Abstract
We demonstrate that in zebrafish, the microRNA miR-451 plays a crucial role in promoting erythroid maturation, in part via its target transcript gata2. Zebrafish miR-144 and miR-451 are processed from a single precursor transcript selectively expressed in erythrocytes. In contrast to other hematopoietic mutants, the zebrafish mutant meunier (mnr) showed intact erythroid specification but diminished miR-144/451 expression. Although erythropoiesis initiated normally in mnr, erythrocyte maturation was morphologically retarded. Morpholino knockdown of miR-451 increased erythrocyte immaturity in wild-type embryos, and miR-451 RNA duplexes partially rescued erythroid maturation in mnr, demonstrating a requirement and role for miR-451 in erythrocyte maturation. mnr provided a selectively miR-144/451-deficient background, facilitating studies to discern miRNA function and validate candidate targets. Among computer-predicted miR-451 targets potentially mediating these biologic effects, the pro-stem cell transcription factor gata2 was an attractive candidate. In vivo reporter assays validated the predicted miR-451/gata2-3′UTR interaction, gata2 down-regulation was delayed in miR-451-knockdown and mnr embryos, and gata2 knockdown partially restored erythroid maturation in mnr, collectively confirming gata2 down-regulation as pivotal for miR-451-driven erythroid maturation. These studies define a new genetic pathway promoting erythroid maturation (mnr/miR-451/gata2) and provide a rare example of partial rescue of a mutant phenotype solely by miRNA overexpression.
Introduction
MicroRNAs (miRNAs) are 19-25 nucleotide, single stranded RNAs that regulate gene expression by binding target mRNAs, modulating their translation and turnover.1-3 However, the biologic role of most individual miRNAs and the targets they modulate remain to be determined.
miRNA expression profiling has been a useful starting point for selecting miRNAs that may influence tissue- or cell-specific processes such as hematopoiesis. High or increasing levels of an miRNA in a specific cell lineage may promote differentiation by repressing transcripts whose persistence or presence would impede cellular commitment (eg, transcripts that promote an alternative cell lineage). On the other hand, high miRNA levels may act to preserve the cellular status quo, as has been proposed in a model of miRNA action in CD34+ hematopoietic progenitor cells (HPC).4 Conversely, miRNAs of low or falling abundance may be acting to release repression of a set of transcripts necessary to drive changes of cellular state. In view of these various possible mechanisms of action, functional studies are required to understand how individual miRNAs actually influence biologic processes and to determine the molecular mechanisms by which each one acts.
Erythropoiesis is a process of progression from stem cells, characterized by pluripotency and self-renewal, through intermediate states retaining proliferative capacity but of increasingly restricted potential, through to lineage-specific cells that undergo a morphologically recognizable pattern of terminal differentiation. This process requires cells to possess mechanisms for progressive modification of their transcriptomes to acquire new capabilities and to discard residual now-redundant or deleterious transcripts from a former cellular state. miRNAs provide a potential mechanism for achieving this.
Several miRNAs are down-regulated as erythropoiesis proceeds, removing constraints that act to preserve a more immature stem or progenitor cellular phenotype. In erythropoietic-driven cultures of human CD34+ HPCs, miR-221 and miR-222 are down-regulated.5 This contributes to erythroid expansion by derepression of at least one of their targets, KIT, through a mechanism verified by HPC transduction with miRNA duplexes or lentiviral overexpression vectors.5 Likewise, miR-24 has higher expression in HPCs and is down-regulated during erythroid differentiation.4,6 miR-24 represses HPC erythroid differentiation in part through repression of its target, the type I activin receptor (ALK4), thereby gate-keeping the pro-erythropoietic action of activin (in conjunction with erythropoietin) mediated by this receptor.6
In contrast, several other miRNAs are strongly up-regulated as erythrocytes mature. Broad expression profiling surveys have recurrently identified miR-144 and miR-451 for their high erythropoietic-restricted expression in several species, including zebrafish,7,8 mice,9-11 and humans.11-15 In mice, they arise from a single transcript.9 Overexpression or loss of expression of miR-451 in murine erythroleukemia (MEL) cells promoted or impaired erythrocyte differentiation, respectively, assessed by β-globin transcript abundance and hemoglobinization,11 consistent with miR-451 being a positive regulator of erythroid maturation. A recent study has shown the murine miR-144/451 locus is transcriptionally regulated by Gata1.9 Loss of miR-144/451 expression in the zebrafish gata1-mutant vlad tepes was interpreted as further evidence for Gata1-dependent activation of miR-144/451 locus.9 This study also examined loss and gain of miR-451 function in zebrafish embryos, concluding that miR-451 was required for either the maintenance/survival or late-stage maturation of committed erythrocytes.
We independently examined the function of the miR-144/451 in zebrafish hematopoiesis by an alternate experimental approach. We exploited a new zebrafish mutant, meunier (mnr), a nonanemic mutant expressing gata1 normally but found to have miR-144/451–deficient erythrocytes. mnr freed miR-144/451 from their otherwise epistatic relationship with gata1 and provided a unique, genetically stable, miR-144/451–deficient but erythrocyte-replete background for experimentation. Using rigorously validated reagents, we demonstrate that miR-451 (but not miR-144) functions to accelerate the kinetics of erythrocyte maturation in zebrafish. Furthermore, we validated gata2 as one bona fide target of miR-451 in zebrafish, and demonstrate that miR-451–mediated clearance of gata2 is a crucial influence on the rate of zebrafish erythrocyte maturation.
Methods
Zebrafish
Zebrafish strains used were: AB*, cloche (clom39),16 Tg(fli1:GFP)y1,17 meunier (mnrgl9, a novel mutant isolated in our ethylnitrosourea mutagenesis screen18 for its lack of myeloperoxidase [mpx] expression) and Tg(mpx:EGFP)i114.19 Fish were housed in the Ludwig Institute for Cancer Research Aquarium using standard husbandry practices. All experiments were approved by the Walter and Eliza Hall Institute Animal Ethics Committee. Zebrafish gene, protein, and mutant naming follows the recommended conventions.20
Microinjections
Fertilized 1- to 2-cell embryos were microinjected with 1 to 2 nL synthetic mRNA (50 μg/μL in H2O), miRNA morpholino oligonucleotides (MOs; 100-500 μmol/L in H2O; Gene Tools, Philomath, OR), control or gata2 MOs21 (130 μmol/L in H2O; Gene_Tools) and synthetic miRNA duplexes (2-20 μmol/L in H2O; Sigma-Proligo, St Louis, MO), traced where appropriate by mixing 1:1 with 5% rhodamine dextran (in 0.2 mol/L KCl). We customarily delivered different nucleic acid reagents by separate microinjections to avoid ex vivo mixing. As summarized schematically (Figures 3,Figure 4,Figure 5–6 and Figure S8, available on the Blood website; see the Supplemental Materials link at the top of the online article), we selected reagent combinations according to experimental purpose. Some assays required optimization and the specific reagent concentrations used were (1) miRNA duplex/mRNA 3′untranslated region (UTR) validation (Figure S8B,D,E): 50 μg/μL RNA, 2 μmol/L synthetic miRNA duplex; (2) MO validation (Supplementary Figure 8C,F,G): 50 μg/μL RNA, 2 μmol/L synthetic miRNA duplex, 500 μmol/L MO; and (3) miRNA rescue experiments (Figure 3) and gata2 target validation tests (Figures 4,6), 20 μmol/L synthetic miRNA duplex.
Oligonucleotides and constructs
Table S1 lists oligonucleotide sequences. The green fluorescent protein (GFP) reporter/sensor constructs were derived from pCS2+GFP-F by standard cloning techniques22 and linearized with NotI for in vitro transcription of capped mRNA using the mMESSAGE mMACHINE kit (Ambion, Austin, TX). Triplicate miRNA binding sites were constructed by annealing complementary oligonucleotides. The gata2 miR-451 binding sites were mutated by sequential inverse polymerase chain reaction (PCR) and ligation via an acquired restriction enzyme site. All constructs were validated by sequencing.
Gene expression analysis
Whole mount in situ hybridization (WISH) was performed by using standard techniques and in vitro transcribed digoxigenin- or fluorescein-labeled antisense gata1, gata2, hbbe3, lcp1, and mpx riboprobes23-26 and 4-nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate or fast red for detection. miRNA WISH with locked nucleic acid (LNA) probes (Exiqon, Vedbaek, Denmark) was performed as described previously8 and with reference to the manufacturer's instructions, using hybridization at 35°C for miR-144, 50°C for miR-451, and 53°C for miR-206 LNA probes. We used standard techniques for double WISH (Figure 2B), probing first with the hbbe3 riboprobe at 68°C followed by stringent washing and reprobing with the miR-144 LNA probe at 35°C. For adult tissues, dissected organs were diced (1 mm fragments) and processed for WISH as for embryos, with proteinase K (20 μg/mL) treatment for 30 minutes. Organ fragments were aligned in 1% agarose, sectioned, and counterstained with nuclear fast red.
The miR144/451 locus RT-PCR used a Superscript III One-Step reverse transcription (RT)-PCR kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, primers as in Table S1, 30 amplification cycles at 94°C, 60°C, and 68°C for 15, 30, and 30 seconds, respectively, and displaying PCR products by 2% agarose gel electrophoresis.
Imaging
Low-power images were collected using a Nikon SMZ1500 or Nikon 90i fluorescence microscope equipped with a DXM1200c camera and Nis-Elements AR software (Nikon, Tokyo, Japan), and high-power images with a Nikon Optiphot-2 microscope with a Zeiss AxioCam MRc5 digital camera and AxioVision AC (Release 4.5) software (Zeiss, Welwyn Garden City, United Kingdom). Images were imported into Adobe Photoshop CS2 9.0.2 or Illustrator CS2 12.0.1 (Adobe Systems, Mountain View, CA) for orientation and figure preparation. To ensure valid documentation of comparative assessments of fluorescence intensity, arrays of multiple embryos were photographed together in a single image.
