Abstract
B7-H4 is an immunoglobulin superfamily molecule and shown to be inhibitory for T-cell responses. To explore physiologic roles of B7-H4, we created B7-H4–deficient (KO) mice by genetic targeting. B7-H4KO mice are healthy and their T- and B-cell responses to polyclonal antigens are in normal range. However, B7-H4KO mice are more resistant to infection by Listeria monocytogenes than their littermates. Within 3 days after infection, bacterial colonies in livers and spleens are significantly lower than the controls, suggesting a role of B7-H4 in enhancing innate immunity. Further studies demonstrate that neutrophils increase in peripheral organs of B7-H4KO mice more so than their littermates but their bactericidal functions remain unchanged. Augmented innate resistance is completely dependent on neutrophils, even in the absence of adaptive immunity. In vitro B7-H4 inhibits the growth of bone marrow–derived neutrophil progenitors, suggesting an inhibitory function of B7-H4 in neutrophil expansion. Our results identify B7-H4 as a negative regulator of the neutrophil response to infection and provide a new target for manipulation of innate immunity.
Introduction
Neutrophils are a major component of the host innate defense against infection and also contribute to autoimmune pathogenesis and chronic inflammation. During infection, neutrophils rapidly migrate to sites of inflammation, become activated, and initiate a cascade of defense mechanisms including phagocytosis, killing, and degradation of microorganisms by antimicrobial and proteolytic proteins, along with the generation of reactive oxygen species. Neutrophils also participate in tissue breakdown, remodeling, wound healing, and modulation of other inflammatory and adaptive immune components.1 Due to their short life span, neutrophils have to be resupplied continuously during infection and inflammation by expansion from myeloid progenitor cells in the bone marrow.2
B7-H4 (B7x and B7S1) is a member of the B7-CD28 cosignaling molecule family/immunoglobulin superfamily with signature IgV and IgC regions in the extracellular domain, a single transmembrane domain, and an intracellular domain.3 Although the mRNA for B7-H4 is broadly detectable in the majority of normal tissues and cell types, its cell surface expression is relatively rare.4-8 Expression of B7-H4 protein, however, can be induced in vitro on hematopoietic cells.4,9 In contrast to its limited distribution in normal tissues, constitutive cell surface and cytoplasmic expression of B7-H4 is found on various cultured cancer lines and freshly isolated human carcinomas of breast, lung, ovarian, and kidney.7-14 Previous in vitro studies demonstrate that B7-H4 fusion protein could inhibit polyclonal T-cell responses including growth and cytokine secretion,4-6 as well as antigen-induced stimulation of transgenic T cells in vivo.4 Blockade of B7-H4 by monoclonal antibody (mAb) was shown to partially inhibit antigen-specific T-cell responses4 and accelerate autoimmune disease progression in vivo.5 The receptor for B7-H4, which mediates inhibitory functions on T cells, remains unknown now. Indeed, the inhibitory activity of B7-H4 on T cells, combined with its up-regulation on many human cancers7-14 and tumor-infiltrating macrophages from the ascites of ovarian cancer patients, has implicated this molecule in tumor immune evasion.9 Interestingly, up-regulation of B7-H4 on antigen-presenting cells (APCs) has been previously proposed as a potential inhibitory mechanism of regulatory T cells.15 These studies provide direct evidence that B7-H4 may contribute to the suppression of adaptive immunity in the tumor microenvironment.
The current study reveals a novel activity of B7-H4 as an endogenous negative regulator for neutrophil responsiveness to Listeria infection, done so by directly inhibiting expansion of neutrophil progenitors from bone marrow (BM). As a result, B7-H4–deficient mice are highly resistant to Listeria infection resulting from enhanced neutrophil-mediated innate immunity.
Methods
Antibodies, recombinant proteins, and flow cytometric analysis
Primary and secondary antibodies against murine Gr-1 and CD11b, which are directly conjugated with FITC, PE, or APC, were purchased from BD Pharmingen (San Diego, CA) or eBiosciences (San Diego, CA). Nonconjugated primary antibodies were purified from hybridoma culture supernatant. All cells were stained using standard protocols as previously described and were analyzed on a FACSCalibur flow cytometer (BD Pharmingen).4 The data were analyzed with Software CellQuest (BD Pharmingen) or FlowJo (TreeStar, Ashland, OR). For in vivo studies, mAbs were prepared and purified as previously described.4 Anti–Gr-1 hybridoma (RB6-8C5) was a generous gift from Dr Hans Schreiber at the University of Chicago (Chicago, IL). Depleting antibodies against natural killer (NK) cells (clone NK1.1) and plasmatoid dendritic cells (clone PDCA1) were described previously.16,17 Control mouse IgG, rat IgG, and hamster IgG were purchased from Sigma-Aldrich (St Louis, MO) and further purified as previously described.4 Carrageenan was purchased from Sigma-Aldrich. All cell culture media and antibiotics were purchased from Cellgro (Herndon, VA). Fetal bovine serum (FBS) was from Hyclone (Logan, UT).
Mice
Six- to 8-week-old B6 mice were obtained from The Jackson Laboratory (Bar Harbor, ME). RAG-1 KO mice were purchased from Taconic Farms (Germantown, NY). Both female and male mice were used for the experiments. All mice were housed under specific pathogen-free conditions in the Johns Hopkins Animal Facility with all protocols approved by the Institutional Animal Care and Use Committee.
The general strategy to generate gene KO mice by homologous recombination was previously described.18,19 To generate B7-H4KO mice, a 5.09-kb DNA fragment upstream of the IgV domain (exon 3) of the murine B7-H4 genomic DNA was polymerase chain reaction (PCR) amplified from a 129SvJ bacterial artificial chromosome (BAC) library (Invitrogen, Carlsbad, CA) and was cloned into the 5′-arm position of the pKOscrambler vector NTKV-1907 (Stratagene, La Jolla, CA). A 5.57-kb DNA fragment downstream of the IgC domain (exon 4) of B7-H4 genomic DNA was PCR amplified from the same library and was cloned into the 3′-arm position of the same vector to generate a targeting plasmid resulting in removing IgV and IgC domains from the B7-H4 gene (Figure 1A). The targeting fragment containing the 5′-arm and the 3′-arm sequences of the B7-H4 gene, a positive selection marker NEO, and a negative selection marker TK was transfected into 129Sv/E embryonic stem (ES) cells. ES cell transfectants underwent neomycin drug selection. The targeted clones were identified by Southern blot analysis using a 3′ external probe (Figure 1B). Chimeric mice were produced by injection of targeted ES cells into blastocysts of B6 hosts. Heterozygous B7-H4 (+/−) mice were obtained from breeding chimeric mice with B6 mice. PCR analysis was used to distinguish the wild-type (WT) and deficient B7-H4 allele. The sequences of the 3 PCR primers are (1) 5′-GTTAGATAGGGTCTCACTGGGTAGC, (2) 5′-CCTACAGCCTTCAGTATGCCAGAGA, and (3) 5′-AGACTAGTGAGACGTGCTACTTCCA. Homozygous mice were produced by backcrossing to B6 for more than 10 generations before use for further analysis. B7-H4 KO/RAG-1 KO mice were obtained by backcrossing B7-H4 KO and RAG-1 KO mice.
