Abstract
Activation of the nuclear factor–κB pathway by Epstein-Barr virus–encoded latent membrane protein-1 (LMP-1) leads to an up-regulation of the major histocompatibility complex class I antigen–processing pathway. Paradoxically, LMP-1 itself induces a subdominant CD8+ T-cell response and appears to have evolved to avoid immune recognition. Here we show that, although expression of LMP-1 in human cells dramatically enhanced the trans-presentation of CD8+ T-cell epitopes, cis-presentation of LMP-1–derived epitopes was severely impaired. Testing of a series of LMP-1 mutants revealed that deletion of the first transmembrane domain of LMP-1, which prevented self-aggregation, significantly enhanced cis-presentation of T-cell epitopes from this protein, whereas it lost its ability to up-regulate trans-presentation. Interestingly, we also found that cis-presentation of LMP-1 epitopes was rescued by blocking the proteasome function. Taken together, these results delineate a novel mechanism of immune evasion, which renders a virally encoded oncogene inaccessible to the conventional major histocompatibility complex class I pathway limiting its cis-presentation to effector cells.
Introduction
The specificity of interaction between T cells and antigen-presenting cells is determined by 2 distinct components, namely, major histocompatibility complex (MHC)–restricted presentation of a peptide epitope and a heterodimeric αβ cell-surface protein called the T-cell receptor. Viruses that establish persistent infection or are associated with malignant transformation have evolved unique mechanisms to interfere with this interaction to evade antiviral T-cell responses.1,2 These escape mechanisms include down-regulation of antigen processing, limiting expression of immunodominant viral proteins, viral replication in immune-privileged tissues, and genetic variation affecting peptide binding to MHC class I or recognition by the T cells.3-5 Epstein-Barr virus (EBV) exploits many immune evasive strategies to successfully establish a latent infection in B cells.6 Analysis of the role of individual EBV latent antigens in the regulation of antigen-processing genes indicated that latent membrane protein-1 (LMP-1) was sufficient to up-regulate expression of transporters associated with antigen processing and trans-presentation of MHC class I–restricted epitopes in B cells.3 Subsequent studies demonstrated that the immunomodulatory effects of LMP-1 are mediated through C-terminal activator regions (CTAR1 and CTAR2),7 which are involved in the induction of the nuclear factor–κB (NF-κB) family protein Rel-B.8 Paradoxically, CD8+ T-cell responses to LMP-1 are generally very low and rarely detected in healthy virus carriers, suggesting that LMP-1 may limit its cis-presentation through the MHC class I pathway.9,10 We have delineated a mechanism by which LMP-1 limits its self-presentation without compromising its ability to modulate trans-presentation of CD8+ T-cell epitopes.
Methods
Establishment and maintenance of cell lines
EBV-transformed lymphoblastoid cell lines (LCLs) transformed with B95.8 virus isolate were used in this study. In addition, 174xCEM.T1 (T1) and antigen-processing defective 174xCEM.T2 cells (T2),11 human cervical carcinoma epithelial cell line C33A,12 a human nasopharyngeal carcinoma cell line C666.1,13 and a B-cell lymphoma cell line, BJAB, with or without LMP-1 (referred to as BJAB.MTL6 and BJAB.gpt),3 were also used in this study. This study was approved by the Queensland Institute of Medical Research Human Research and Ethics Committee.
Generation of EBV- and HCMV-specific T-cell lines
LMP-1 expression vectors and transient transfection
Intracellular cytokine assay
Fluorescence microscopy
C33A cells were transfected with LMP-1 expression vectors, fixed and stained with DAPI and phalloidin, and analyzed using a DeltaVision Core DV (Applied Precision, Issaquah, WA).