Morphometry
Embryonic zebrafish erythrocytes were collected by transecting tails of 6 to 8 embryos in 600 μL of balanced salt solution with 5 mmol/L ethylenediaminetetraacetic acid (EDTA) and 4% fetal calf serum. Cytospin slides were prepared, stained with May-Grünwald/Giemsa stain, and examined at high power. Nuclear and cytoplasmic areas of 20 to 30 randomly selected erythrocytes were measured using either the outline tool of AxioVision AC software or NIS Elements software, and the nuclear/cytoplasmic (N:C) area ratio was calculated. O-dianisidine staining for hemoglobin was as described previously.23
Genotyping
clo embryos were recognized as a Mendelian proportion (Figure 1E). mnr embryos were recognized at 48 hours postfertilization (hpf) by their characteristic syndrome of small eyes, cerebral degeneration, and thinner yolk extension. Younger mnr embryos were PCR-genotyped at the closely linked (3 centimorgans) simple sequence length polymorphism (SSLP) marker z3984 (oligonucleotides, Table S1; 10 μL reactions; Taq polymerase [New England Biolabs, Ipswich, MA] with supplied buffer; 40 cycles at 95°C, 60°C, and 72°C for 20, 20, and 30 seconds, respectively; PCR products were separated by 2% agarose gel electrophoresis).
Expression of the miR-144/451 locus in wild-type embryonic and adult zebrafish erythroid cells. (A) miR144/451 locus expression (22 hours postfertilization [hpf]) by RT-PCR using primers confirming that both miRNAs originate from a single precursor transcript. Schema shows primer binding sites in green. RT indicates reverse transcriptase; Gen, genomic DNA control. (B) Whole mount in situ hybridization (WISH) analysis of miR-144 at the 3 embryonic ages shown. miR-144 expression (blue) initiates at 18 hpf in the hematopoietic intermediate cell mass (ICM), becomes confluent across all cells of the ICM (20 hpf), and disperses with erythrocytes at the onset of circulation, pooling over the yolk during fixation for WISH (30 hpf). Lateral views, anterior to left; inset (bottom panel) is ventral view. (C) Sagittal section of 24-hpf WISH embryos showing miR-144 expression (blue) in cytoplasm of erythrocytes of the axial ICM. NC indicates notochord, Y, yolk. Scale bar = 20 μm. (D) Double WISH showing concordant miR-144 (blue) and hbbe3 (red) expression. Left panel, bright field; right panel, fluorescence microscopy highlighting hbbe3 expression. (E) miR-451 and gata1 expression (blue) in WT and clo embryos at 24 hpf, showing concordant loss of gata1 and miR-451 expression in clo. (F) miR-144 expression (blue) in hematopoietic cells nestled between renal tubules (RT) of adult kidney (i, white arrowheads) and the red pulp of the spleen (ii, white arrowheads) in nuclear fast red-counterstained sections cut from tissue fragment in situ hybridization. Detail (iii) shows miR-144 expression localized to the cytoplasm of circulating adult erythrocytes. Scale bar = 10 μm.
Expression of the miR-144/451 locus in wild-type embryonic and adult zebrafish erythroid cells. (A) miR144/451 locus expression (22 hours postfertilization [hpf]) by RT-PCR using primers confirming that both miRNAs originate from a single precursor transcript. Schema shows primer binding sites in green. RT indicates reverse transcriptase; Gen, genomic DNA control. (B) Whole mount in situ hybridization (WISH) analysis of miR-144 at the 3 embryonic ages shown. miR-144 expression (blue) initiates at 18 hpf in the hematopoietic intermediate cell mass (ICM), becomes confluent across all cells of the ICM (20 hpf), and disperses with erythrocytes at the onset of circulation, pooling over the yolk during fixation for WISH (30 hpf). Lateral views, anterior to left; inset (bottom panel) is ventral view. (C) Sagittal section of 24-hpf WISH embryos showing miR-144 expression (blue) in cytoplasm of erythrocytes of the axial ICM. NC indicates notochord, Y, yolk. Scale bar = 20 μm. (D) Double WISH showing concordant miR-144 (blue) and hbbe3 (red) expression. Left panel, bright field; right panel, fluorescence microscopy highlighting hbbe3 expression. (E) miR-451 and gata1 expression (blue) in WT and clo embryos at 24 hpf, showing concordant loss of gata1 and miR-451 expression in clo. (F) miR-144 expression (blue) in hematopoietic cells nestled between renal tubules (RT) of adult kidney (i, white arrowheads) and the red pulp of the spleen (ii, white arrowheads) in nuclear fast red-counterstained sections cut from tissue fragment in situ hybridization. Detail (iii) shows miR-144 expression localized to the cytoplasm of circulating adult erythrocytes. Scale bar = 10 μm.
Statistics
Single images are representative of at least 30 embryos or representative of x of y embryos as stated. Quantitative data derives from at least 3 independent experiments; descriptive statistics are either medians or mean plus or minus SD of data for n individuals, or mean plus or minus SE of n independent experiments. Prism 4.0c (GraphPad Software, San Diego, CA) was used for analytical statistics, using unpaired, 2-tailed t tests for normally distributed continuous variables. P less than .05 indicated statistically significant difference.
Results
Expression of the zebrafish miR144/451 locus in embryonic and adult erythrocytes
miR-144 and miR-451 form a clustered pair in the genome of zebrafish, separated by only 65 nucleotides (Figure S1). We demonstrated, using RT-PCR, that miR-144 and miR-451 were present on a single transcript (Figure 1A). To determine the anatomical and temporal constraints on their direct function, we examined their expression pattern by whole-mount in situ hybridization. miR-144/451 expression in zebrafish hematopoietic tissues has been previously noted.7-9 The locus was first expressed at 18 hpf in the cytoplasm of individual erythrocytes in the intermediate cell mass (ICM), the axial hematopoietic tissue of the early zebrafish embryo (Figure 1B,C). As expected, embryonic miR-144 expression overlapped with miR-451 expression (Figure 1A,E; data not shown). Early miR-144/451 expression tracked exactly with other genes expressed in erythroid-lineage cells (eg, the erythroid-determining transcription factor gata1 and globin [hbbe3]23,26 [Figure 1D,E]) and required the presence of erythrocytes. For example, cloche (clo) is an early hematopoietic/vascular-failure mutant with absent/reduced expression of all known hematopoietic genes.16,27,28 miR-144/451 expression was absent in clo (Figure 1E), confirming their restricted expression in hemovascular derivatives. The restricted expression of this locus is maintained into adulthood in clusters of cells in the hematopoietic organs (spleen and kidney; Figure 1Fi,ii) and the cytoplasm of circulating erythrocytes (Figure 1Fiii).
meunier, a nonanemic mutant with reduced miR-144/451 expression
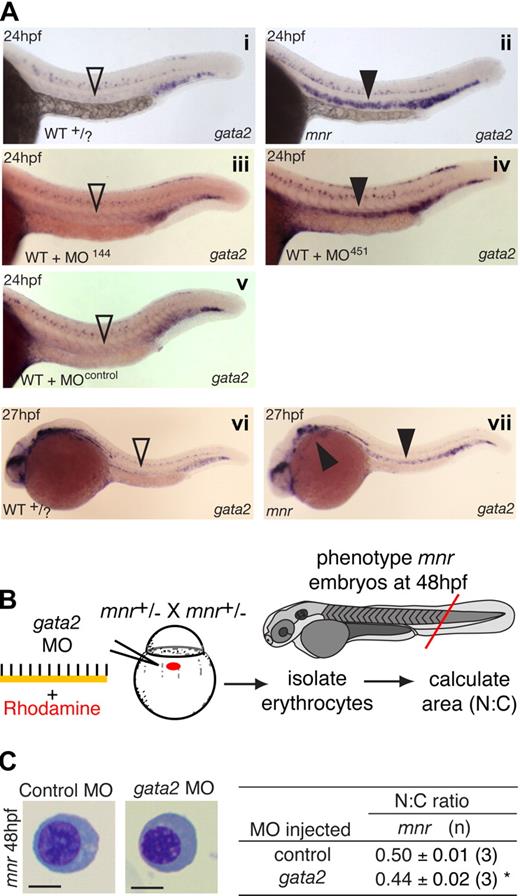
The expression of the miR-144/451 locus and other erythroid genes (gata1, hbbe3) was surveyed in a panel of other hematopoietic-failure mutants. In 8 of 9 mutants from the Melbourne Myeloid screen,18 there was concordance between miR-144/451 locus and other erythroid gene expression. The single exception was meunier (mnr), which showed greatly reduced (but not absent) miR-144/451 expression by WISH and semiquantitative RT-PCR despite normal gata1 and hbbe3 expression (Figure 2A,B; Figure S2) and normal O-dianisidine hemoglobin staining (Figure 2C; Figure S3A).