Generation of B7-H4 KO mice. (A) Strategy for disruption of the B7-H4 gene. A 4.7-kb DNA fragment containing exons encoding the IgV and IgC domains of murine B7-H4 gene is substituted by a 1.7-kb fragment encoding the neomycin resistant (Neo) gene. Closed boxes represent B7-H4 coding exons. Lines between exons represent intron sequences. Open boxes represent untranslated exons. The Neo is represented by a shaded box. (B) Screen of targeted ES cells by Southern blot analysis. Genomic DNA of ES cells were digested with SpeI and probed with a fragment (probe) as indicated in panel A. (C) Lack of B7-H4 gene expression in B7-H4 KO mice. Liver RNAs were prepared from B7-H4 KO (−/−) and littermates (+/+). RT-PCR was performed with primers corresponding to the IgV domain of B7-H4 gene. RT-PCR of actin gene was used as positive control in the analysis.
Generation of B7-H4 KO mice. (A) Strategy for disruption of the B7-H4 gene. A 4.7-kb DNA fragment containing exons encoding the IgV and IgC domains of murine B7-H4 gene is substituted by a 1.7-kb fragment encoding the neomycin resistant (Neo) gene. Closed boxes represent B7-H4 coding exons. Lines between exons represent intron sequences. Open boxes represent untranslated exons. The Neo is represented by a shaded box. (B) Screen of targeted ES cells by Southern blot analysis. Genomic DNA of ES cells were digested with SpeI and probed with a fragment (probe) as indicated in panel A. (C) Lack of B7-H4 gene expression in B7-H4 KO mice. Liver RNAs were prepared from B7-H4 KO (−/−) and littermates (+/+). RT-PCR was performed with primers corresponding to the IgV domain of B7-H4 gene. RT-PCR of actin gene was used as positive control in the analysis.
Cell depletion in vivo
WT or B7-H4KO mice 6 to 9 weeks old were used for all experiments. All depletion reagents and control reagents were administrated by intraperitoneal injection in 0.5-mL volume. Depletion of subset of cells was confirmed by flow cytometry analysis with subset-specific antibody. At day 0, mice were given 106 CFUs Listeria by intraperitoneal injection. Two days after infection, mice were terminated and the liver Listeria load was evaluated by colony plating assay. For neutrophil depletion, 150 μg/mouse anti–Gr-1 antibody (clone RB6-8C5) or rat IgG was used at day −1 before Listeria infection (day 0). For macrophage depletion, carrageenan at 2 mg/mouse or 0.5 mL PBS was applied on days −3 and −1. For NK-cell depletion, 500 μg/mouse anti-NK1.1 antibody (clone PK136) or mouse IgG was administered on day −1. For plasmatoid dendritic cell depletion, 250 μg/mouse anti-pDC antibody (clone PDCA-1) or rat IgG was injected on days −6, −3, and −1.
Listeria infection and colony counting
Listeria monocytogenes strain DP-L4056 was kindly provided by Dr Thomas W. Dubensky Jr from Cerus (Concord, CA). To prepare Listeria stock, Listeria cells were grown in DIFCO Listeria Enrichment Broth (Becton Dickinson, Sparks, MD) to 0.8 to 1 at OD 600 nm. Culture was harvested by centrifugation and was washed twice with PBS. Pellets were then resuspended in stock solution (PBS with 15%-20% glycerol) and aliquoted to 200 μL per microtube for storage at −80°C. The colony-forming units (CFUs) of Listeria stock were determined by counting colonies of series dilutions of the aliquots growing on BBL CHROMagar Listeria plates (Becton Dickinson). Prior to infection, Listeria stock was thawed and diluted in PBS to appropriate concentration of CFUs/mL and applied to mice or cells as indicated. Six- to 8-week-old mice were infected by intraperitoneal or intravenous (for colony formation assay only) injection of indicated CFUs Listeria. At indicated time points after infection, a piece of mouse liver or spleen was cut, weighed, and ground in PBS. The liver or spleen suspension was plated on BBL CHROMagar Listeria plates or on agar plates of Listeria enrichment broth. Colonies were counted 2 days after plating, and adjusted to CFUs per gram of liver or spleen.
Listeria infection of neutrophils in vitro
Neutrophils were isolated in a similar manner to the methods previously described.20 Briefly, mice were injected intraperitoneally with 3% thioglycollate broth. Four to 5 hours after injection, peritoneal cavities of each mouse were washed with 5 mL PBS and cells were harvested by centrifugation. By this method, more than 90% harvested cells are Gr-1+CD11b+ neutrophils. Neutrophils (106) were incubated with 108 CFUs LM for 10 minutes at 37°C. The cultures were terminated by adding penicillin-streptomycin (Cellgro). Subsequently, cells were harvested by centrifugation, plated in 96-well plates. The plates were incubated at 37°C and harvested at indicated time points. Cells were lysed immediately by resuspending in 1 mL sterile water. Cell lysates or diluted cell lysates were plated on agar plates of Listeria enrichment broth for colony counting.
Respiratory burst and phagocytosis of neutrophils
Neutrophil phagocytic activity and oxidative burst activity were measured as described.21,22 Briefly, 1 × 106 neutrophils were incubated with 5 × 107 red-fluorescent microbeads (Polysciences, Warrington, PA) and 25 μM H2DCFDA (2′,7′,-dihydrochlorofluoresein diacetate; Sigma-Aldrich) for 30 to 60 minutes at 37°C. Cells were washed twice with fluorescence-activated cell sorting (FACS) buffer (1% FBS in PBS) and fixed in 1% paraformaldehyde in PBS. Viable cells were first gated and the respiratory burst activity was quantified by green mean fluorescence intensity (MFI) and phagocytotic capacity was quantified by red MFI. The number in upper right gate represents percentage of cells with both phagocytic and oxidative capacity.