Results and discussion
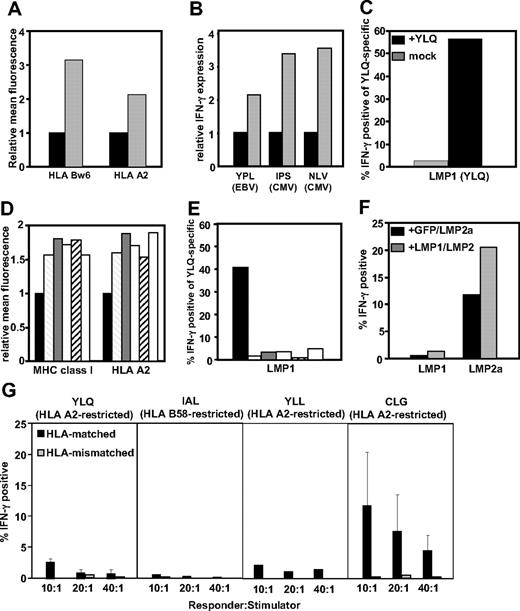
To assess the impact of LMP-1 expression on cis- and trans-presentation of MHC class I–restricted epitopes, BJAB-MTLM6 (LMP-1+) and BJAB-gpt (LMP-1−) cells, which display a marked difference in surface MHC expression (Figure 1A), were infected with recombinant vaccinia virus encoding the EBNA-3 or the HCMV-encoded, pp65, and then incubated with CD8+ T cells specific for EBNA-3, pp65, or LMP-1. As previously reported,3 a 2- to 3-fold higher stimulation of EBNA-3 and pp65-specific T cells was observed when exposed to BJAB.MTLM6 cells compared with BJAB-gpt cells (Figure 1B). In contrast, activation of T cells specific for an human leukocyte antigen (HLA) A2–restricted LMP-1 epitope (YLQQNWWTL),9,10 after incubation with BJAB-MTLM6 cells, was only evident after presensitization of these cells with peptide (Figure 1C). It is important to stress here that poor endogenous recognition of LMP-1 was not the result of low avidity of LMP-1–specific T cells (data not shown).
Cis- and trans-presentation of CD8+ T-cell epitopes in LMP-1–expressing cells. (A) LMP-1− BJAB-gpt (■) and LMP-1+ BJAB-MTLM6 ( ) cells were assessed for HLA class I expression by flow cytometery. Data represent the relative mean fluorescence after staining with anti-HLA-Bw6 and anti-HLA-A2–specific monoclonal antibodies. (B) HLA B35–restricted YPLHEQHGM (EBV-encoded EBNA3), HLA B35–restricted IPSINVHHY (HCMV-encoded pp65), and HLA A2–restricted NLVPMVATV (HCMV-encoded pp65)–specific T cells were incubated with BJAB-gpt (■) and BJAB-MTLM6 (
) cells were assessed for HLA class I expression by flow cytometery. Data represent the relative mean fluorescence after staining with anti-HLA-Bw6 and anti-HLA-A2–specific monoclonal antibodies. (B) HLA B35–restricted YPLHEQHGM (EBV-encoded EBNA3), HLA B35–restricted IPSINVHHY (HCMV-encoded pp65), and HLA A2–restricted NLVPMVATV (HCMV-encoded pp65)–specific T cells were incubated with BJAB-gpt (■) and BJAB-MTLM6 ( ) cells infected with recombinant vaccinia viruses encoding EBNA3 or pp65, in the presence of brefeldin A. Cells were incubated with MHC-peptide pentamers and anti-CD8 antibody and then assessed for intracellular IFN-γ production. Data represent the relative increase in IFN-γ+ cells after incubation with BJAB-MTLM6 infected cells compared with BJAB-gpt infected cells. (C) CD8+ T cells specific for a HLA A2–restricted epitope, YLQQNWWTL (EBV-encoded LMP-1), were incubated in the presence of brefeldin A with BJAB-MTLM6 cells pulsed with and without 1 μg/mL YLQQNWWTL peptide (the first 3 amino acids of the sequences are underlined to indicate the abbreviation used for each epitope in the figure). Data represent the percentage of YLQQNWWTL-specific T cells producing IFN-γ after incubation with BJAB-MTLM6 cells at a responder-to-stimulator ratio of 10:1. (D) HLA class I expression in C33A cells after expression of LMP-1. C33A cells were transfected with EGFP-tagged expression vectors encoding GFP alone (■) or different LMP-1 sequences, including B95.8-LMP-1 (