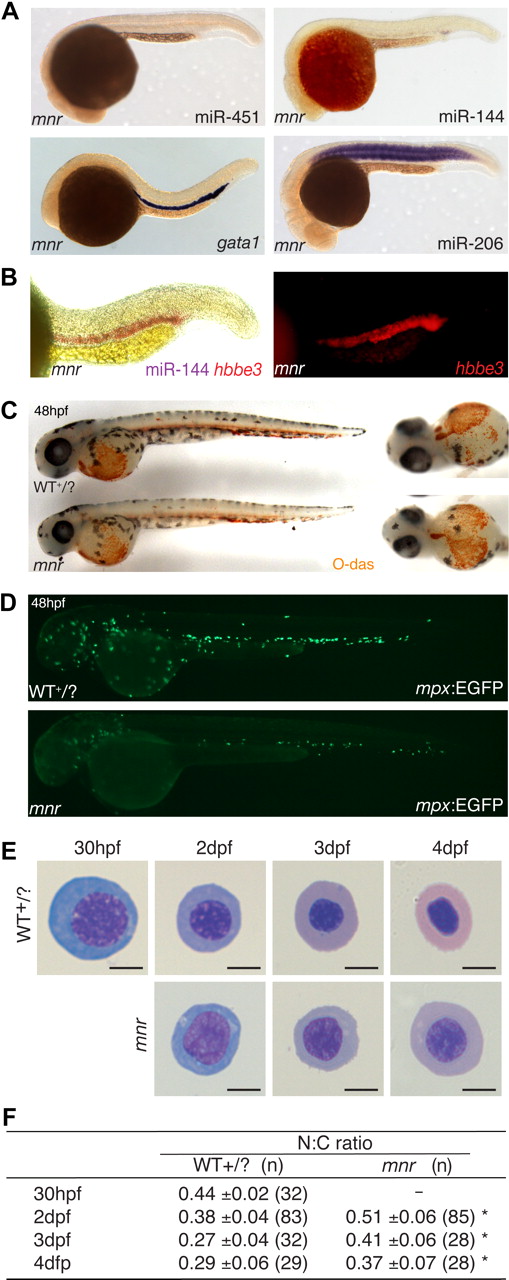
meunier (mnr) has miR-144/451 deficiency and an erythroid maturation defect despite normal erythroid specification. (A) Whole-mount in situ hybridization (WISH) expression analysis of miR-144, miR-451, miR-206, and gata1 at 24 hpf. mnr has normal gata1 expression (compared with wild-type Figure 1E) but is selectively deficient in the 2 erythroid miRNAs. Top panel mnr embryos were PCR genotype-confirmed; bottom panel is representative of 57 of 57 (gata1) and 32 of 34 (miR-206) embryos resulting from a mnr heterozygote incross. (B) Double WISH demonstrating that mnr has loss of miR-144 (loss of blue) despite retaining hbbe3 (red) expression; for comparison with WT, see Figure 1D. Left panel, bright field; right panel, fluorescence microscopy highlighting hbbe3 expression. (C) Normal expression of hemoglobin (brown) in mnr demonstrated by O-dianisidine (O-das) staining. At 48 hpf, mnr is recognizable by its smaller eye and head. Representative embryos are shown in lateral (left) and ventral (right) views to demonstrate equivalent O-das staining despite variation in the pattern of blood pooling at fixation. Embryos are representative of at least 30 embryos/genotype (see also Figure S3A). (D) Deficiency of mpx-expressing cells in mnr, displayed by a reduced number of EGFP-expressing cells in mnr embryos carrying the Tg(mpx:EGFP) reporter transgene (bottom panel) compared with wild type (top panel). Figure S4 shows expression of other hematopoietic and myeloid cell markers in mnr determined by WISH (tal1, spi1, lcp1, lyz, mpll). (E) Series of representative circulating erythrocytes from progressively older wild-type (WT + /?, top) and mnr (bottom) embryos, demonstrating the progressive maturation of WT erythrocytes and the persistent immature morphology of mnr erythrocytes. Erythrocytes are representative of the mean of groups tablulated in panel F. May-Grünwald/Giemsa stain; scale bars = 5 μm. (F) Tabulation of the N:C area ratio in WT and mnr embryos from 30 hpf to 4 dpf. The N:C ratio declines in both genotypes but is always greater in mnr than in WT. Data are mean plus or minus SD for n embryos, collected over 1 to 4 independent experiments. *P < .001 for WT compared with mnr. Figure S6 further demonstrates the reproducibility of these data by presenting them as scatterplots and displaying the interassay variation for the 48-hpf timepoint.
meunier (mnr) has miR-144/451 deficiency and an erythroid maturation defect despite normal erythroid specification. (A) Whole-mount in situ hybridization (WISH) expression analysis of miR-144, miR-451, miR-206, and gata1 at 24 hpf. mnr has normal gata1 expression (compared with wild-type Figure 1E) but is selectively deficient in the 2 erythroid miRNAs. Top panel mnr embryos were PCR genotype-confirmed; bottom panel is representative of 57 of 57 (gata1) and 32 of 34 (miR-206) embryos resulting from a mnr heterozygote incross. (B) Double WISH demonstrating that mnr has loss of miR-144 (loss of blue) despite retaining hbbe3 (red) expression; for comparison with WT, see Figure 1D. Left panel, bright field; right panel, fluorescence microscopy highlighting hbbe3 expression. (C) Normal expression of hemoglobin (brown) in mnr demonstrated by O-dianisidine (O-das) staining. At 48 hpf, mnr is recognizable by its smaller eye and head. Representative embryos are shown in lateral (left) and ventral (right) views to demonstrate equivalent O-das staining despite variation in the pattern of blood pooling at fixation. Embryos are representative of at least 30 embryos/genotype (see also Figure S3A). (D) Deficiency of mpx-expressing cells in mnr, displayed by a reduced number of EGFP-expressing cells in mnr embryos carrying the Tg(mpx:EGFP) reporter transgene (bottom panel) compared with wild type (top panel). Figure S4 shows expression of other hematopoietic and myeloid cell markers in mnr determined by WISH (tal1, spi1, lcp1, lyz, mpll). (E) Series of representative circulating erythrocytes from progressively older wild-type (WT + /?, top) and mnr (bottom) embryos, demonstrating the progressive maturation of WT erythrocytes and the persistent immature morphology of mnr erythrocytes. Erythrocytes are representative of the mean of groups tablulated in panel F. May-Grünwald/Giemsa stain; scale bars = 5 μm. (F) Tabulation of the N:C area ratio in WT and mnr embryos from 30 hpf to 4 dpf. The N:C ratio declines in both genotypes but is always greater in mnr than in WT. Data are mean plus or minus SD for n embryos, collected over 1 to 4 independent experiments. *P < .001 for WT compared with mnr. Figure S6 further demonstrates the reproducibility of these data by presenting them as scatterplots and displaying the interassay variation for the 48-hpf timepoint.
mnr is an ethylnitrosourea-induced recessive mutant initially identified for its reduced number of myeloperoxidase (mpx)–expressing cells (Figure 2D). Expression of myeloid markers that are less lineage-specific (lcp1, lyz) is reduced, but early hematopoietic transcription factors tal1 and spi1 are normal (Figure S4).25 mnr develops later nonhematopoietic defects, most significantly cerebral degeneration leading to death at 4 to 5 days postfertilization (dpf). However, until approximately 38 hpf, mnr embryos are morphologically indistinguishable from wild-type (WT) siblings (confirmed by longitudinal observation), after which they can be reliably identified by reduced head and eye size (Figure 2C).
The loss of miR-144/451 expression in mnr did not merely reflect globally reduced miRNA expression/processing, because expression of the somite-specific miR-206 was normal (Figure 2A),8,29 and there was no accumulation of the primary miRNA transcript detected by RT-PCR (Figure S2). Therefore, mnr uniquely dissociates miR-144/451 locus expression from its otherwise tight epistatic relationship to gata19 without being a global miRNA biogenesis mutant and provides an erythrocyte-replete, genetically stable, miR-144/451–deficient background for further experimentation.
mnr itself is the independent positive regulator of the miR-144/451 locus
Two observations suggest that the miR-144/451–deficiency and mpx-deficiency phenotypes of mnr are not caused by 2 independent mutations. First, offspring from 3 of 3 independent mnr heterozgyote pairs genotyped on the basis of their mpx deficiency phenotype all showed reduced miR-144/451 expression. Second, embryos sorted by either phenotype showed tight linkage to the chromosome 14 SSLP marker z3984 (Figure S5). This tight linkage also means that mnr is not a mutation of the miR-144/451 locus itself, because the miR-144/451 locus is on chromosome 5 (Zebrafish Genome Assembly Zv730 ).
Erythrocyte maturation is delayed in mnr
We hypothesized that mnr erythrocytes may exhibit an abnormal phenotype that would point to the nature of their requirement for miR-144/451, and we therefore carefully scrutinized erythroid development in mnr mutant embryos. During zebrafish development, there is a characteristic change in erythrocyte morphology associated with developmental age.31,32 Proerythroblasts, characterized by deeply basophilic cytoplasm and large finely granular nucleus, mature through stages displaying an increasingly acidic cytoplasm, progressively denser nuclear chromatin, and a decreasing N:C area ratio despite an overall concomitant reduction in erythrocyte diameter (Figure 2E).
Comparing erythrocyte morphology between mnr and its WT siblings revealed delayed erythroid maturation in mnr. Although evident morphologically (mnr erythrocytes were larger, had more basophilic cytoplasm, and more open nuclear chromatin), the N:C ratio (48 hpf) provided a robust objective numeric variable reflecting this delay that was statistically different between mnr and WT erythrocytes (Figure 2E,F; Figure S6).
The immaturity of mnr erythrocytes was not due to a population skewing effect from loss of mature erythrocytes by apoptosis or from an erythropoietic drive resulting from accelerated erythrocyte destruction, because acridine orange staining failed to demonstrate excess death of ICM hematopoietic cells or later erythroid cells (Figure S7). Overall, mnr shows that miR-144/451 are not essential for erythroid specification, early development including hemoglobinization, and survival but suggests their involvement in promoting erythroid maturation.
miR-451 deficiency alone is sufficient to retard erythrocyte maturation
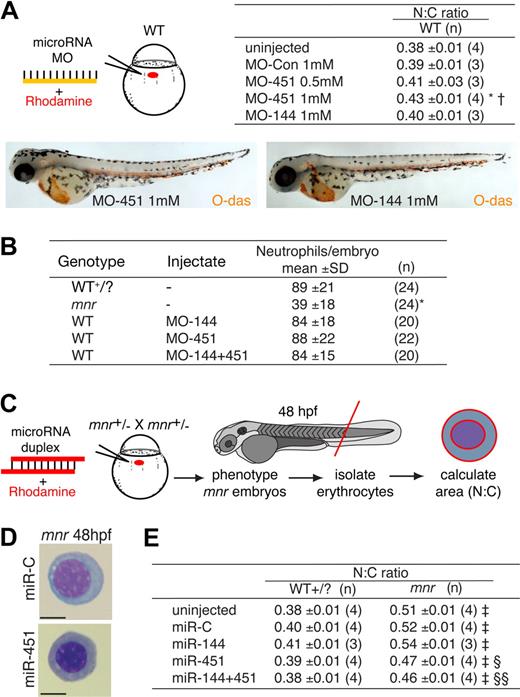
To determine whether miR-144 and/or miR-451 absence alone was sufficient to cause erythrocyte immaturity observed in mnr, we used antisense MO knockdown of miR-144 and miR-451.9,29 The capability and efficacy of each MO to intercept repressive miRNA/3′UTR interactions was demonstrated by their activity in an in vivo reporter assay based on microinjection of combinations of MO, synthetic miRNA duplexes, and mRNA encoding enhanced green fluorescent protein (EGFP) with a 3′UTR containing canonical miRNA target-sites (Figure S8). MO-451 (but not MO-144) resulted in a statistically significant dose-dependent increase in N:C ratio reflecting morphologic erythrocyte immaturity (Figure 3A; Figure S9). However, despite maximal MO doses, this phenotype was not as marked as that observed in mnr. MO-451 specifically affected erythrocyte development; MO knockdown of miR144/451 did not affect neutrophil number (Figure 3B), nor did it recapitulate the nonhematologic mnr phenotypes. A second MO, MO-451B (of the “multiblocking” type, also interfering with miRNA biogenesis), proved toxic at a dose achieving only modest inhibition in the reporter assay (data not shown). MO-451 affected the rate of erythrocyte maturation but did not abort it, because O-dianisidine staining demonstrated wild-type hemoglobin levels at 48 hpf (Figures 3A, S3B).