Assays for neutrophil growth inhibition
BM cells were aspirated and prepared as previously described.23 Cells were then incubated in 2 μM carboxyfluorescein diacetate succinimidyl ester (CFSE; Invitrogen) in PBS for 10 minutes. After extensive washing, cells were plated at 2.5 × 105/well. Cells were harvested at indicated time points and cell numbers were counted with Beckman Coulter Counter (Fullerton, CA). The numbers of dividing neutrophils in each sample were decided by gating Gr-1+ mAb and CFSE dilution using flow cytometric analysis as shown previously.4 Lin− BM cells were prepared as described previously.24 Briefly, BM cells were collected from WT B6 mice (8-10 weeks old). Lineage+ cells were removed by PE-labeled anti–Gr-1, anti-CD11c, anti-Ter119, anti-CD11c, anti-CD3, anti-B220, and anti-CD19 (BD PharMingen) followed by magnetic-activated cell sorting (MACS) anti-PE microbeads and LD columns (Miltenyi Biotec). Lin− BM cells were then labeled with CFSE and were plated in alpha-MEM supplemented with 20% FBS, 100 ng/mL SCF, and 10 ng/mL G-CSF. Recombinant B7-H4 fusion proteins (B7-H4Ig) were added at the indicated concentrations. Cells were harvested daily for cell counting and flow cytometric analysis.
Statistical analysis
Statistical analysis was performed by Student t test method for 2 parameters and Anova test for multiple parameters.
Results
Generation of B7-H4KO mice
We generated B7-H4KO mice by homologous recombination in 129 ES cells by deleting the entire Ig V and Ig C regions of the B7-H4 gene to completely eliminate their interaction with any potential receptor. Exons encoding both the Ig V and Ig C domains of the B7-H4 gene were replaced with a Neo gene cassette (Figure 1A). Targeted recombination of ES cells was confirmed by Southern blot analysis and the data from 4 independent ES clones are shown. The B7-H4+ allele is predicted to have a 12.25-kb SpeI fragment and the B7-H4− allele has an 8.9-kb SpeI fragment. The clones 2 and 3 with both fragments indicate a recombination (Figure 1B). Chimeric male mice were derived from these ES clones by standard procedures. They were backcrossed to C57BL/6 (B6) females and heterozygous mutant mice were established from 2 independently targeted ES clones. Heterozygous or homozygous B7-H4 mutant mice were then identified by PCR analysis of genomic DNA isolated from tail biopsies. Southern blot analysis confirmed the replacement of genomic DNA. Reverse-transcription (RT)–PCR analysis demonstrated that B7-H4 mRNA was not expressed in livers of B7-H4–deficient mice (Figure 1C). B7-H4KO mice develop normally and give normal litter numbers. These mice were backcrossed to the B6 background for 10 generations before they were used in the studies described below.
B7-H4KO mice have enhanced neutrophil-mediated resistance to Listeria infection
B7-H4KO mice display normal numbers and ratios of T, B, NK, and NKT cells and macrophages using specific monoclonal antibodies (mAbs) in flow cytometric analysis. In addition, T cells from B7-H4KO mice do not have obvious alterations compared with littermates in responding to polyclonal stimuli, judged by in vitro proliferation of purified T cells by CD3 cross-linking, allogeneic antigen stimulation, or cytolytic T-cell response to alloantigens (G.Z., L.C., unpublished data, January 2008). These results indicate that the lack of B7-H4 does not globally affect T-cell responses to antigens. Consistent with these in vitro findings, we also found that B7-H4KO mice have normal responses to Con A–induced hepatitis,18 hapten-induced hypersensitivity,25 and OVA-induced airway inflammation.26 B7-H4KO mice were also found to be comparable with littermate control in OT-I and OT-II cell expansion to OVA proteins upon transfer,4 CD4-Vβ8.1/8.2 T-cell expansion to superantigens,19 and cytotoxic T lymphocyte (CTL) responses to allogeneic antigens in vivo.27 Normal B-cell responses in B7-H4KO mice were also observed after immunization by TNP-KLH.28 Finally, B7-H4KO mice do not develop spontaneous autoimmune diseases up to 1.5 years under specific pathogen–free environment (G.Z., L.C., unpublished data, January 2008).
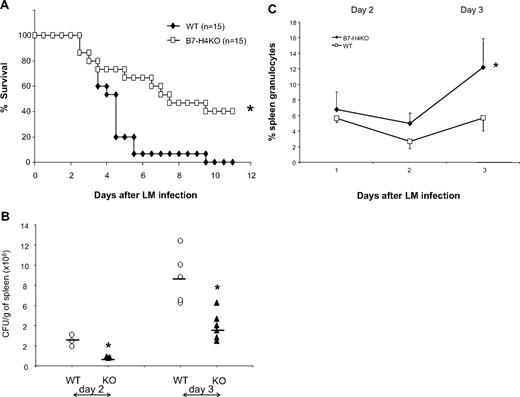
Although our data indicate that B7-H4 plays a minimal role in antigen-driven T- and B-cell responses, especially polyclonal responses, in our assays, these responses were conducted in the absence of active infection, which usually requires a much more sophisticated coordination between innate and adaptive immunity. To test this possibility, we next evaluated the effect of B7-H4 ablation in mice infected with L monocytogenes (LM) to examine whether B7-H4 contributes to immune responses against infection. Mice were challenged with an intraperitoneal dose (2 × 106 CFUs) of LM sufficient to induce lethality. The survival of these mice was then subsequently evaluated. B7-H4KO mice were significantly more resistant to LM infection: B7-H4KO mice survived much longer than their B7-H4 WT littermates and up to 40% of mice cleared bacteria and lived indefinitely, whereas all littermates died around day 9 (Figure 2A). This effect correlates with decreased Listeria numbers in the spleen (Figure 2B) and liver (data not shown) of B7-H4KO mice. Interestingly, the majority of mice succumbed to infection within 3 to 4 days, time points at which adaptive immunity is usually not yet developed. Our results thus suggest a role for B7-H4 in altering the context of the innate immune response.
B7-H4 KO mice are resistant to Listeria infection, accompanied with increased neutrophils. (A) B7-H4 KO mice are more resistant to Listeria infection than WT mice. Fifteen WT or B7-H4 KO mice were intraperitoneally injected with 2 × 106 CFUs Listeria. Survival of mice was monitored daily. Data represent a pool of 2 independent experiments. *P < .05. (B) Decreased Listeria colony formation in spleens from B7-H4KO mice. WT or B7-H4KO mice (KO) were intravenously injected with 105 CFUs Listeria. Days 1 to 3 after infection, whole spleens were weighted and homogenized in 5 mL PBS, and tissue lysates were plated on agar plates of Listeria enrichment broth for colony counting. The results from individual mice are presented. Data represent at least 3 independently performed experiments. *P < .05. (C) Spleen neutrophils increase after Listeria infection in B7-H4 KO mice. Three B7-H4KO or littermate control mice were grouped and intravenously injected with 105 CFUs Listeria. Splenocytes were isolated 1, 2, and 3 days after infection and were stained with Gr-1 and CD11b mAb. The same numbers of littermates were included as controls. Each point represents results from pool of 3 mice. Data represent at least 3 independent experiments. *P < .05. Error bars indicate SD (n = 3 mice).