) cells infected with recombinant vaccinia viruses encoding EBNA3 or pp65, in the presence of brefeldin A. Cells were incubated with MHC-peptide pentamers and anti-CD8 antibody and then assessed for intracellular IFN-γ production. Data represent the relative increase in IFN-γ+ cells after incubation with BJAB-MTLM6 infected cells compared with BJAB-gpt infected cells. (C) CD8+ T cells specific for a HLA A2–restricted epitope, YLQQNWWTL (EBV-encoded LMP-1), were incubated in the presence of brefeldin A with BJAB-MTLM6 cells pulsed with and without 1 μg/mL YLQQNWWTL peptide (the first 3 amino acids of the sequences are underlined to indicate the abbreviation used for each epitope in the figure). Data represent the percentage of YLQQNWWTL-specific T cells producing IFN-γ after incubation with BJAB-MTLM6 cells at a responder-to-stimulator ratio of 10:1. (D) HLA class I expression in C33A cells after expression of LMP-1. C33A cells were transfected with EGFP-tagged expression vectors encoding GFP alone (■) or different LMP-1 sequences, including B95.8-LMP-1 ( ), CAO-LMP-1 (
), CAO-LMP-1 ( ), HS6-LMP-1 (
), HS6-LMP-1 ( ), NPC9-LMP-1 (
), NPC9-LMP-1 ( ), and QC-LMP-1 (□). Data represent the relative mean fluorescence, compared with cells transfected with GFP-expression vector, after staining with anti-HLA class I and anti-HLA-A2–specific antibodies. (E) YLQQNWWTL-specific T cells were incubated in the presence of brefeldin A with C33A cells transfected with B95.8-LMP-1 (
), and QC-LMP-1 (□). Data represent the relative mean fluorescence, compared with cells transfected with GFP-expression vector, after staining with anti-HLA class I and anti-HLA-A2–specific antibodies. (E) YLQQNWWTL-specific T cells were incubated in the presence of brefeldin A with C33A cells transfected with B95.8-LMP-1 ( ), CAO-LMP-1 (
), CAO-LMP-1 ( ), HS6-LMP-1 (
), HS6-LMP-1 ( ), NPC9-LMP-1 (
), NPC9-LMP-1 ( ), and QC-LMP-1 (□) or B95.8-LMP-1 sensitized with synthetic YLQQNWWTL peptide epitope (■). Cells were incubated with MHC peptide pentamers and anti-CD8 and then assessed for intracellular IFN-γ production. Data represent the percentage of IFN-γ+ YLQ-specific T cells. (F) CD8+ T cells specific for HLA A2–restricted YLQQNWWTL epitope (EBV-encoded LMP-1) and HLA A2–restricted CLGGLLTMV epitope (EBV-encoded LMP-2A) were incubated with C33A cells cotransfected with vectors encoding GFP and LMP-2A-GFP or LMP-1-GFP and LMP-2A-GFP and then assessed for intracellular IFN-γ production. Data represent the percentage of pentamer-positive cells producing IFN-γ at a responder-to-stimulator ratio of 10:1. (G) CD8+ T cells specific for HLA A2–restricted YLQQNWWTL epitope (EBV-encoded LMP-1), HLA B58–restricted IALYLQQNW epitope (EBV-encoded LMP-1 protein), HLA A2–restricted YLLEMWRL epitope (EBV-encoded LMP-1 protein), and HLA A2–restricted CLGGLLTMV epitope (EBV-encoded LMP-2A) were incubated with HLA-matched and -mismatched LCLs in the presence of brefeldin A and then assessed for intracellular IFN-γ production by flow cytometry. Data represent the mean IFN-γ–producing cells after stimulation with 3 different LCLs.