Functional studies of the role of miR-451 in erythroid maturation. (A) miR-451 deficiency, but not miR-144 deficiency, is sufficient to cause erythroid immaturity. MO miRNA antagonists or control MO was microinjected, tracing delivery by rhodamine (schematic diagram) and the N:C area ratio computed as an indicator of maturation (table). O-diansidine staining confirmed normal hemoglobinization in MO-injected embryos at 48 hpf (compare Figure 2C); embryos in panels are representative of at least 16 embryos/group (see Figure S3B). MO reagent validation experiments are in Figure S2. Only MO-451 increased the N:C ratio, reflecting delayed erythrocyte maturation, in a dose-dependent manner. Data are mean plus or minus SE for n independent groups from 3 separate experiments. *P = .036; †P = .048 for the comparisons of MO-451-injected with control-injected and MOmiR-144-injected groups, respectively, 2-tailed t test. Figure S9 further demonstrates the reproducibility of these data by presenting them as scatterplots and providing additional fields of representative cells. (B) Knockdown of miR-144/451 does not affect neutrophil numbers and does not replicate the deficiency of mpx-expressing cells in mnr. Experiments were performed using WT or mnr embryos on the Tg(mpx:EGFP) background. Concentration of each MO injected was 1 mmol/L. Neutrophils were quantified by counting EGFP-positive cells/embryo at 48 hpf. *P < .0001 for comparison of mnr with all 4 other groups. (C-E) Overexpression of miR-451, but not miR-144, is sufficient to partially rescue the erythroid maturation block in mnr, as evident morphologically (D) and quantitatively (E) using the N:C area ratio. (C) A schema of the experimental design, which required genotyping embryos by nonhematologic phenotypic features at 48 hpf. Data are mean plus or minus SE for n independent experiments; ‡P < .0025 for line-by-line comparisons of WT to mnr, and §P = .035 and §§P = .003 for the indicated comparisons of miRNA-injected mnr with miR-C-injected mnr, 2-tailed t test. Scale bars = 5 μm. Figure S10 further demonstrates the reproducibility of these data by presenting them as scatterplots and providing additional fields of representative cells.
Functional studies of the role of miR-451 in erythroid maturation. (A) miR-451 deficiency, but not miR-144 deficiency, is sufficient to cause erythroid immaturity. MO miRNA antagonists or control MO was microinjected, tracing delivery by rhodamine (schematic diagram) and the N:C area ratio computed as an indicator of maturation (table). O-diansidine staining confirmed normal hemoglobinization in MO-injected embryos at 48 hpf (compare Figure 2C); embryos in panels are representative of at least 16 embryos/group (see Figure S3B). MO reagent validation experiments are in Figure S2. Only MO-451 increased the N:C ratio, reflecting delayed erythrocyte maturation, in a dose-dependent manner. Data are mean plus or minus SE for n independent groups from 3 separate experiments. *P = .036; †P = .048 for the comparisons of MO-451-injected with control-injected and MOmiR-144-injected groups, respectively, 2-tailed t test. Figure S9 further demonstrates the reproducibility of these data by presenting them as scatterplots and providing additional fields of representative cells. (B) Knockdown of miR-144/451 does not affect neutrophil numbers and does not replicate the deficiency of mpx-expressing cells in mnr. Experiments were performed using WT or mnr embryos on the Tg(mpx:EGFP) background. Concentration of each MO injected was 1 mmol/L. Neutrophils were quantified by counting EGFP-positive cells/embryo at 48 hpf. *P < .0001 for comparison of mnr with all 4 other groups. (C-E) Overexpression of miR-451, but not miR-144, is sufficient to partially rescue the erythroid maturation block in mnr, as evident morphologically (D) and quantitatively (E) using the N:C area ratio. (C) A schema of the experimental design, which required genotyping embryos by nonhematologic phenotypic features at 48 hpf. Data are mean plus or minus SE for n independent experiments; ‡P < .0025 for line-by-line comparisons of WT to mnr, and §P = .035 and §§P = .003 for the indicated comparisons of miRNA-injected mnr with miR-C-injected mnr, 2-tailed t test. Scale bars = 5 μm. Figure S10 further demonstrates the reproducibility of these data by presenting them as scatterplots and providing additional fields of representative cells.
miR-451 replenishment partially rescues erythroid maturation in mnr
The selective miR-144/451 deficiency of mnr provided a scenario for testing whether miRNA replenishment was sufficient to rescue erythrocyte maturation in an endogenously miRNA-deficient environment. Overexpression was achieved by microinjecting a synthetic RNA duplex that mimics the endogenous dicer processed product. The activity of synthetic duplexes was confirmed by an in vivo reporter assay (Figure S8). Overexpression of miR-451 (but not miR-144) partially rescued the mnr erythrocyte maturation defect (Figure 3C-E), reflected morphologically and by a statistically significant reduction in the N:C ratio. This indicates that the mnr maturation defect was at least partially due to miR-451 deficiency. In a WT background, overexpression of miR-144/451 did not affect erythrocyte maturation or gross embryo morphology (Figure 3E; Figure S10; data not shown). Furthermore, several nonhematopoietic mnr phenotypes were not altered by miR-144/451 overexpression (data not shown), suggesting that they resulted from a role of the mnr gene product independent of its role in miR-144/451 up-regulation.
Together, these observations indicate a specific requirement for miR-451 (but not miR-144) in promoting erythroid maturation. The MO-451 knockdown and miR-deficient mnr phenotypes indicate a necessity for miR-451 for normal erythrocyte maturation. The partial rescue by miR-451 replenishment in mnr indicates that miR-451 is sufficient to promote erythroid maturation in this sensitized genetic background. In all these assays, miR-451 makes a much more critical contribution to erythrocyte maturation than miR-144.
In zebrafish, gata2 transcripts are a target of miR-451
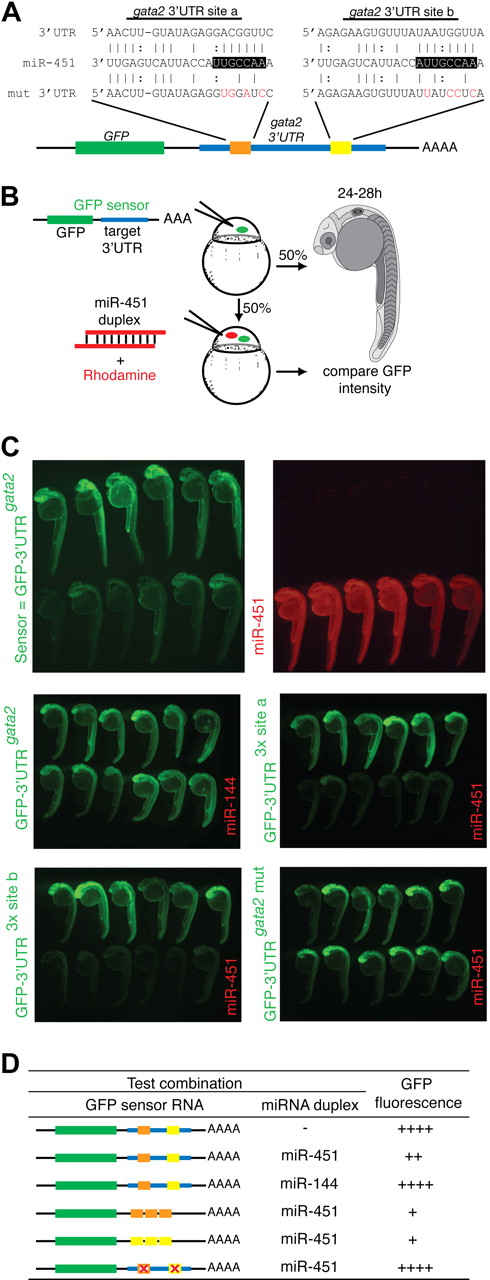
We used database searching to generate lists of potential mRNA targets of miR-451 that might mediate these biologic effects.33 miRNA target specificity is conferred by complementary base pairing of its “seed sequence” (nucleotides 2-7) to target mRNAs, and the inhibitory effect is amplified as the number of binding sites in an individual transcript increases.34 As with most miRNAs, miR-451 has numerous potential targets. However, mRNAs encoding transcription factors expressed in the ICM before or during miR-144/miR-451 expression, whose 3′UTR carried 2 or more predicted miRNA binding sites, seemed reasonable candidate targets. Using these criteria, gata2, known to preserve the immaturity of hematopoietic precursor cells,35 stood out among 631 predicted targets as an attractive potential target of miR-451 (Figure 4A).