B7-H4 KO mice are resistant to Listeria infection, accompanied with increased neutrophils. (A) B7-H4 KO mice are more resistant to Listeria infection than WT mice. Fifteen WT or B7-H4 KO mice were intraperitoneally injected with 2 × 106 CFUs Listeria. Survival of mice was monitored daily. Data represent a pool of 2 independent experiments. *P < .05. (B) Decreased Listeria colony formation in spleens from B7-H4KO mice. WT or B7-H4KO mice (KO) were intravenously injected with 105 CFUs Listeria. Days 1 to 3 after infection, whole spleens were weighted and homogenized in 5 mL PBS, and tissue lysates were plated on agar plates of Listeria enrichment broth for colony counting. The results from individual mice are presented. Data represent at least 3 independently performed experiments. *P < .05. (C) Spleen neutrophils increase after Listeria infection in B7-H4 KO mice. Three B7-H4KO or littermate control mice were grouped and intravenously injected with 105 CFUs Listeria. Splenocytes were isolated 1, 2, and 3 days after infection and were stained with Gr-1 and CD11b mAb. The same numbers of littermates were included as controls. Each point represents results from pool of 3 mice. Data represent at least 3 independent experiments. *P < .05. Error bars indicate SD (n = 3 mice).
To address mechanisms of this resistance, we examined the cell composition of both the innate and adaptive immune response. Mice were infected with Listeria and T, B, NK, macrophages, and neutrophils in peripheral blood and in lymphoid organs were examined by specific mAbs. Although there were no significant differences in the numbers of NK, macrophages, T cells, and B cells of spleens within the first 3 days after LM infection (G.Z., L.C., unpublished data, January 2008), significantly more neutrophils in composition (Figure 2C) and in absolute numbers (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) were found from LM-infected B7-H4KO mice than identically infected WT littermates. Similar results were also obtained in neutrophils isolated from livers and in peripheral blood after infection (G.Z., L.C., unpublished data, January 2008).
To exclude the possibility that the increases in neutrophil counts are due to other genetic modifications in B7-H4KO mice, and are not limited to LM infection, we first attempted to validate this finding by blocking B7-H4 in normal B6 mice. Injection of previously described anti–murine B7-H4 mAb4 showed a small or minimal effect, consistent with weak neutralizing ability of this mAb. We then took an alternative approach to express a soluble decoy B7-H4 in B6 mice as a means of blocking endogenous B7-H4. We first constructed 2 plasmids encoding only an extracellular portion of murine B7-H4 with deletion of transmembrane and intracellular domains (B7-H4VC). In addition, we also constructed another plasmid with only Ig V domain (B7-H4V). Hydrodynamic injection29 (also see Figure S1 legend) of these plasmids in B6 mice led to rapid appearance of soluble B7-H4 in sera as detected by enzyme-linked immunosorbent assay (ELISA). Soluble B7-H4 peaked up to 2 μg/mL sera at day 1 and dropped to basal level at day 7 after injection (T.A., L.C., unpublished data, January 2008). We subsequently tested the effect of soluble B7-H4 blockade in the neutrophil response to LPS in a pouch assay, in which neutrophils in a skin air pouch could be quantified after LPS challenge. Expression of soluble B7-H4 (either B7-H4VC or B7-H4V) significantly increased the number of neutrophils in the pouches (Figure S1) and inhibited growth of Listeria in spleen (Figure S2). This result also indicates that the V region of B7-H4 is sufficient to compete for binding of endogenous B7-H4. Consistent with this, pouches created in B7-H4KO mice had significantly increased neutrophils relative to littermate controls (Figure S3). Our results thus indicate that B7-H4 can modulate the innate immune response by regulating the number of peripheral neutrophils in response to bacterial infection.
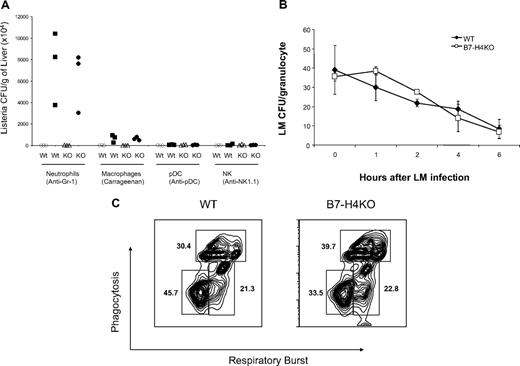
To determine whether neutrophils are required for the resistance of LM infection in B7-H4KO mice, we depleted neutrophils by in vivo injection of Gr-1 mAb. Injection of Gr-1 mAb led to the rapid decline of neutrophils to undetectable levels at day 2 in spleens (G.Z., L.C., unpublished data, January 2008). Depletion of Gr-1+ neutrophils led to a significant increase of LM load in livers from B7-H4KO mice, in comparison with those treated with either PBS or isotype-matched control mAb (Figure 3A). Plasmatoid dendritic cells (pDCs) are shown to express Gr-1 and also have antibacterial function.30 Elimination of pDCs by specific mAb against PDCA1, however, did not increase LM colonies. Depletion of NK cells by NK1.1 mAb also did not affect LM colony formation in liver, whereas depletion of macrophages by carrageenan increased LM colonies to a moderate but less significant level compared with Gr-1+ cell depletion (Figure 3A). Our experiments also demonstrate that treatment by these methods eliminated specific subsets up to 2 weeks. Our results thus support the notion that Gr-1+ neutrophils play a critical role in the resistance to LM infection in the absence of B7-H4.
