Mutations in Notch are known to play a critical role in the development of T-ALL.1 In this issue of Blood, Rosati and colleagues provide compelling evidence that Notch signaling also plays an important role in cell survival in B-CLL.
The Notch signaling pathway, first described in Drosophila and Caenorhabditis elegans, has long been recognized as a way to regulate many developmental cell fate decisions. Mammalian Notch was first identified in 1991 in a translocation that brought the Notch1 gene under the transcriptional control of the T-cell receptor β locus in a T-cell acute lymphoblastic leukemia (T-ALL) cell line.2 This early observation of Notch translocation in T-cell leukemia led many to propose that Notch translocations may have a causative role in the development of this disease. However, as many groups searched widely for evidence of Notch translocations, it became apparent that translocation of Notch genes in these and other leukemias was a rare event. The breakthrough in our understanding of the contribution of Notch to T-ALL came with the exciting results of Weng et al, demonstrating that over 60% of human T-ALL patients carried activating mutations in the Notch1 gene.1 This observation rekindled interest in how Notch facilitates leukemia development. In this issue, the report by Rosati et al implicates Notch1 and Notch2 signaling in B-cell chronic lymphocytic leukemia (B-CLL) and suggests a different mechanism for Notch activation than that observed by Weng et al.1
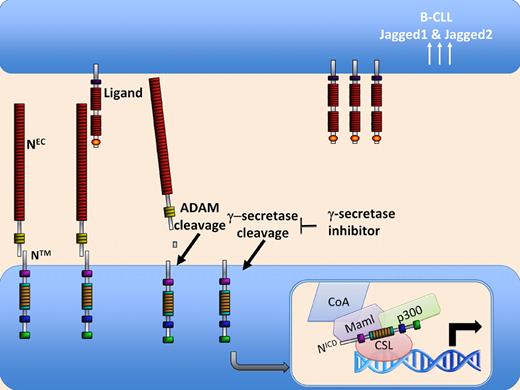
Notch signaling is initiated by interaction of NEC with ligand on a neighboring cell. This results in cleavage of NTM by ADAM protease followed quickly by γ-secretase cleavage, allowing the release and translocation of intracellular Notch to the nucleus. Rosati et al suggest that, in B-CLL cells, increased levels of the ligands Jagged1 and Jagged2 lead to increased Notch signaling, resulting in enhanced survival of these cells.
Notch signaling is initiated by interaction of NEC with ligand on a neighboring cell. This results in cleavage of NTM by ADAM protease followed quickly by γ-secretase cleavage, allowing the release and translocation of intracellular Notch to the nucleus. Rosati et al suggest that, in B-CLL cells, increased levels of the ligands Jagged1 and Jagged2 lead to increased Notch signaling, resulting in enhanced survival of these cells.
To describe the differences between these 2 mechanisms, it is important to understand Notch signaling. In mammals, 4 Notch genes have been described and it is thought that all 4 signal in a similar fashion (reviewed in Bray3 ). Notch is synthesized as a large 300 kD intracellular protein. During transport to the cell surface in the trans-Golgi, Notch is cleaved by a furin protease into an extracellular domain (NEC) and a transmembrane domain (NTM). This heterodimer associates and inserts into the cell membrane. Upon interaction of NEC with a ligand (there are 5 mammalian ligands; Jagged 1, Jagged 2, Dll1, Dll3, and Dll4), a conformational change in NTM renders it susceptible to cleavage by extracellular ADAM proteases, resulting in the release of NEC. This second cleavage event itself induces a conformational change in NTM, which allows access by an intramembraneous protease complex known as γ-secretase. Cleavage by γ-secretase results in the release of the intracellular Notch domain, NICD. Unlike many signaling pathways that require numerous steps to gain access to the nucleus, release of NICD results in rapid and direct activation of genes normally suppressed by the ubiquitous transcription factor, CSL. When Notch is inactive in cells, CSL is assembled on DNA in a complex with transcriptional repressors. However, upon Notch activation by ligand, the entry of NICD into the nucleus results in its rapid displacement of these repressors and the recruitment of key transcriptional activators, such as p300 and Mastermind. Thus, with a single cleavage event mediated by γ-secretase, genes normally repressed by CSL are rapidly activated. We now know that this signaling event is key to the development of a large number of cell lineages during embryonic and postembryonic development in mammals. But tight regulation of this pathway is required since uncontrolled Notch signaling is linked to many forms of cancer (reviewed in Rizzo et al4 ).
Returning to the work of Weng et al, this group described mutations in Notch1 that render it susceptible to cleavage by ADAM proteases in the absence of ligand interactions. In other words, these mutations result in constitutive activation of Notch signaling in T-ALL. Importantly, this group also demonstrated that in vitro treatment of T-ALL cell lines with γ-secretase inhibitors (GSI), block Notch activation and induce cell cycle arrest and apoptosis. Other mutations that presumably inhibit the proteolysis of NICD also were described by this group. In the current paper, Rosati et al describe another possible mechanism for activation of Notch in B-CLL. In this study, the authors show that B-CLL cells taken from 25 patients have increased levels both of N1ICD and N2ICD, indicating increased activation of these proteins in this type of leukemia. They then show that, not only do these cells express high levels of activated Notch1 and Notch2, they also express high levels of the ligands, Jagged1 and Jagged2, as well as proteins associated with cell survival, such as XIAP, c-IAP2 and active NF-κB. Furthermore, they provide compelling evidence that blocking Notch activation with GSI or Notch2 siRNA induces spontaneous apoptosis in freshly-isolated B-CLL cells from several patients in this study. Conversely, they demonstrated increased survival of freshly isolated B-CLL cells when cultured on immobilized soluble Jagged1 ligand, suggesting that Jagged1 engagement of Notch enhances survival. These data do not rule out the possibility that activating mutations in Notch1 and/or Notch2 are present in cells from B-CLL patients; in fact this possibility was not explored in this report. However, the data do provide yet another mechanism, namely increased expression of ligand, by which enhanced Notch signaling may result in increased cell survival and leukemic transformation. Collectively, these data suggest the possibility that, in addition to the use of GSIs as a treatment for T-ALL and B-CLL, the latter might also be targeted by compounds designed to block Notch/ligand interaction.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■