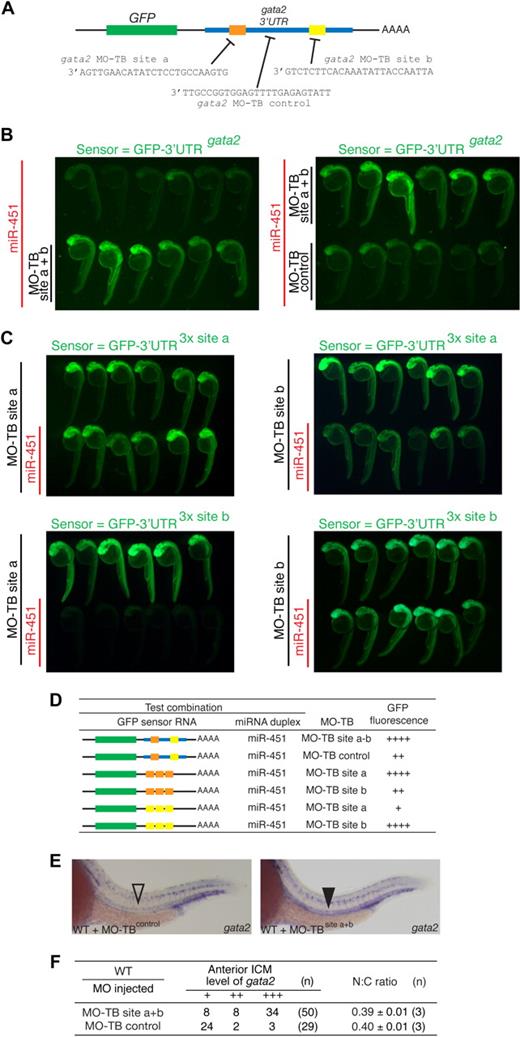
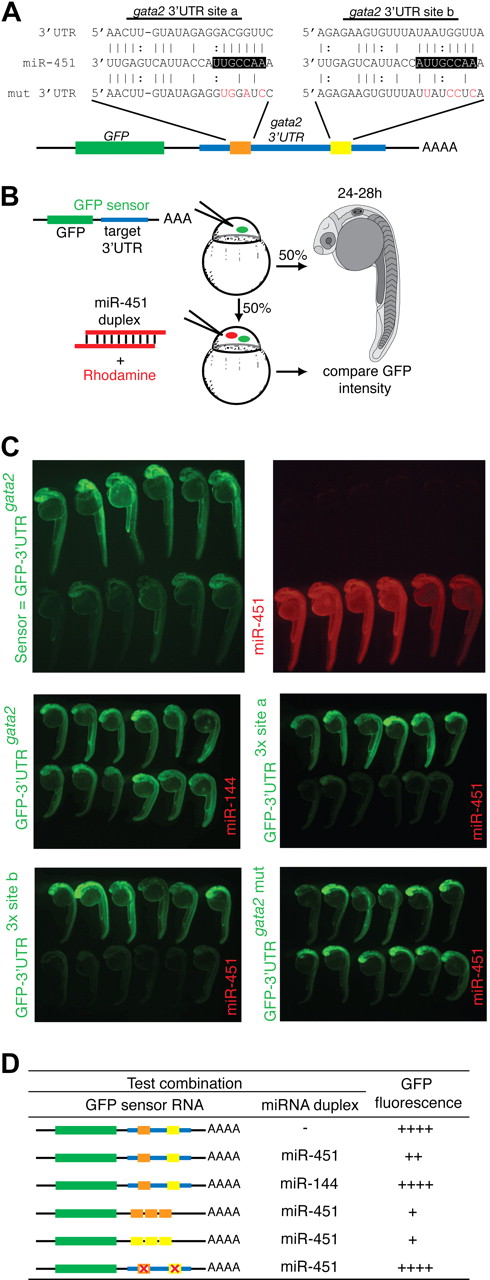
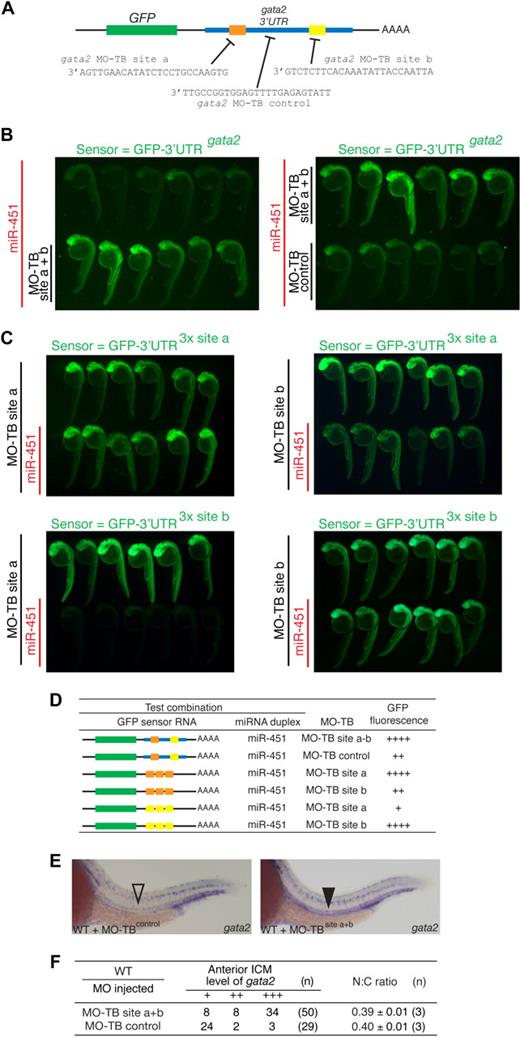
gata2 is a bona fide target of miR-451. (A) Schematic diagram of the zebrafish gata2 3′UTR (blue, nts 128-782), which contains 2 predicted miR-451 binding sites (site a, orange; site b, yellow). Predicted seed complementarity sequences are boxed. Red nucleotides in “mut 3′UTR” indicate those mutated to destroy seed sequence binding. (B) Schema of a reporter assay to evaluate the gata2-3′UTR for interaction with miR-451. Microinjection of 1-cell embryos with a series of sensor mRNAs encoding GFP fused to various test 3′UTRs (as tabulated in panel D) was followed by separate injection of 50% of embryos with miR-451 (or mir-144 or control) duplex, tracing miRNA delivery by rhodamine, and the impact on GFP fluorescence intensity assessed at 24 to 28 hpf. (C) miR-451 negatively regulates the sensor GFP-3′UTRgata2 mRNA. Top: compared with embryos receiving only sensor mRNA (top row in each panel), GFP-fluorescence brightness was reduced in those also injected with miR-451 (traced by rhodamine, red fluorescence in bottom row). Middle and bottom: results of embryos similarly arranged testing various GFP-gata2-3′UTR sensor RNA and miRNA combinations as labeled. Arrays of representative embryos were photographed together in a single image to ensure valid comparison of relative green fluorescence intensity between the 2 groups. (D) Summary of reporter assay outcomes using the series of GFP sensor RNA and miRNA combinations, using a comparative scale for GFP fluorescence (+ to ++++). The color-coding of the gata2-3′UTR variants in the sensor mRNA column refers to panel A; the red “X” indicates a mutated miR-451 site. Assays validating the bioactivity and specificity of the morpholino and duplex oligonucleotides used are presented in Figure S8.
gata2 is a bona fide target of miR-451. (A) Schematic diagram of the zebrafish gata2 3′UTR (blue, nts 128-782), which contains 2 predicted miR-451 binding sites (site a, orange; site b, yellow). Predicted seed complementarity sequences are boxed. Red nucleotides in “mut 3′UTR” indicate those mutated to destroy seed sequence binding. (B) Schema of a reporter assay to evaluate the gata2-3′UTR for interaction with miR-451. Microinjection of 1-cell embryos with a series of sensor mRNAs encoding GFP fused to various test 3′UTRs (as tabulated in panel D) was followed by separate injection of 50% of embryos with miR-451 (or mir-144 or control) duplex, tracing miRNA delivery by rhodamine, and the impact on GFP fluorescence intensity assessed at 24 to 28 hpf. (C) miR-451 negatively regulates the sensor GFP-3′UTRgata2 mRNA. Top: compared with embryos receiving only sensor mRNA (top row in each panel), GFP-fluorescence brightness was reduced in those also injected with miR-451 (traced by rhodamine, red fluorescence in bottom row). Middle and bottom: results of embryos similarly arranged testing various GFP-gata2-3′UTR sensor RNA and miRNA combinations as labeled. Arrays of representative embryos were photographed together in a single image to ensure valid comparison of relative green fluorescence intensity between the 2 groups. (D) Summary of reporter assay outcomes using the series of GFP sensor RNA and miRNA combinations, using a comparative scale for GFP fluorescence (+ to ++++). The color-coding of the gata2-3′UTR variants in the sensor mRNA column refers to panel A; the red “X” indicates a mutated miR-451 site. Assays validating the bioactivity and specificity of the morpholino and duplex oligonucleotides used are presented in Figure S8.
The validity of this prediction was tested using a reporter assay based on monitoring GFP fluorescence in embryos microinjected with mRNA encoding GFP fused to the gata2-3′UTR (GFP-3′UTRgata2), in the presence or absence of miRNA duplex (Figure 4B-D). In this assay, a decrease in GFP-fluorescence in the presence of duplex indicated miRNA-mediated repression. Embryos sequentially injected with GFP-3′UTRgata2 reporter and miR-451 showed suppressed GFP-fluorescence, whereas miR-144 had no effect on fluorescence intensity (Figure 4C). This confirmed that the gata2-3′UTR was targeted by miR-451 but not by miR-144. The 2 predicted miR-451 binding sites in the gata2-3′UTR were validated as bona fide target sequences by further modifications of this assay in which (1) embryos microinjected with mRNA encoding GFP fused to each individual recognition site in triplicate (GFP-3′UTR3x site a and GFP-3′UTR3x site b) displayed strong miR-451–induced suppression of GFP-fluorescence (Figure 4C); (2) miR-451–mediated repression on the gata2-3′UTR was abolished when both seed sequences were mutated (GFP-3′UTRgata2-mut), shown by comparable GFP-fluorescence of this reporter construct alone and in the presence of miR-451 (Figure 4C,D). Collectively, these data prove the capability of miR-451 to target the gata2-3′UTR via the 2 predicted recognition sequences.
gata2 expression is sustained in mnr and partly responsible for delayed erythrocyte maturation
We sought in vivo evidence in support of the hypothesis that miR-451 negatively regulates gata2, which predicts an inverse correlation between the presence of gata2 transcript and miR-451 activity.