Increased neutrophils in B7-H4KO mice are required for resistance to infection but the bactericidal functions of neutrophils are normal. (A) Wild-type (WT) B6 mice or B7-H4KO (KO) mice of 6 to 9 weeks old at groups of 3 were used for all experiments. The mice were injected intraperitoneally with anti–Gr-1, anti-NK1.1, and anti-pDC mAb to deplete neutrophils, NK cells, and plasmatoid cells, respectively. Injection of carrageenan was used to deplete macrophages. Depletion of subset of cells was confirmed by flow cytometry analysis. After depletion, mice were challenged with 106 CFUs Listeria by intraperitoneal injection. Two days after Listeria infection, mice were terminated and the liver Listeria load was evaluated by colony plating assay. Listeria colonies in each mouse were shown. The data are expressed as CFUs/gram of liver. Data are representative of at least 3 independent experiments for each treatment. Open symbols indicate control reagents; closed symbols, treatment by antibody or carrageenan. (B) Neutrophils from B7-H4 KO mice have normal capacity to intake and digest Listeria. Three mice of B7-H4 KO or littermate control mice were intraperitoneally injected with l mL 3% thioglycollate. Mice were terminated 4 to 5 hours after injection and peritoneal cells were harvested and incubated with Listeria for 10 minutes. Listeria infection was stopped by washing cells with medium containing antibiotics in large volumes. Neutrophils were then cultured. At indicated time points, cultured cells were lysed and the cell lysates were plated for CFU counting as described in “Listeria infection of neutrophils in vitro.” Eror bars indicate SD (n = 3 samples). (C) Normal respiratory burst and phagocytosis of neutrophils from B7-H4KO mice. Neutrophils were harvested as described in panel B. Neutrophils (1 × 106) were incubated with 5 × 107 of red-fluorescent microbeads and 25 μM H2DCFDA for 30 to 60 minutes at 37°C. Red and green fluorescence were analyzed by flow cytometry and the results were expressed as percentage of cells with oxidative and/or phagocytic capacity. The data are representative of 2 separate experiments. There is no statistically significant difference between neutrophils with both oxidative and phagocytic capacity from WT and B7-H4KO mice.
Increased neutrophils in B7-H4KO mice are required for resistance to infection but the bactericidal functions of neutrophils are normal. (A) Wild-type (WT) B6 mice or B7-H4KO (KO) mice of 6 to 9 weeks old at groups of 3 were used for all experiments. The mice were injected intraperitoneally with anti–Gr-1, anti-NK1.1, and anti-pDC mAb to deplete neutrophils, NK cells, and plasmatoid cells, respectively. Injection of carrageenan was used to deplete macrophages. Depletion of subset of cells was confirmed by flow cytometry analysis. After depletion, mice were challenged with 106 CFUs Listeria by intraperitoneal injection. Two days after Listeria infection, mice were terminated and the liver Listeria load was evaluated by colony plating assay. Listeria colonies in each mouse were shown. The data are expressed as CFUs/gram of liver. Data are representative of at least 3 independent experiments for each treatment. Open symbols indicate control reagents; closed symbols, treatment by antibody or carrageenan. (B) Neutrophils from B7-H4 KO mice have normal capacity to intake and digest Listeria. Three mice of B7-H4 KO or littermate control mice were intraperitoneally injected with l mL 3% thioglycollate. Mice were terminated 4 to 5 hours after injection and peritoneal cells were harvested and incubated with Listeria for 10 minutes. Listeria infection was stopped by washing cells with medium containing antibiotics in large volumes. Neutrophils were then cultured. At indicated time points, cultured cells were lysed and the cell lysates were plated for CFU counting as described in “Listeria infection of neutrophils in vitro.” Eror bars indicate SD (n = 3 samples). (C) Normal respiratory burst and phagocytosis of neutrophils from B7-H4KO mice. Neutrophils were harvested as described in panel B. Neutrophils (1 × 106) were incubated with 5 × 107 of red-fluorescent microbeads and 25 μM H2DCFDA for 30 to 60 minutes at 37°C. Red and green fluorescence were analyzed by flow cytometry and the results were expressed as percentage of cells with oxidative and/or phagocytic capacity. The data are representative of 2 separate experiments. There is no statistically significant difference between neutrophils with both oxidative and phagocytic capacity from WT and B7-H4KO mice.
We next determined whether the augmented clearance of Listeria by B7-H4KO mice was caused by alterations in neutrophil function. B7-H4–deficient neutrophils displayed normal growth inhibition of LM colony formation in culture, which is comparable with littermate neutrophils (Figure 3B), suggesting that B7-H4–deficient neutrophils have normal bactericidal functions. To confirm this, we evaluated phagocytosis and oxygen burst response of B7-H4KO neutrophils to LM infection. To do so, neutrophils from WT or B7-H4KO mice were incubated with dichlorodihydrofluorescein diacetate (H2DCFDA) and PE-labeled latex beads. H2DCFDA is a cell-permeable green fluorogenic probe that could localize to the cytosol, where the H2DCFDA molecules can be oxidized to green fluorescent dichlorofluorescein (DCF), and thus measures oxidative burst. PE-labeled latex beads measure phagocytosis capacity. As shown, both respiratory burst and phagocytosis by B7-H4KO neutrophils are within the normal range (Figure 3C), supporting that B7-H4–deficient neutrophils are functionally comparable with WT neutrophils. Therefore, increased resistance to LM infection in B7-H4KO mice could not be attributed to function modification of neutrophils, but likely mediated by an increased number of neutrophils.
Neutrophil-mediated innate resistance in B7-H4KO mice is independent of adaptive immunity
Although our data support that resistance of B7-H4KO mice to LM infection requires neutrophils, it is still possible that adaptive immunity also contributes to neutrophil growth. To exclude this possibility, we backcrossed B7-H4KO mice to the RAG-1KO background to eliminate T and B cells. Unlike RAG-1KO (RKO) mice, which possess small spleens, double- knockout (DKO) mice display enlarged spleens (Figure 4A) accompanied by increased spleen weight (Figure 4B). Further analysis of cell components in spleens, peripheral blood, livers, and bone marrows revealed that Gr-1+CD11b+ neutrophils increased dramatically in percentages (Figure 4C) and in the absolute number (Table S1) of DKO mice compared with those of RKO mice.
Phenotypes of B7-H4 × RAG-1 double KO mice (DKO). (A,B) Enlarged and increased weight of spleens from the DKO mice. Spleens were obtained from 6- to 8-week-old mice and the representatives of their appearance (A) and weights of average of 4 spleens (B) are shown. DKO indicates B7-H4 × RAG-1 double KO; RKO, RAG-1 KO; B7-H4 KO, B7-H4 KO mice in B6 background; and WT, normal B6. *P < .05. Error bars indicate SD (n = 3 mice). (C) Flow cytometric analysis of Gr-1+CD11b+ neutrophils in the absence of B7-H4. Cells were isolated from bone marrows, spleens, livers, and peripheral blood mononuclear cells (PBMCs) of indicated mice and stained with Gr-1 and CD11b mAb. Data were analyzed by flow cytometry.
Phenotypes of B7-H4 × RAG-1 double KO mice (DKO). (A,B) Enlarged and increased weight of spleens from the DKO mice. Spleens were obtained from 6- to 8-week-old mice and the representatives of their appearance (A) and weights of average of 4 spleens (B) are shown. DKO indicates B7-H4 × RAG-1 double KO; RKO, RAG-1 KO; B7-H4 KO, B7-H4 KO mice in B6 background; and WT, normal B6. *P < .05. Error bars indicate SD (n = 3 mice). (C) Flow cytometric analysis of Gr-1+CD11b+ neutrophils in the absence of B7-H4. Cells were isolated from bone marrows, spleens, livers, and peripheral blood mononuclear cells (PBMCs) of indicated mice and stained with Gr-1 and CD11b mAb. Data were analyzed by flow cytometry.
