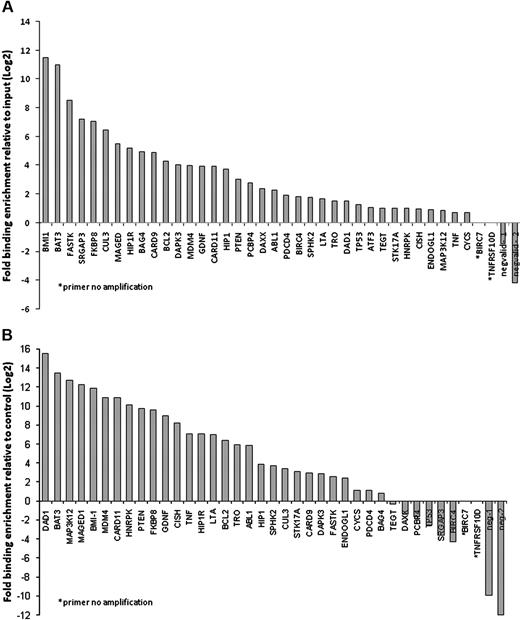
On page 808 in the August 1, 2008, issue, there are errors in Figure 3A and related text. The quantitative Q-RT-PCR results for SALL4 ChIP enrichment for 38 apoptosis gene regions erroneously listed the first 26 genes in alphabetical order instead of the actual order of their relative gene expression.
ChIP-Q-RT-PCR of apoptosis genes associated with SALL4 binding. Thirty-eight apoptosis genes identified by ChIP-chip were subjected to ChIP-Q-RT-PCR to confirm SALL4 binding by an alternative method. These genes are candidates for SALL4 involvement in leukemogenesis. (A) In NB4 cells, 36 of the 38 genes identified by ChIP-chip were confirmed as bound by SALL4 using ChIP-PCR. (B) Identification of SALL4 binding to apoptosis genes in CD34+ cells sorted from human bone marrow samples. CD34+ cells are likely progenitor cells to the promyelocytic cell line NB4 within these lineages. Identified SALL4 target genes had more than 2-fold enrichment compared with the input control. The position of negative control primers is presented in Figure S1. In Figure 3B, primer neg-2 failed to amplify the experimental DNA but did amplify the input DNA, indicating the absence of DNA template in the experimental group. This is represented by a fold change of less than 0.001.
ChIP-Q-RT-PCR of apoptosis genes associated with SALL4 binding. Thirty-eight apoptosis genes identified by ChIP-chip were subjected to ChIP-Q-RT-PCR to confirm SALL4 binding by an alternative method. These genes are candidates for SALL4 involvement in leukemogenesis. (A) In NB4 cells, 36 of the 38 genes identified by ChIP-chip were confirmed as bound by SALL4 using ChIP-PCR. (B) Identification of SALL4 binding to apoptosis genes in CD34+ cells sorted from human bone marrow samples. CD34+ cells are likely progenitor cells to the promyelocytic cell line NB4 within these lineages. Identified SALL4 target genes had more than 2-fold enrichment compared with the input control. The position of negative control primers is presented in Figure S1. In Figure 3B, primer neg-2 failed to amplify the experimental DNA but did amplify the input DNA, indicating the absence of DNA template in the experimental group. This is represented by a fold change of less than 0.001.
In “Results,” in the seventh sentence of the first paragraph under “SALL4 directly regulates expression of apoptosis genes,” reference to “35 of 38 apoptosis genes” is incorrect. The sentence should read: The resulting data established that 36 of 38 apoptosis genes were indeed targets of SALL4 in NB4 cells (Figure 3A).
In the ninth sentence of the same paragraph, reference to “28 of 37 apoptosis genes” is in error. The sentence should read: Using this cell population, we demonstrate that 29 of 37 apoptosis genes are indeed targets of SALL4 in the CD34+ fraction (Figure 3B).
In the third sentence of the Figure 3 legend, instead of “35 of the 38 genes,” the sentence should read: (A) In NB4 cells, 36 of the 38 genes identified by ChIP-chip were confirmed as bound by SALL4 using ChIP-PCR.
In the penultimate sentence of the Figure 3 legend, reference to Figure 2B is incorrect. The sentence should read: In Figure 3B, primer neg-2 failed to amplify the experimental DNA but did amplify the input DNA, indicating the absence of DNA template in the experimental group.
Figure 3, showing the correct gene expression levels, and the corrected figure legend are shown below.