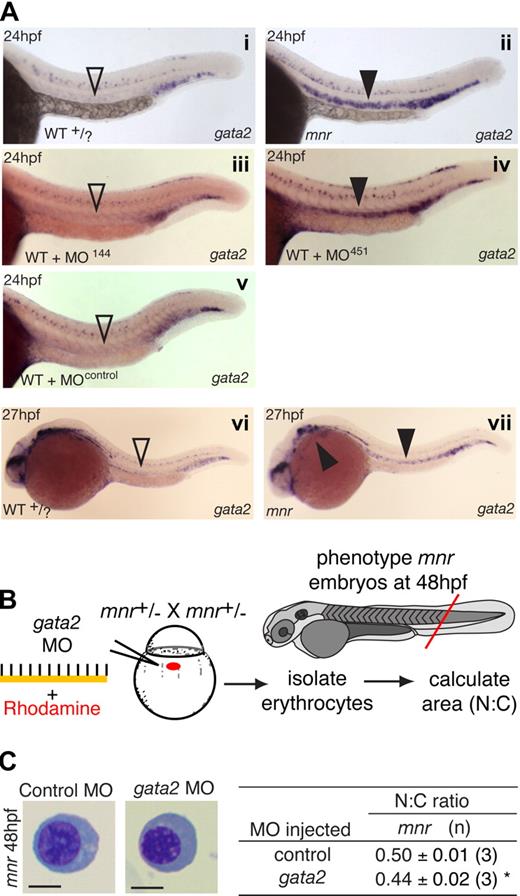
First, such an inverse correlation is observed in the wild-type expression dynamics of gata2 and miR-451. In zebrafish, gata2 caudal hematopoietic expression initiates in the lateral-plate mesoderm at 10 hpf.23,36 These cells give rise to the anterior ICM directly above the yolk extension and the posterior ICM37,38 distal to the yolk. After 19 hpf, gata2 expression decreases substantially in the anterior ICM but is maintained in the posterior ICM.23 The dynamics of this change in gata2 expression at 19 hpf coincides with the activation of the miR-144/451 locus in the anterior but not posterior ICM. By 24 hpf, gata2 expression is nearly completely lost in the strongly miR-451 expressing anterior ICM (comparing miR-451, Figure 1E with gata2, Figure 5A).
gata2 and miR-451 interact in vivo to regulate erythrocyte maturation. (A) gata2 expression (blue) in 24- and 27-hpf embryos by WISH arranged for side-by-side comparison. Left panels (i, iii, v): in wild-type (WT), MO-144- and MO-control-injected embryos, gata2 expression is waning in the anterior intermediate cell mass at 24 hpf (▿). Right panels (ii, iv); in miR-451-deficient mnr and MO-451-injected morphants, gata2 expression in the anterior intermediate cell mass persists at 24 hpf (▾). (vi,vii) To examine the duration of the persistence of gata2 expression, a time course of gata2 expression was examined by WISH at 27-, 30-, 36- and 48-hpf timepoints. At the onset of circulation (27 hpf), sustained gata2 expression (> WT) was still evident in mnr erythocytes in the anterior intermediate cell mass and over the yolk (vii, solid arrowheads). i and ii are unmanipulated age-matched siblings and representative of 19 of 19 and 11 of 12 PCR-genotype confirmed WT and mnr embryos, respectively. Subpanel i, ii, vi, and vii embryos are PCR-genotype confirmed. Subpanels iii through vii are representative of more than 40 age-matched embryos. See Figure 6E legend for further details about controls for comparing the level of gata2 expression. (B) Schematic diagram of an experiment testing if gata2 knockdown by a gata2 MO affects erythrocyte maturation. (C) Results of the experiment in panel B, showing that gata2 knockdown in mnr is sufficient to partially restore erythrocyte maturation (both morphologically and as measured by N:C area ratio). Data are mean plus or minus SE for n independent experiments. *P = .04, 2-tailed t test. Scale bar = 5 μm. Figure S11 further demonstrates the reproducibility of these data by presenting them as scatterplots and providing additional fields of representative cells.
gata2 and miR-451 interact in vivo to regulate erythrocyte maturation. (A) gata2 expression (blue) in 24- and 27-hpf embryos by WISH arranged for side-by-side comparison. Left panels (i, iii, v): in wild-type (WT), MO-144- and MO-control-injected embryos, gata2 expression is waning in the anterior intermediate cell mass at 24 hpf (▿). Right panels (ii, iv); in miR-451-deficient mnr and MO-451-injected morphants, gata2 expression in the anterior intermediate cell mass persists at 24 hpf (▾). (vi,vii) To examine the duration of the persistence of gata2 expression, a time course of gata2 expression was examined by WISH at 27-, 30-, 36- and 48-hpf timepoints. At the onset of circulation (27 hpf), sustained gata2 expression (> WT) was still evident in mnr erythocytes in the anterior intermediate cell mass and over the yolk (vii, solid arrowheads). i and ii are unmanipulated age-matched siblings and representative of 19 of 19 and 11 of 12 PCR-genotype confirmed WT and mnr embryos, respectively. Subpanel i, ii, vi, and vii embryos are PCR-genotype confirmed. Subpanels iii through vii are representative of more than 40 age-matched embryos. See Figure 6E legend for further details about controls for comparing the level of gata2 expression. (B) Schematic diagram of an experiment testing if gata2 knockdown by a gata2 MO affects erythrocyte maturation. (C) Results of the experiment in panel B, showing that gata2 knockdown in mnr is sufficient to partially restore erythrocyte maturation (both morphologically and as measured by N:C area ratio). Data are mean plus or minus SE for n independent experiments. *P = .04, 2-tailed t test. Scale bar = 5 μm. Figure S11 further demonstrates the reproducibility of these data by presenting them as scatterplots and providing additional fields of representative cells.
Second, if miR-451 were involved in the clearance of gata2 transcripts in the anterior ICM, then in a miR-451–deficient environment such as mnr or miR-451 morphant embryos, gata2 transcripts would persist beyond this transition point. Consistent with this hypothesis, although anterior ICM gata2 expression is normally nearly completely down-regulated in WT at 24 hpf, in genotyped mnr embryos it persisted to 27 hpf and in miR-451 morphants was still present at 24 hpf (Figure 5A). The negative controls (WT siblings, miR-144 morphants, and control-MO–injected embryos) all displayed comparable loss of anterior ICM gata2 expression at 24 hpf. Up until 48 hpf, erythrocyte gata2 was not reinstated in mnr, miR-451 morphants, WT, or miR-144 morphant controls (data not shown).
Third, we hypothesized that if mnr erythroid maturation is delayed because of persistent gata2 expression, then gata2 knockdown should rescue the maturation defect of mnr (Figure 5B). Strikingly, MO knockdown of gata2 translation21 accelerated erythroid maturation in mnr both morphologically and by a significant reduction in N:C ratio (Figure 5C; Figure S11).
Finally, we sought to determine whether gata2 was the sole miR-451 target responsible for the erythrocyte maturation delay of mnr. This was achieved by using the newly described target-blocking MOs,39 which we submitted to a rigorous efficacy and specificity evaluation. MOs complementary to each of the gata2-3′UTR miR-451 binding sites (sites a or b as in Figures 4A and 6A; designated MO-TB site a and MO-TB site b) were designed to selectively block endogenous miR-451 binding to the gata2-3′UTR, without interfering with the interaction of miR-451 with other target 3′UTRs. The efficacy of these MOs at protecting their respective miR-451 binding site was demonstrated in a set of appropriately designed GFP-fluorescence reporter assays (Figure 6B-D). As a baseline, embryos injected with GFP-3′UTRgata2 reporter and then miR-451 showed suppressed GFP-fluorescence as previously demonstrated (Figures 4C,6B). In contrast, in the presence of both target blocking MOs (MO-TB-sites a and b), GFP-fluorescence was not suppressed by the presence of miR-451, as expected from blocking the miR-451/gata2-3′UTR interaction (Figure 6B). This protection was not due simply to the MO-TBs binding to the gata2-3′UTR and thereby stabilizing the transcript, because the MO-TB control (designed to bind to the gata2-3′UTR between the miR-451 binding sites) did not block the miR-451 suppression of GFP-fluorescence. Further demonstration of the sequence specificity of the MO-TBs and of their inability to bind miR-451 duplexes themselves came from assays in which the GFP-3′UTR3x site a sensor mRNA was protected only by MO-TB site a but not MO-TB site b, and vice versa (Figure 6C). Together, these evaluations demonstrate that the 2 MO-TBs together block the miR-451/gata2-3′UTR interaction in a sequence-specific manner.
gata2 is not the only miR-451 target implicated in regulating erythrocyte maturation (A) Diagram of target blocking (TB) MO designed to protect the 2 gata2-3′UTR miR-451 binding sites a and b, and a control MO corresponding to sequences between sites a and b. (B,C) Results of a modification of the reporter assay outlined in Figure 4, for the purpose of validating the efficacy and specificity of the target blocking MOs. For each panel, the sensor RNA was as shown in green. Labels to left of each panel indicate rhodamine-traced miRNA (red labels) and the target blocking MO (MO-TB) delivered to the corresponding rows of embryos. Relative fluorescence intensity between the 2 rows of embryos in each panel assays for miRNA-mediated GFP-sensor-RNA repression. (B) Left panel: a combination of MO-TBs to sites a + b (bottom row of embryos) protect the GFP- gata2-3′UTR sensor RNA from miR-451-mediated repression (top row of embryos). Right panel: MO-TB control does not protect against miR-451-mediated repression. (C,D) Panels illustrating other experimental outcomes for different GFP sensor RNA, miRNA and MO-TB combinations as summarized in the Table (D), using a comparative scale for GFP fluorescence (+ to ++++). The color-coding of the gata2-3′UTR variants in the sensor mRNA column refers to panel A. (E,F) gata2 is not the only miR-451 target responsible for the erythrocyte immaturity of mnr. gata2 target-blocking MOs interfere with the gata2-3′UTR/miR-451 interaction in vivo, verified by persistence of gata2 transcripts in the anterior intermediate cell mass (ICM) of 24-hpf embryos (▾ compared with control ▿). Note that there are 3 controls for comparing the level of gata2 expression: (i) the parallel-processed embryos that received TB-MO-control; (ii,iii) 2 internal controls within each embryo provided by (ii) the comparison of expression in the anterior ICM versus the posterior ICM, and (iii) the comparison between ICM gata2 expression (which in the anterior ICM is exposed to endogenous miR-451 regulation), and the neuronal gata2 expression (which is not exposed to endogenous miR-451 expression). (F) Tabulates the anterior ICM level of gata2 expression by categories (+++, anterior ICM expression as strong as posterior ICM expression; ++, anterior ICM expression less than posterior ICM expression; +, anterior ICM expression absent or less than 30% of posterior ICM expression). n = number of embryos. Despite the effect on gata2 expression, gata2 target-blocking MOs did not affect erythrocyte maturation as assessed either morphologically or by the N:C area ratio. N:C ratio data are mean plus or minus SE for n independent experiments. Figure S12 further demonstrates the reproducibility of these data by presenting them as scatterplots and providing additional fields of representative cells.