RKO and DKO mice were then challenged by the administration of a lethal dose of LM to examine the efficacy of their respective innate immune resistance. Infection of RKO mice by LM induced massive growth of LM within the livers (Figure 5A,B) and spleens (Figure 5C) as well as 100% mortality by day 4 (Figure 5D). In sharp contrast, DKO mice had significantly less bacterial load in the livers (Figure 5A,B) and spleens (Figure 5C) and the majority of the mice were able to survive more than 10 days after LM challenge (Figure 5D). In contrast to long-term survival of a significant fraction of infected B7-H4KO mice with B6 background (Figure 2A), all DKO mice eventually died of infection on day 15. Although these results confirm an important role of adaptive immunity in providing long-term anti-Listeria immunity, they show that the acute enhancement of anti-Listeria immunity in B7-H4KO mice occurs in the absence of an adaptive immune system.
Innate resistance against Listeria infection in B7-H4KO mice is independent of adaptive immunity. (A) Altered colony formation of Listeria in liver from B7-H4 × RAG-1 DKO mice. Mice were intraperitoneally injected with 3 × 106 CFUs Listeria. Forty-eight hours after infection, liver tissues at 0.2 mg from each mouse were homogenized in 10 mL PBS. Tissues (50 μL) were plated on agar plates of Listeria enrichment broth for colony counting. Data represent at least 3 independently performed experiments. DKO indicates B7-H4 × RAG-1 double KO; RKO, RAG-1 KO littermate. (B,C) Enumeration of Listeria colonies in livers (B) and spleens (C) on agar plates. Livers and spleens were prepared from RKO or DKO mice and cultivated as described in panel A to quantify Listeria colonies. Data represent 5 independently performed experiments. At day 3, differences of Listeria colonies in the organs from RKO and DKO are significant. *P < .05. (D) Resistance of DKO mice to Listeria infection. Five male RKO and 8 male DKO were intraperitoneally injected with 4 × 106 CFUs Listeria. Survival of mice was monitored daily for 15 days. Data represent 2 independently performed experiments. P < .05. (E,F) Gr-1+ cell depletion eliminated resistance to Listeria infection equally in both RKO and DKO mice. Three mice of RKO or DKO were intraperitoneally injected with 250 μg anti–Gr-1 mAb or isotype control rat IgG 24 hours before Listeria infection. Mice were then intravenously injected with 0.1 × 106 CFUs Listeria. Twenty-four hours after infection, mice were terminated and Listeria in liver was counted as described previously. The data are expressed as CFUs per gram of liver (E) and spleen (F). Data represent 2 independent experiments. * indicates significantly different from the control (Cont mAb; P < .05); **, significantly different from other control Ab group. No significant differences were found in anti–Gr-1 mAb-treated groups between RKO and DKO (P > .05). Error bars in panels B, C, E, and F indicate SD (n = 3 mice).
Innate resistance against Listeria infection in B7-H4KO mice is independent of adaptive immunity. (A) Altered colony formation of Listeria in liver from B7-H4 × RAG-1 DKO mice. Mice were intraperitoneally injected with 3 × 106 CFUs Listeria. Forty-eight hours after infection, liver tissues at 0.2 mg from each mouse were homogenized in 10 mL PBS. Tissues (50 μL) were plated on agar plates of Listeria enrichment broth for colony counting. Data represent at least 3 independently performed experiments. DKO indicates B7-H4 × RAG-1 double KO; RKO, RAG-1 KO littermate. (B,C) Enumeration of Listeria colonies in livers (B) and spleens (C) on agar plates. Livers and spleens were prepared from RKO or DKO mice and cultivated as described in panel A to quantify Listeria colonies. Data represent 5 independently performed experiments. At day 3, differences of Listeria colonies in the organs from RKO and DKO are significant. *P < .05. (D) Resistance of DKO mice to Listeria infection. Five male RKO and 8 male DKO were intraperitoneally injected with 4 × 106 CFUs Listeria. Survival of mice was monitored daily for 15 days. Data represent 2 independently performed experiments. P < .05. (E,F) Gr-1+ cell depletion eliminated resistance to Listeria infection equally in both RKO and DKO mice. Three mice of RKO or DKO were intraperitoneally injected with 250 μg anti–Gr-1 mAb or isotype control rat IgG 24 hours before Listeria infection. Mice were then intravenously injected with 0.1 × 106 CFUs Listeria. Twenty-four hours after infection, mice were terminated and Listeria in liver was counted as described previously. The data are expressed as CFUs per gram of liver (E) and spleen (F). Data represent 2 independent experiments. * indicates significantly different from the control (Cont mAb; P < .05); **, significantly different from other control Ab group. No significant differences were found in anti–Gr-1 mAb-treated groups between RKO and DKO (P > .05). Error bars in panels B, C, E, and F indicate SD (n = 3 mice).
To also confirm that increased innate resistance against LM in DKO mice is due to neutrophils but not other factors, we compared the effect of neutrophil depletion in DKO versus RKO mice. Injection of Gr-1 mAb led to comparable formation of LM colonies in livers (Figure 5E) and spleens (Figure 5F) of both RKO and DKO mice. Our results thus suggest that the lack of B7-H4 confers enhanced innate immunity against LM infection, which is largely mediated through increased neutrophils.
Absence of B7-H4 in bone marrow–derived cells promotes proliferation of Gr-1+CD11b+ neutrophil progenitors
We next examined growth potential of neutrophil progenitors in the presence and absence of B7-H4. To do so, total BM cells, which contain large numbers of neutrophil precursors, from WT, B7-H4KO, RKO, and DKO mice, were labeled with CFSE and cultured in vitro. Cell division was then monitored in these cultures by CFSE dilution. Flow cytometric analysis of cultured BM cells at day 3 showed that the majority of live cells in the culture (> 95%) were Gr-1+CD11b+ (data not shown). Importantly, 70% of Gr-1+CD11b+ cells from B7-H4KO mice (B6) had diluted CFSE, whereas only 56% of Gr-1+CD11b+ cells from WT B6 mice exhibited division. Mice with the RAG-1KO background exhibit similar changes: 86% of Gr-1+CD11b+ cells from DKO mice divided, whereas only 64.8% of Gr-1+CD11b+ cells from RKO mice had diluted CSFE (Figure 6A). Similar results were also obtained by comparing proliferative potential of purified lineage-negative (Lin−) bone marrow progenitor cells from RKO versus DKO mice in the presence of G-CSF (data not shown). These results suggest that B7-H4 is a potent inhibitor for the growth of neutrophils, which may be responsible, at least in part, for the decreased response of innate immunity against bacterial infection in WT mice.