), and QC-LMP-1 (□) or B95.8-LMP-1 sensitized with synthetic YLQQNWWTL peptide epitope (■). Cells were incubated with MHC peptide pentamers and anti-CD8 and then assessed for intracellular IFN-γ production. Data represent the percentage of IFN-γ+ YLQ-specific T cells. (F) CD8+ T cells specific for HLA A2–restricted YLQQNWWTL epitope (EBV-encoded LMP-1) and HLA A2–restricted CLGGLLTMV epitope (EBV-encoded LMP-2A) were incubated with C33A cells cotransfected with vectors encoding GFP and LMP-2A-GFP or LMP-1-GFP and LMP-2A-GFP and then assessed for intracellular IFN-γ production. Data represent the percentage of pentamer-positive cells producing IFN-γ at a responder-to-stimulator ratio of 10:1. (G) CD8+ T cells specific for HLA A2–restricted YLQQNWWTL epitope (EBV-encoded LMP-1), HLA B58–restricted IALYLQQNW epitope (EBV-encoded LMP-1 protein), HLA A2–restricted YLLEMWRL epitope (EBV-encoded LMP-1 protein), and HLA A2–restricted CLGGLLTMV epitope (EBV-encoded LMP-2A) were incubated with HLA-matched and -mismatched LCLs in the presence of brefeldin A and then assessed for intracellular IFN-γ production by flow cytometry. Data represent the mean IFN-γ–producing cells after stimulation with 3 different LCLs.
Cis- and trans-presentation of CD8+ T-cell epitopes in LMP-1–expressing cells. (A) LMP-1− BJAB-gpt (■) and LMP-1+ BJAB-MTLM6 ( ) cells were assessed for HLA class I expression by flow cytometery. Data represent the relative mean fluorescence after staining with anti-HLA-Bw6 and anti-HLA-A2–specific monoclonal antibodies. (B) HLA B35–restricted YPLHEQHGM (EBV-encoded EBNA3), HLA B35–restricted IPSINVHHY (HCMV-encoded pp65), and HLA A2–restricted NLVPMVATV (HCMV-encoded pp65)–specific T cells were incubated with BJAB-gpt (■) and BJAB-MTLM6 (
) cells were assessed for HLA class I expression by flow cytometery. Data represent the relative mean fluorescence after staining with anti-HLA-Bw6 and anti-HLA-A2–specific monoclonal antibodies. (B) HLA B35–restricted YPLHEQHGM (EBV-encoded EBNA3), HLA B35–restricted IPSINVHHY (HCMV-encoded pp65), and HLA A2–restricted NLVPMVATV (HCMV-encoded pp65)–specific T cells were incubated with BJAB-gpt (■) and BJAB-MTLM6 ( ) cells infected with recombinant vaccinia viruses encoding EBNA3 or pp65, in the presence of brefeldin A. Cells were incubated with MHC-peptide pentamers and anti-CD8 antibody and then assessed for intracellular IFN-γ production. Data represent the relative increase in IFN-γ+ cells after incubation with BJAB-MTLM6 infected cells compared with BJAB-gpt infected cells. (C) CD8+ T cells specific for a HLA A2–restricted epitope, YLQQNWWTL (EBV-encoded LMP-1), were incubated in the presence of brefeldin A with BJAB-MTLM6 cells pulsed with and without 1 μg/mL YLQQNWWTL peptide (the first 3 amino acids of the sequences are underlined to indicate the abbreviation used for each epitope in the figure). Data represent the percentage of YLQQNWWTL-specific T cells producing IFN-γ after incubation with BJAB-MTLM6 cells at a responder-to-stimulator ratio of 10:1. (D) HLA class I expression in C33A cells after expression of LMP-1. C33A cells were transfected with EGFP-tagged expression vectors encoding GFP alone (■) or different LMP-1 sequences, including B95.8-LMP-1 (