gata2 is not the only miR-451 target implicated in regulating erythrocyte maturation (A) Diagram of target blocking (TB) MO designed to protect the 2 gata2-3′UTR miR-451 binding sites a and b, and a control MO corresponding to sequences between sites a and b. (B,C) Results of a modification of the reporter assay outlined in Figure 4, for the purpose of validating the efficacy and specificity of the target blocking MOs. For each panel, the sensor RNA was as shown in green. Labels to left of each panel indicate rhodamine-traced miRNA (red labels) and the target blocking MO (MO-TB) delivered to the corresponding rows of embryos. Relative fluorescence intensity between the 2 rows of embryos in each panel assays for miRNA-mediated GFP-sensor-RNA repression. (B) Left panel: a combination of MO-TBs to sites a + b (bottom row of embryos) protect the GFP- gata2-3′UTR sensor RNA from miR-451-mediated repression (top row of embryos). Right panel: MO-TB control does not protect against miR-451-mediated repression. (C,D) Panels illustrating other experimental outcomes for different GFP sensor RNA, miRNA and MO-TB combinations as summarized in the Table (D), using a comparative scale for GFP fluorescence (+ to ++++). The color-coding of the gata2-3′UTR variants in the sensor mRNA column refers to panel A. (E,F) gata2 is not the only miR-451 target responsible for the erythrocyte immaturity of mnr. gata2 target-blocking MOs interfere with the gata2-3′UTR/miR-451 interaction in vivo, verified by persistence of gata2 transcripts in the anterior intermediate cell mass (ICM) of 24-hpf embryos (▾ compared with control ▿). Note that there are 3 controls for comparing the level of gata2 expression: (i) the parallel-processed embryos that received TB-MO-control; (ii,iii) 2 internal controls within each embryo provided by (ii) the comparison of expression in the anterior ICM versus the posterior ICM, and (iii) the comparison between ICM gata2 expression (which in the anterior ICM is exposed to endogenous miR-451 regulation), and the neuronal gata2 expression (which is not exposed to endogenous miR-451 expression). (F) Tabulates the anterior ICM level of gata2 expression by categories (+++, anterior ICM expression as strong as posterior ICM expression; ++, anterior ICM expression less than posterior ICM expression; +, anterior ICM expression absent or less than 30% of posterior ICM expression). n = number of embryos. Despite the effect on gata2 expression, gata2 target-blocking MOs did not affect erythrocyte maturation as assessed either morphologically or by the N:C area ratio. N:C ratio data are mean plus or minus SE for n independent experiments. Figure S12 further demonstrates the reproducibility of these data by presenting them as scatterplots and providing additional fields of representative cells.
Wild-type embryos microinjected with MO-TB sites a and b together showed prolonged gata2 expression at 24hpf in the anterior ICM (ie, replicating the mnr gata2 phenotype; Figure 6E), providing evidence both for their functionality in vivo and supporting a direct interaction between endogenous miR-451 and gata2 in vivo. However, these embryos did not show altered erythrocyte morphology or N:C ratio compared with control embryos (Figure 6F; Figure S12). This suggests that in mnr and miR-451 morphant embryos, at least one other miR-451 target needs to be dysregulated to impair erythroid maturation.
Discussion
The specific biologic functions of individual miRNAs are now emerging through reverse genetic studies, revealing important roles in development, physiology and disease, including hematopoiesis.3,40 Several murine miRNA loci have recently been disrupted by gene targeting with resultant hematopoietic phenotypes (eg, mice lacking miR-155, a lymphoid-restricted miRNA, have defective immune responses41,42 ; overexpression of miR-155 in murine bone marrow cells drives a myeloproliferative syndrome43 ; mice lacking miR-223, a granulocyte-lineage-restricted miRNA, have granulocytosis and impaired granulocyte function44 ).
Zebrafish offer several advantages over mice for the study of miRNA function, particularly in developmental processes. In zebrafish, loss- and gain-of-function studies can be undertaken quickly using transient genetic approaches: overexpression by microinjecting a synthetic RNA duplex and miRNA knockdown by injecting an MO antagonist. Predicted miRNA/transcript interactions can be evaluated using fluorescent reporter assays. Interactions between endogenous miRNAs and their targets can be specifically interrupted by target-blocking MOs. Because these approaches are transient, the efficacy and specificity of reagents can be rigorously evaluated quickly and efficiently in vivo in a manner not possible in mice.
However, in wild-type backgrounds, miRNA knockdown/overexpression in zebrafish appears only occasionally to result in any phenotype,29,45 and this approach can be technically challenging, particularly to adequately control for spurious effects. A relevant mutant, particularly those that are not epistatic to the pathway of interest, offers advantages such as genetic stability over these transient approaches (eg, the utility of the miRNA biogenesis mutant dicer in elucidating the role of miR-430 in early development).22,34 However, biogenesis mutants such as this lack numerous miRNAs and have complex later phenotypes22,46 that complicate discerning the specific roles of later-expressed miRNAs. A mutant selectively deficient in a particular miRNA but retaining the cell type in which it is expressed would be particularly useful.
We exploited zebrafish reverse genetic techniques to examine the function of the erythroid-restricted miRNAs miR-144 and miR-451, showing that miR-451 accelerated the rate of erythrocyte maturation, an action mediated in part by repression of gata2. Our studies were aided by the mutant mnr, which dissociated the otherwise invariable link between the presence of erythrocytes and the presence of these 2 miRNAs. mnr provided an miR-144/451–deficient model that could independently corroborate observations from other loss-of-function studies and provided a genetically stable miR-144/451–deficient background “sensitized” for the study of the function of these miRNAs. Indeed, the subtle and somewhat variable effects of miR-451 MO knockdown on erythrocyte maturation were strongly corroborated by the stronger, more stable, immature erythrocyte phenotype of mnr, and the biologic effects of miR-451 overexpression were discernible only on the mnr background. mnr is not a general miRNA biogenesis mutant, nor is it a mutation of the miR-144/451 locus itself; exactly how the mnr gene product acts as a strong positive regulator of miR144/451 is unknown and will require the identification of the mnr mutation.
A recently published study demonstrated that miR-144/451 expression is directly controlled by GATA1, and this was interpreted as GATA1-dependence, based on a comprehensive analysis of the murine locus promoter, and on the loss of miR-144/451 expression in the zebrafish gata1 mutant vlad tepes.9 However, vlad tepes has markedly defective precirculation ICM erythropoiesis with abundant cell death and is “bloodless” at the onset of circulation32,47 ; hence, we interpret its loss of miR-144/451 expression to largely reflect the loss of the cells expressing these 2 miRNAs. In contrast, mnr expresses ICM gata1 normally and is erythrocyte-replete but miR-144/451–deficient, thus severing the epistatic relationship between gata1-dependent erythrocyte production and miR-144/451 expression. Furthermore, mnr indicates that Gata1 requires at least one other gene product, mnr, to activate the miR-144/451 promoter. The genetic location of mnr on chromosome (Chr) 14 precludes mnr from being either gata1 itself (Chr 11) or several other known or potential components of GATA1-containing transcriptional complexes30,48 : zfpm1/fog1 (Friend-of-gata-1), Chr 18; klf4 (Krüppel-like factor 4) Chr 2; and lmo2 (Lim domain only 2) Chr 18.
Our data are consistent with the previous interpretation of miR-451 MO knockdown experiments in zebrafish (ie, that miR-451 is not required for erythroid specification).9 However, in contrast to this previous study, we did not observe severe anemia in miR-451 morphant embryos at the onset of circulation or at 48 hpf. The late-onset anemia observed in this other study was interpreted as indicating a requirement for miR-451 in erythrocyte maturation or survival. The reasons for these different outcomes are unclear, particularly because the miR-451 MO sequences were identical, but may reflect subtle differences in MO dose, preparations, or other technical differences. However, our MO knockdown observations are independently corroborated by the erythroid phenotype of mnr, which does not show postcirculation erythroid failure, but only delayed erythroid maturation.
Maturation of a population of cells could result either from the progressive maturation of a synchronized cohort of cells, or alternately from the progressive addition of newly produced cells of increasing maturity. Cell survival studies demonstrated that the first cohort of ICM-derived erythrocytes was undiluted by newly produced erythrocytes until 4 dpf32 ; hence, we hypothesize that miR-451–dependent promotion of erythrocyte maturation up to 48 hpf is a cell autonomous effect driving the maturation of individual cells.
In mammalian hematopoiesis, persistent production of GATA2 maintains a stem cell phenotype and needs to be down-regulated to allow erythropoiesis to proceed normally.35 One known mechanism for down-regulating GATA2 is by GATA1 directly binding to and repressing its promoter.49 An important outcome of our studies was the identification of a mechanism for miR-451 promotion of erythroid maturation in zebrafish: the clearance of gata2 transcripts. To prove that the prolonged expression of gata2 in mnr and miR-451-morphant erythrocytes was not merely secondary to erythroid immaturity, but directly due to miR-451 loss, we evaluated the gata2-3′UTR/miR-451 interaction outside the context of erythrocyte maturation in a direct in vivo reporter assay. This confirmed that a functional interaction occurred. We also evaluated the cause/effect biologic consequences of perturbing this interaction: rerepressing gata2 in miR-451–deficient mnr with MO-gata2 and relieving gata2 alone of miR-repression in WT with the target-blocking MOs.
Repression of GATA2 by this miRNA locus may be conserved in mammals, although it may be achieved through miR-144 rather than miR-451, because human, murine, and rat GATA2-3′UTRs all contain several predicted miR-144 binding sites but no predicted miR-451 sites (Table S2). The in vivo GFP reporter assay provides a method for evaluating the functional validity of these potential miRNA/transcript interactions.
Gata1 displaces Gata2 from the murine miR-144/451 promoter to activate promoter function.9 Hence, in zebrafish, at least, there is scope for a 2-pronged Gata1-driven regulatory mechanism for relaxing Gata2 repression of erythroid maturation; Gata1 turns off further gata2 transcription and activates transcription of a miRNA locus that down-regulates extant gata2 transcripts.
We interpret the failure of MOs exclusively interfering with the miR-451/gata2 interaction to repress erythrocyte maturation to indicate that miR-451 targets other than gata2 are also important in erythrocyte maturation. This indicates that there is redundancy and robustness in miR-451–driven erythroid maturation. Reporter assays and mnr-based functional assays provide a basis for evaluating other candidate target transcripts carrying putative miR-451 binding sites for their role in erythrocyte maturation. An alternate hypothesis is that the target blocking MOs alter the dose of Gata2 but not as much as in mnr or miR-451 morphants.