B7-H4 inhibits growth of neutrophil progenitors from BM. (A) B7-H4–deficient neutrophil progenitors have increased cell division. BM cells (2 × 106) from indicated mice were labeled with CFSE and cultured for 3 days. Cells were harvested and doubly stained with anti–Gr-1/CD11b mAb. The dilution of CFSE in gated Gr-1+CD11b+ neutrophils was analyzed by flow cytometry. Data represent at least 3 independently performed experiments. (B) BM was harvested by flushing both femoral bones of WT B6 mice. Lineage+ cells were removed by PE-labeled anti–Gr-1, anti-CD11c, anti-Ter119, anti-CD11c, anti-CD3, anti-B220, and anti-CD19 (BD PharMingen) followed by MACS anti-PE microbeads and LD-negative selection columns (Miltenyi Biotec). The Lin− cells in the BM were quantified and normally with 75% to 85% purity. Gr-1+/CD11b+ cells were completely depleted and were plated with 100 ng/mL SCF, 10 ng/mL G-CSF, and 40 μg/mL B7-H4Ig. Cells were harvested on 1 to 3 days for cell counting. Results are presented as triplicates of mean numbers with standard deviation. Data are a representative of 4 independent experiments. *P < .05. Error bars indicate SD (n = 3 wells of cultured cells). (C) Lin− BM cells were prepared and in vitro differentiated as described in panel B. Cells were harvested from day 1 to day 3 and analyzed by staining with anti–Gr-1 and anti-CD11b mAb. (Top panel) Treated by control Ig. (Middle panel) Treated by B7-H4Ig. Cell division was also monitored by flow cytometric analysis of CFSE dilution (bottom panel). Closed symbol indicates control Ig; open symbol, B7-H4Ig.
B7-H4 inhibits growth of neutrophil progenitors from BM. (A) B7-H4–deficient neutrophil progenitors have increased cell division. BM cells (2 × 106) from indicated mice were labeled with CFSE and cultured for 3 days. Cells were harvested and doubly stained with anti–Gr-1/CD11b mAb. The dilution of CFSE in gated Gr-1+CD11b+ neutrophils was analyzed by flow cytometry. Data represent at least 3 independently performed experiments. (B) BM was harvested by flushing both femoral bones of WT B6 mice. Lineage+ cells were removed by PE-labeled anti–Gr-1, anti-CD11c, anti-Ter119, anti-CD11c, anti-CD3, anti-B220, and anti-CD19 (BD PharMingen) followed by MACS anti-PE microbeads and LD-negative selection columns (Miltenyi Biotec). The Lin− cells in the BM were quantified and normally with 75% to 85% purity. Gr-1+/CD11b+ cells were completely depleted and were plated with 100 ng/mL SCF, 10 ng/mL G-CSF, and 40 μg/mL B7-H4Ig. Cells were harvested on 1 to 3 days for cell counting. Results are presented as triplicates of mean numbers with standard deviation. Data are a representative of 4 independent experiments. *P < .05. Error bars indicate SD (n = 3 wells of cultured cells). (C) Lin− BM cells were prepared and in vitro differentiated as described in panel B. Cells were harvested from day 1 to day 3 and analyzed by staining with anti–Gr-1 and anti-CD11b mAb. (Top panel) Treated by control Ig. (Middle panel) Treated by B7-H4Ig. Cell division was also monitored by flow cytometric analysis of CFSE dilution (bottom panel). Closed symbol indicates control Ig; open symbol, B7-H4Ig.
To further evaluate the role of B7-H4 in the inhibition of neutrophil progenitors, we examined the effect of exogenous B7-H4. We first purified Lin− bone marrow (BM) cells to enrich hematopoietic stem cells. In the presence of G-CSF, these stem cells were differentiated to Gr-1+CD11b+ neutrophils. Recombinant B7-H4Ig fusion protein was then included in the beginning of culture. The cultures treated with B7-H4Ig had significantly lower cell numbers than those treated with medium or isotype-matched control Ig in day 2 and day 3 (Figure 6B) and in a dose-dependent fashion (data not shown). In addition, there was no significant difference on cell death between control versus B7-H4Ig treatment (data not shown). As shown in Figure 6C, normal BM cells contain approximately 50% Gr-1+CD11b+ myeloid cells (46.5%) and depletion of Lin+ cells by mix of mAbs efficiently removed the majority of these cells (from 46.5% to 1.3%; left top panel). Inclusion of G-CSF in the culture induced significant increase of Gr-1+CD11b+ cells; up to 83.1% of cells in the culture at day 3 were Gr-1+CD11b+ cells (Figure 6C top panel). Addition of B7-H4Ig in the culture significantly inhibited this effect (Figure 6C middle panel). This inhibition is associated with decreased cell division as indicated by dilution of CFSE (Figure 6C bottom panel). Our results provide direct evidence that B7-H4 inhibits expansion of neutrophils from their progenitors.
Discussion
During analysis of the phenotypes in a newly generated B7-H4KO mouse strain, we find that these mice are resistant to challenge of otherwise lethal dose of LM. This resistance reveals itself as early as 2 to 3 days after infection, indicating that innate but not adaptive immunity plays a key role. This is further supported by the resistance remaining in B7-H4KO mice in the absence of RAG-1 gene, in which lack of recombination-associated gene-1 leads to developmental deficiencies in T cells, B cells, and NKT cells. Enhanced innate immunity in B7-H4KO mice requires neutrophils because depletion of Gr-1+ completely abolished the resistance to the infection. Therefore, our data support B7-H4 as a checkpoint molecule in negative control of innate immunity through growth inhibition of neutrophil progenitors.
In B7-H4KO mice, the gene encoding the majority of the extracellular portion of B7-H4 protein, including entire IgV and IgC domains, is deleted to ensure complete elimination of interactions between endogenous B7-H4 and its putative receptor. As predicted from our knockout construct, the recombined murine B7-H4 gene could encode only 47 amino acids in the N-terminus, including the 20–amino acid signal peptide (Figure 1). Therefore, it is unlikely that residual 27–amino acid B7-H4 polypeptide could signal any B7-H4 receptor. To our surprise, ablation of B7-H4 does not have a profound effect on T-cell responses to either polyclonal or allogeneic antigen stimulation in vitro and in vivo. Similar observations have been made in a recent study.31 These findings appear to be different from previous studies showing that neutralizing mAb to murine B7-H4 could enhance T-cell responses to several antigens.4,5 A potential interpretation for these seemingly contradicting data is that B7-H4 does not influence global T-cell response but it affects specific effector functions downstream of T-cell responses. To support this notion, although B7-H4KO mice responded normally to several types of airway inflammatory responses, as well as LCMV and influenza infection, they have slightly enhanced T-cell immune responses to Leishmania major infection. This was attributed to a mildly enhanced Th1 response.31 However, responses of innate immunity in this knockout system were not examined. Our experiments indicate that a dominant role of B7-H4 in Listeria infection is to suppress neutrophil-mediated innate immunity and this effect could also be observed in the absence of T, NKT, and B cells. Our findings thus reveal a new function of B7-H4 as a negative regulator for innate immunity against bacterial infection.