) cells infected with recombinant vaccinia viruses encoding EBNA3 or pp65, in the presence of brefeldin A. Cells were incubated with MHC-peptide pentamers and anti-CD8 antibody and then assessed for intracellular IFN-γ production. Data represent the relative increase in IFN-γ+ cells after incubation with BJAB-MTLM6 infected cells compared with BJAB-gpt infected cells. (C) CD8+ T cells specific for a HLA A2–restricted epitope, YLQQNWWTL (EBV-encoded LMP-1), were incubated in the presence of brefeldin A with BJAB-MTLM6 cells pulsed with and without 1 μg/mL YLQQNWWTL peptide (the first 3 amino acids of the sequences are underlined to indicate the abbreviation used for each epitope in the figure). Data represent the percentage of YLQQNWWTL-specific T cells producing IFN-γ after incubation with BJAB-MTLM6 cells at a responder-to-stimulator ratio of 10:1. (D) HLA class I expression in C33A cells after expression of LMP-1. C33A cells were transfected with EGFP-tagged expression vectors encoding GFP alone (■) or different LMP-1 sequences, including B95.8-LMP-1 ( ), CAO-LMP-1 (
), CAO-LMP-1 ( ), HS6-LMP-1 (
), HS6-LMP-1 ( ), NPC9-LMP-1 (
), NPC9-LMP-1 ( ), and QC-LMP-1 (□). Data represent the relative mean fluorescence, compared with cells transfected with GFP-expression vector, after staining with anti-HLA class I and anti-HLA-A2–specific antibodies. (E) YLQQNWWTL-specific T cells were incubated in the presence of brefeldin A with C33A cells transfected with B95.8-LMP-1 (
), and QC-LMP-1 (□). Data represent the relative mean fluorescence, compared with cells transfected with GFP-expression vector, after staining with anti-HLA class I and anti-HLA-A2–specific antibodies. (E) YLQQNWWTL-specific T cells were incubated in the presence of brefeldin A with C33A cells transfected with B95.8-LMP-1 ( ), CAO-LMP-1 (
), CAO-LMP-1 ( ), HS6-LMP-1 (
), HS6-LMP-1 ( ), NPC9-LMP-1 (
), NPC9-LMP-1 ( ), and QC-LMP-1 (□) or B95.8-LMP-1 sensitized with synthetic YLQQNWWTL peptide epitope (■). Cells were incubated with MHC peptide pentamers and anti-CD8 and then assessed for intracellular IFN-γ production. Data represent the percentage of IFN-γ+ YLQ-specific T cells. (F) CD8+ T cells specific for HLA A2–restricted YLQQNWWTL epitope (EBV-encoded LMP-1) and HLA A2–restricted CLGGLLTMV epitope (EBV-encoded LMP-2A) were incubated with C33A cells cotransfected with vectors encoding GFP and LMP-2A-GFP or LMP-1-GFP and LMP-2A-GFP and then assessed for intracellular IFN-γ production. Data represent the percentage of pentamer-positive cells producing IFN-γ at a responder-to-stimulator ratio of 10:1. (G) CD8+ T cells specific for HLA A2–restricted YLQQNWWTL epitope (EBV-encoded LMP-1), HLA B58–restricted IALYLQQNW epitope (EBV-encoded LMP-1 protein), HLA A2–restricted YLLEMWRL epitope (EBV-encoded LMP-1 protein), and HLA A2–restricted CLGGLLTMV epitope (EBV-encoded LMP-2A) were incubated with HLA-matched and -mismatched LCLs in the presence of brefeldin A and then assessed for intracellular IFN-γ production by flow cytometry. Data represent the mean IFN-γ–producing cells after stimulation with 3 different LCLs.
