition, the in vivo recruitment experiments in jam-a−/− mice indicate that JAM-A in general is also required for inhibition by exogenously added soluble JAM-A, implying a function of homophilic interactions. Therefore, these observations suggest that the inhibition of neutrophil migration by JAM-A is the result of both interference with LFA-1 functions on the neutrophil surface as well as with JAM-A–JAM-A interactions, thus disrupting the functional interplay between heterophilic and homophilic interactions, which constitute the molecular zipper at interendothelial junctions.1
In conclusion, our data indicate that JAM-A is shed by ADAM17 and ADAM10 at sites of vascular inflammation, which has multiple consequences: at the endothelial cell surface, shedding would release a soluble antagonist interacting with adhesion molecules on neutrophils and thereby down-regulate their transmigration. Moreover, within the endothelial junctions, shedding of JAM-A may also function to increase endothelial permeability. Inhibition of vascular shedding enzymes could represent a therapeutic strategy to control vascular inflammation. However, it would be important to carefully consider shedding of other well-known substrates of these proteases, including TNF-α, TNFR2, IL6R, L-selectin, transmembrane chemokines, and VCAM-1, which are all implicated in inflammatory processes within the vasculature.15,16
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Tanja Kogel, Melanie Esser, Sabine Winkler, Dennis Suylen, and Freek Bouwman for expert technical assistance.
This work was supported in part by the Interdisciplinary Center for Clinical Research Biomat of the Rheinisch-Westfaelische Technische Hochschule Aachen and by the Deutsche Forschungsgemeinschaft (FOR809/P5 and P6, SFB 542/A12, SFB415, and postdoctoral grant SO876/1-1) and the Center of Excellence Inflammation at Interfaces at the University of Kiel.
Authorship
Contribution: J.P. designed and performed experiments, analyzed data, and made the figures; R.R.K. provided substantial intellectual input and reagents; O.S, L.F., A.Z., A.S., and N.S. performed experiments and analyzed data; K.R. provided vital reagents and intellectual input; T.M.H., L.L., and C.W. provided substantial techniques and intellectual input; and A.L. designed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andreas Ludwig, Institute for Pharmacology and Toxicology, RWTH Aachen University, Pauwelsstrasse 30, 52074 Aachen, Germany; e-mail: aludwig@ukaachen.de.
References
Author notes
*R.R.K. and J.P. contributed equally to this article.