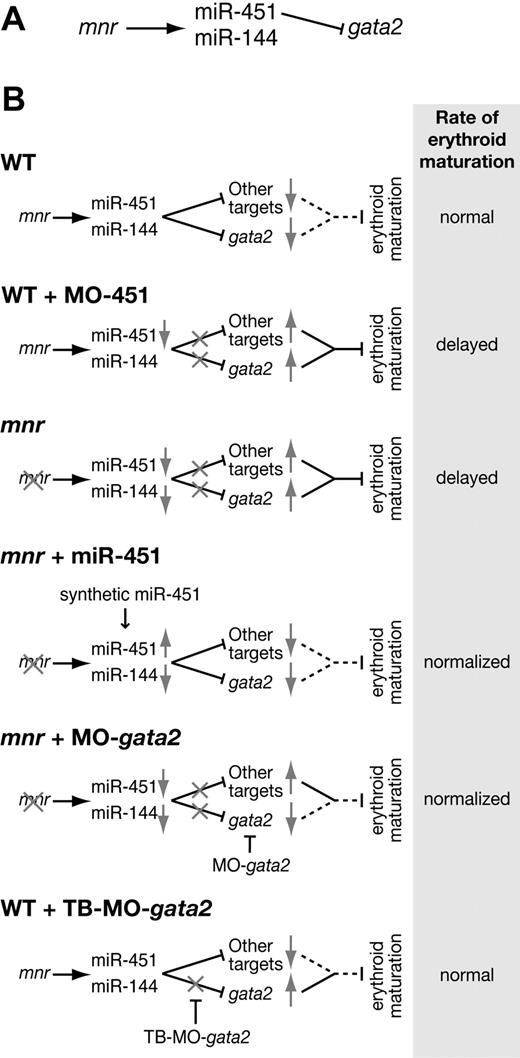
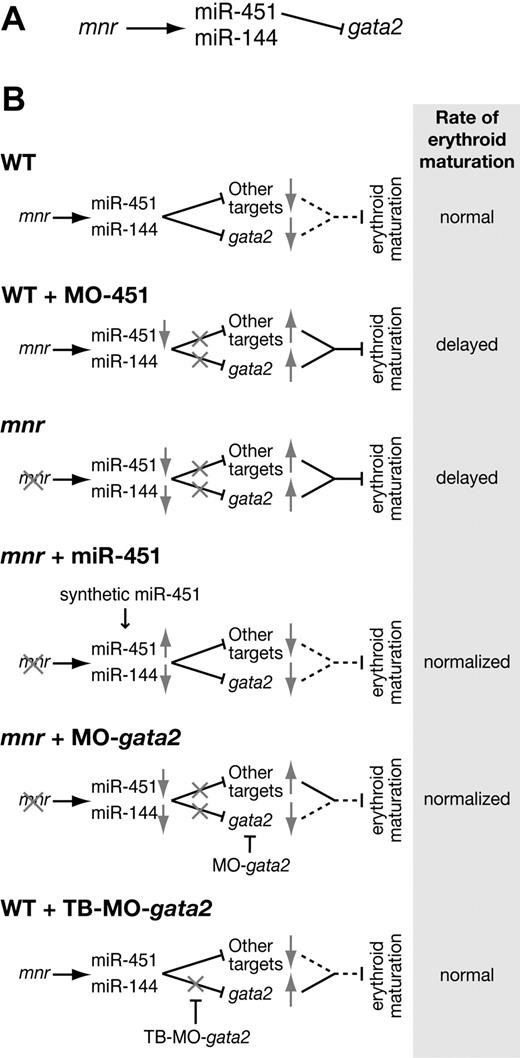
Together, our results assemble a new genetic pathway that regulates the pace of erythroid maturation: mnr activating miR-451, which in turn represses gata2 (Figure 7A). This genetic pathway informs a biologic model (Figure 7B) consistent with our experimental observations. In general, once committed to an erythroid fate, cells require mnr to use miR-451 to down-regulate repressors of erythroid maturation, gata2 being one of them. miR-451 deficiency (either MO-451 morphants or mnr mutants) retards the pace of erythroid maturation by permitting gata2 and other target transcripts to persist longer than usual. In mnr, normalizing levels either of miR-451 alone or of its target gata2 (by restoring miR-451 levels or by gata2 knockdown) is sufficient to partially rescue the rate of red cell maturation. However, because gata2 target-blocking MOs are insufficient to retard erythrocyte maturation in wild-type embryos, there must be other miR-451 targets that need to be codysregulated to result in the erythrocyte immaturity of mnr or miR-451 morphants. Finally, these experiments exemplify an hypothesized miRNA role in sharpening developmental transitions,34,50 in this case from a hematopoietic progenitor cell into a maturing erythrocyte.
miR-144 and miR-451 in zebrafish erythroid development. (A) Genetic pathway connecting the mutation meunier (mnr), the microRNAs miR-144 and miR-451, and the miR-451 target gata2. (B) Models of the biologic impact of this genetic pathway on erythroid maturation, in wild-type, and in various perturbed scenarios. Gray arrows indicate reduced (↓) or increased (↑) net level of activity at steps of the genetic pathway. Gray crosses (X) indicate the level of block in the pathway in the individual scenarios. For the net biologic outcome of the genetic pathway on repression of erythroid maturation, a dotted line indicates a normal (ie, wild-type) level of repression, and a continuous line indicates a stronger-than-normal repressive effect. TB-MO indicates target-blocking morpholino oligonucleotide.
miR-144 and miR-451 in zebrafish erythroid development. (A) Genetic pathway connecting the mutation meunier (mnr), the microRNAs miR-144 and miR-451, and the miR-451 target gata2. (B) Models of the biologic impact of this genetic pathway on erythroid maturation, in wild-type, and in various perturbed scenarios. Gray arrows indicate reduced (↓) or increased (↑) net level of activity at steps of the genetic pathway. Gray crosses (X) indicate the level of block in the pathway in the individual scenarios. For the net biologic outcome of the genetic pathway on repression of erythroid maturation, a dotted line indicates a normal (ie, wild-type) level of repression, and a continuous line indicates a stronger-than-normal repressive effect. TB-MO indicates target-blocking morpholino oligonucleotide.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the following for helpful discussions, encouragement, support, and/or technical help: Warren Alexander, Tony Burgess, Stephen Cody, David Curtis, John Hayman, Joan Heath, Ben Hogan, Stephen Jane, Luke Kapitany, Anne Lagendijk, Nick Nicola, Sony Varma, and colleagues in the Colon Molecular and Cell Biology Group at the Ludwig Institute and the Bone Marrow Research Laboratory (Royal Melbourne Hospital). We particularly thank Belinda Phipson and Gordon Smyth for statistical advice and Stephen Renshaw for supplying the Tg(mpx:EGFP) line. We thank several anonymous reviewers for constructive criticism and suggestions that improved the manuscript.
This work was supported in part by grants from the National Institutes of Health (Bethesda, MD; R01-HL079545) and National Health and Medical Research Council (NHMRC 461222 and 461208) to G.L. L.P. was supported by an Australian Postgraduate Award and CSIRO enhancement stipend and received travel support from the ARC/NHMRC Network in Genes and Environment in Development (NGED).
National Institutes of Health
Authorship
Contribution: L.P. designed, conducted, and interpreted the experiments and wrote the manuscript; J.E.L. generated the mutant meunier and initiated its mapping; W.P.K. performed miR-451 and miR-206 in situ hybridizations; D.C. performed some in situ hybridizations and made early contributions to mnr mapping and erythrocyte morphometrics; P.M.W. helped plan the studies and cosupervised L.P.; and G.J.L. designed and interpreted the experiments, supervised L.P., and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Graham J. Lieschke, Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, VIC, Australia 3050; e-mail: lieschke@wehi.edu.au.

![Figure 1. Expression of the miR-144/451 locus in wild-type embryonic and adult zebrafish erythroid cells. (A) miR144/451 locus expression (22 hours postfertilization [hpf]) by RT-PCR using primers confirming that both miRNAs originate from a single precursor transcript. Schema shows primer binding sites in green. RT indicates reverse transcriptase; Gen, genomic DNA control. (B) Whole mount in situ hybridization (WISH) analysis of miR-144 at the 3 embryonic ages shown. miR-144 expression (blue) initiates at 18 hpf in the hematopoietic intermediate cell mass (ICM), becomes confluent across all cells of the ICM (20 hpf), and disperses with erythrocytes at the onset of circulation, pooling over the yolk during fixation for WISH (30 hpf). Lateral views, anterior to left; inset (bottom panel) is ventral view. (C) Sagittal section of 24-hpf WISH embryos showing miR-144 expression (blue) in cytoplasm of erythrocytes of the axial ICM. NC indicates notochord, Y, yolk. Scale bar = 20 μm. (D) Double WISH showing concordant miR-144 (blue) and hbbe3 (red) expression. Left panel, bright field; right panel, fluorescence microscopy highlighting hbbe3 expression. (E) miR-451 and gata1 expression (blue) in WT and clo embryos at 24 hpf, showing concordant loss of gata1 and miR-451 expression in clo. (F) miR-144 expression (blue) in hematopoietic cells nestled between renal tubules (RT) of adult kidney (i, white arrowheads) and the red pulp of the spleen (ii, white arrowheads) in nuclear fast red-counterstained sections cut from tissue fragment in situ hybridization. Detail (iii) shows miR-144 expression localized to the cytoplasm of circulating adult erythrocytes. Scale bar = 10 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/8/10.1182_blood-2008-05-155812/5/m_zh80010928650001.jpeg?Expires=1765907903&Signature=1n7xBRUGsJRPtYGlLZXbTMiGolgZBJY6SWEaNNF-npXu1uoOrpq9iL2scovv~~3RHDOKTlKHYLJjPaBNt2o1ALAZwDWsl8LNgsydhFXT6TGVuKovPrZdxjZjPoNmhDsq6dLZqtxLgUMu4HyQ4IIDAzZJoGiQpYUdRFttJ-t2sbcdDgtXPVqBX6HArhmC0MsoTLSHtzt6JT53yfwT3Mcqddh4jfnIhPwrNGlDYcQAu7J0T3M1f1ftZZnhM1Qr~Y9IGUmOZPZ-THbPvOsZsFDEIhPHVlmyvA2cPMmr5LJW3KDT8GaSEErkB9WYT1pKtkyQ5b3ncSuTWwfWdpIKqk1cmQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)