), and QC-LMP-1 (□) or B95.8-LMP-1 sensitized with synthetic YLQQNWWTL peptide epitope (■). Cells were incubated with MHC peptide pentamers and anti-CD8 and then assessed for intracellular IFN-γ production. Data represent the percentage of IFN-γ+ YLQ-specific T cells. (F) CD8+ T cells specific for HLA A2–restricted YLQQNWWTL epitope (EBV-encoded LMP-1) and HLA A2–restricted CLGGLLTMV epitope (EBV-encoded LMP-2A) were incubated with C33A cells cotransfected with vectors encoding GFP and LMP-2A-GFP or LMP-1-GFP and LMP-2A-GFP and then assessed for intracellular IFN-γ production. Data represent the percentage of pentamer-positive cells producing IFN-γ at a responder-to-stimulator ratio of 10:1. (G) CD8+ T cells specific for HLA A2–restricted YLQQNWWTL epitope (EBV-encoded LMP-1), HLA B58–restricted IALYLQQNW epitope (EBV-encoded LMP-1 protein), HLA A2–restricted YLLEMWRL epitope (EBV-encoded LMP-1 protein), and HLA A2–restricted CLGGLLTMV epitope (EBV-encoded LMP-2A) were incubated with HLA-matched and -mismatched LCLs in the presence of brefeldin A and then assessed for intracellular IFN-γ production by flow cytometry. Data represent the mean IFN-γ–producing cells after stimulation with 3 different LCLs.
To extend these investigations, we included multiple LMP-1 sequences from both healthy persons and nasopharyngeal carcinoma (NPC) patients in our analysis. Data presented in Figure 1D show that, although all LMP-1 variants up-regulated the expression of MHC class I on a human epithelial cell line, C33A, considerable activation of LMP-1-specific T cells was only evident after incubation with peptide-sensitized cells (Figure 1E). In contrast, LMP-1 expression in C33A cells enhanced trans-presentation of CD8+ T-cell epitopes from another EBV-encoded membrane protein, LMP-2A (Figure 1F). Collectively, these results suggested that, despite its capacity to up-regulate trans-presentation via the MHC class I pathway, LMP-1 limits its self-presentation to CD8+ T cells.
Much of the data presented in Figure 1A through F are based on either stable or transient expression of LMP-1. To confirm these observations, we assessed endogenous presentation of LMP-1 epitopes in EBV-transformed LCLs. Data presented in Figure 1G show that, although activation of LMP-2A specific T cells was evident after incubation with HLA-matched LCLs, very low levels of activation of T cells specific for multiple LMP-1–encoded peptide epitopes was observed. Activation of LMP-1–specific T cells could be detected after incubation with peptide-sensitized LCLs (data not shown).
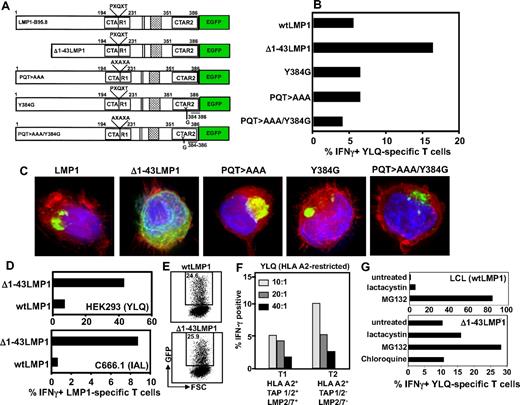
To investigate the mechanism for this immune evasion, we tested a series of LMP-1 expression vectors with mutations in the CTAR1 and/or CTAR2 domain; and a deletion mutant commencing at the second methionine of LMP-1 (Δ1-43LMP-1-GFP), which removes the first trans-membrane domain (Figure 2A). This domain has been shown to have immunomodulatory effect,21 and Δ1-43LMP-1 has been used for expanding LMP-1-specific T cells.22 Although mutations within the CTAR domains had no impact on the presentation of LMP-1 epitopes, deletion of the first trans-membrane domain enhanced endogenous presentation of HLA class I–restricted epitopes (Figure 2B). Interestingly, deletion of the first trans-membrane domain of LMP-1 completely impaired its ability to enhance MHC class I expression (data not shown). Consistent with previous studies,17 intracellular localization analysis using fluorescent microscopy revealed that, although full-length LMP-1 and its CTAR mutants formed large aggregates in the perinuclear region, Δ1-43LMP-1-GFP lost its ability to aggregate (Figure 2C). Subsequent experiments confirmed the data presented in Figure 2A through C, whereby presentation of LMP-1 epitopes9,10 was enhanced in HEK293 and C666.1 cells after expression of Δ1-43LMP-1-GFP (Figure 2D). This enhanced presentation was not the result of increased expression of Δ1-43LMP-1-GFP compared with full-length LMP-1 (Figure 2E). These observations indicate that, although the NF-κB signaling domains had no impact on cis-presentation of LMP-1 epitopes, aggregation of this protein (a critical requirement for trans-presentation) limits its accessibility to the MHC class I processing machinery.