Interestingly, bactericidal functions of neutrophils from B7-H4KO mice are normal, although percentage of neutrophils increased in spleens and other organs after infection, indicating that B7-H4 is not required for effector function of neutrophils. On the other hand, our results indicate that B7-H4 inhibits the expansion of neutrophil progenitors. B7-H4KO neutrophil progenitors have increased division compared with those from within the culture (Figure 6A). In addition, B7-H4Ig inhibited the differentiation of Lin− bone marrow cells to neutrophils in the presence of G-CSF in vitro (Figure 6B). Therefore, B7-H4 appears to directly affect the generation of neutrophils from its progenitors by inhibiting their proliferation. This may contribute, at least partially, to increased resistance to bacterial infection in B7-H4KO mice. However, other factors such as those affecting the entry and exit of neutrophils in lymphoid and peripheral organs could not be excluded now. Shortly after Listeria infection (1-2 days), T and B lymphocytes, as well as neutrophils, undergo rapid apoptosis (data not shown), which may be responsible for the decline of neutrophil numbers after a brief expansion (Table S1). In DKO mice, apoptosis appears to even outweigh expansion. However, the percentage of neutrophils was often increased (data not shown) and may contribute to resistance against Listeria.
Although our studies using Listeria infection model emphasize the critical role of B7-H4 in the negative regulation of neutrophil-mediated innate immunity, the potential role of B7-H4 on T- and B-cell responses could not be completely excluded now. B7-H4KO mice in immunocompetent B6 background have more long-term survivors than DKO mice upon challenge by lethal dose Listeria (Figure 2A vs Figure 5D). This suggests that B6/B7-H4KO mice have enhanced adaptive immunity, in addition to innate immunity. This observation in enhanced adaptive immunity in B7-H4KO mice, however, could also be interpreted by the consequence of enhanced innate immunity, which remains to be examined experimentally. It is also evident that DKO mice have significantly more neutrophils than single KO mice in virtually all organs examined so far, even without infection. It is interesting that one of the major mechanisms of B7-H4 in the adaptive immune system is inhibition of cell-cycle progression.4 It is thus possible that the lack of B7-H4 on T, B, and/or NKT cells (absence in Rag-1−/− mice) affects normal turnover or homeostasis of neutrophils.
Our results indicate that a mechanism behind resistance to Listeria infection in the absence of B7-H4 is due to increased neutrophils in peripheral organs. Although there are slight increases in neutrophil counts in uninfected B6/B7-H4KO mice (Figure 4B), more significant increases of neutrophils occur upon LM infection (Figure 2C; Table S1). A similar increase was also observed in DKO mice (Table S1). In addition, depletion of neutrophils by anti–Gr-1 mAb efficiently eliminated this resistance (Figures 3A and 5E,F). In contrast to the increased number or responses of neutrophils to infection, phagocytosis and respiratory burst, 2 major bactericidal functions of neutrophils, do not have obvious modifications in the absence of B7-H4 (Figure 3C). Therefore, our data support that increased neutrophils in B7-H4KO mice may be a key mechanism for the resistance to Listeria infection.
It has been shown that B7-H4, upon binding to its putative receptor, inhibits cell-cycle progression on T cells.4,9 In our cell culture system, division of neutrophil progenitors in WT bone marrows is clearly slower than those from B7-H4KO mice (Figure 6A). In addition, recombinant B7-H4 inhibited proliferation (Figure 6B) and division (Figure 6C) of Gr-1+CD11b+ cells. These results implicate that B7-H4 also inhibits cell-cycle progression of neutrophils. We did not observe significant changes of cell apoptosis in the culture of BM from B7-H4KO mice up to 5 days (G.Z., L.C., unpublished data, January 2008), excluding the role of cell death. Our data thus support a direct role of B7-H4 in the inhibition of neutrophils. However, the mechanisms and biochemical details underlying this inhibition are yet to be clarified. It is tempting to speculate that neutrophils express the receptor for B7-H4, thus transmitting inhibitory signal directly to neutrophils. However, the receptor for B7-H4 has not yet been identified, therefore preventing further analysis. Although we failed to detect binding of B7-H4Ig fusion protein to the surface of neutrophils (G.Z., L.C., unpublished data, January 2008), B7-H4Ig inhibits proliferation of Lin− neutrophil progenitors from bone marrow (Figure 6B). This could be interpreted by either low-level expression of B7-H4 receptor or low affinity/avidity of B7-H4Ig fusion protein. Neutrophils do not express B7-H4 in our experiments by RT-PCR, flow cytometry, or immunohistochemistry analyses (G.Z., L.C., data not shown), indicating that B7-H4 is provided by other stromal cells in bone marrow.
Neutrophils are in the first line of host defense against infection. Our findings show an increased resistance to Listeria infection in B7-H4KO mice, implicating a new approach to enhance innate immunity against infection by Listeria and possibly other pathogens. Therefore, the method of selectively blocking B7-H4 expression by neutralizing mAb or appropriately engineered B7-H4 protein with antagonistic activity may represent a new approach to increase neutrophils and enhanced innate immunity against pathogen infection.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Drew M. Pardoll for constructive comments; Dr Hans Schreiber for the generous gift of Gr-1 hybridoma; Dr Thomas W. Dubensky Jr for Listeria strains; and Jennifer Osborne for editing.
This study was supported by National Institutes of Health (NIH, Bethesda, MD) grant CA98731, CA113341, and CA97085. M.M.A. was supported by the Niarchos Surgical Research Fellowship.
National Institutes of Health
Authorship
Contribution: G.Z. and M.M.A. participated in designing and performing the research and writing the paper; T.A., L.L., S.Y., S.A., A.C.R., J.H., H.X., A.S.F., S.J.F., and K.T. performed some experiments; J.M.A.D. participated in the designing and performing the construction of genetic knockout mice and wrote the paper; M.C. provided reagents and advised on their usage; and L.C. organized the study, analyzed data, and wrote the paper. All authors checked the final version of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lieping Chen, Johns Hopkins Medical Institutions, 2M07 David H. Koch Building, 1550 Orleans Street, Baltimore, MD 21231; e-mail: lchen42@jhmi.edu.