We next assessed the potential role of peptide transporters (TAP1 and TAP2) and immunoproteasomes in the endogenous processing of CD8+ T-cell epitopes from LMP-1. Data presented in Figure 2F show that TAP1 and TAP2 and immunoproteasome-deficient T2 cells were twice as efficient in activating LMP-1–specific T cells compared with the parental T1 cell line. Further analysis revealed that inhibition of TAP1 and TAP2 function in LCL, using HSV-encoded ICP47, had minimal impact on the presentation of LMP-1 epitopes (data not shown). However, pretreatment of cells expressing wtLMP-1 and Δ1-43LMP-1 with proteasomal inhibitors (particularly MG132) significantly enhanced the endogenous presentation of CD8+ T-cell epitopes from this protein (Figure 2G). These observations are consistent with previous studies on LMP and influenza nucleoprotein, where proteasome-inhibitor treatment enhanced endogenous presentation of CD8+ T-cell epitopes.23-25
Impact of LMP-1 aggregation, signaling, and the immunoproteasome on presentation of LMP-1 epitopes through the MHC class I pathway. (A) Schematic representation of LMP-1 constructs used in the study. Expression constructs encoding either the full-length LMP-1 (referred to as wtLMP-1) or a series of deletion/point mutants expressing different forms of LMP-1 protein were used. The deletion mutant was designed to express LMP-1 protein in which the first transmembrane domain was deleted (referred to as Δ1-43LMP-1). The expression vector with point mutations within the CTAR domains of LMP-1 (referred to as PQT > AAA, Y384G, or PQT > AAA/Y384G) blocks TRAFF/TRADD binding and NF-κB activation.8,18 (B) C33A cells were transiently transfected with the expression vectors encoding wtLMP-1, Δ1-43LMP-1, Y384G, PQT > AAA, and PQT/AAA/Y384G. These cells were incubated with YLQQNWWTL-specific T cells at a responder-to-stimulator ratio of 10:1 and then assessed for intracellular IFN-γ production by flow cytometry (the first 3 amino acids of the sequences are underlined to indicate the abbreviation used for each epitope in the figure). Data represent the percentage of IFN-γ-producing YLQQNWWTL-pentamer–positive cells. (C) Intracellular location was assessed by fluorescent microscopy. (D) HEK293 and C666.1 cells were transiently transfected with vectors encoding wtLMP-1 or Δ1-43LMP-1 fused to GFP. Transfected cells were incubated with either YLQQNWWTL-specific T cells (HEK293 cells) or IALYLQQNW-specific T cells (C666.1) at responder-to-stimulator ratio of 10:1 and then assessed for intracellular IFN-γ production by flow cytometry. Data represent the percentage of IFN-γ-producing YLQ-pentamer–positive cells or IAL-specific cells. (E) LMP-1-GFP expression in epithelial cells transfected with expression vectors encoding wtLMP-1 or Δ1-43LMP-1 fused to GFP. (F) YLQQNWWTL-specific T cells were incubated with T1 and T2 cells at responder-to-stimulator ratios of 10:1, 20:1, and 40:1 and then assessed for intracellular IFN-γ production by flow cytometry. Data represent the percentage of YLQQNWWTL-pentamer–positive cells producing IFN-γ. (G) HLA-matched LCL and Δ1-43LMP-1-transfected C33A cells were treated with the proteasomal inhibitors, lactacystin and MG132 (as a control C33A cells were also treated with the lysosomal inhibitor chloroquine), and then incubated with YLQQNWWTL-specific T cells at a responder-to-stimulator ratio of 10:1. Data represent the percentage of YLQQNWWTL-pentamer–positive cells producing IFN-γ.
Impact of LMP-1 aggregation, signaling, and the immunoproteasome on presentation of LMP-1 epitopes through the MHC class I pathway. (A) Schematic representation of LMP-1 constructs used in the study. Expression constructs encoding either the full-length LMP-1 (referred to as wtLMP-1) or a series of deletion/point mutants expressing different forms of LMP-1 protein were used. The deletion mutant was designed to express LMP-1 protein in which the first transmembrane domain was deleted (referred to as Δ1-43LMP-1). The expression vector with point mutations within the CTAR domains of LMP-1 (referred to as PQT > AAA, Y384G, or PQT > AAA/Y384G) blocks TRAFF/TRADD binding and NF-κB activation.8,18 (B) C33A cells were transiently transfected with the expression vectors encoding wtLMP-1, Δ1-43LMP-1, Y384G, PQT > AAA, and PQT/AAA/Y384G. These cells were incubated with YLQQNWWTL-specific T cells at a responder-to-stimulator ratio of 10:1 and then assessed for intracellular IFN-γ production by flow cytometry (the first 3 amino acids of the sequences are underlined to indicate the abbreviation used for each epitope in the figure). Data represent the percentage of IFN-γ-producing YLQQNWWTL-pentamer–positive cells. (C) Intracellular location was assessed by fluorescent microscopy. (D) HEK293 and C666.1 cells were transiently transfected with vectors encoding wtLMP-1 or Δ1-43LMP-1 fused to GFP. Transfected cells were incubated with either YLQQNWWTL-specific T cells (HEK293 cells) or IALYLQQNW-specific T cells (C666.1) at responder-to-stimulator ratio of 10:1 and then assessed for intracellular IFN-γ production by flow cytometry. Data represent the percentage of IFN-γ-producing YLQ-pentamer–positive cells or IAL-specific cells. (E) LMP-1-GFP expression in epithelial cells transfected with expression vectors encoding wtLMP-1 or Δ1-43LMP-1 fused to GFP. (F) YLQQNWWTL-specific T cells were incubated with T1 and T2 cells at responder-to-stimulator ratios of 10:1, 20:1, and 40:1 and then assessed for intracellular IFN-γ production by flow cytometry. Data represent the percentage of YLQQNWWTL-pentamer–positive cells producing IFN-γ. (G) HLA-matched LCL and Δ1-43LMP-1-transfected C33A cells were treated with the proteasomal inhibitors, lactacystin and MG132 (as a control C33A cells were also treated with the lysosomal inhibitor chloroquine), and then incubated with YLQQNWWTL-specific T cells at a responder-to-stimulator ratio of 10:1. Data represent the percentage of YLQQNWWTL-pentamer–positive cells producing IFN-γ.
In conclusion, data presented in this study indicate that, in addition to constraining its cis-presentation through aggregation, epitopes encoded within LMP-1 protein are most probably destroyed by cellular proteasomes, providing a dual strategy by which LMP-1 limits self-presentation to CD8+ T cells. To our knowledge, this is the first documented evidence of any pathogen-encoded protein that can activate the antigen-processing machinery of the cell while successfully evading a highly efficient scanning process by the components of this machinery for foreign protein.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by funding from the National Health and Medical Research Council (NHMRC) and Cancer Council of Queensland, Australia. R.K. is supported by a Research Fellowship from the NHMRC. C.S. is supported by a Fellowship from the Leukemia Foundation.
Authorship
Contribution: C.S. and R.K. designed this study and wrote the manuscript; and C.S., N.W., T.C., J.P., T.Y., and L.B. conducted various experimental studies.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rajiv Khanna, Queensland Institute of Medical Research, 300 Herston Rd, Brisbane, Australia; e-mail: rajiv.khanna@qimr.edu.au.
References
Author notes
*C.S. and N.W. contributed equally to this study, and the order in which they are listed should be considered arbitrary.