Abstract
Abstract 4424
Recent studies have demonstrated that sequential administration of demethylating or immunomudulator agents have clinical efficacy in patients with myelodysplastic syndromes (MDS). Demethylating agents induce an optimal re-expression of epigenetically silenced tumor suppressor genes. However, the global DNA demethylation observed in malignant cells during treatment doesn't guarantee a better prognosis, suggesting the presence of others important unknown factors. On the other hand, global DNA hypomethylation of CD4+ T-cells have been related with autoimmune pathology diseases like systemic lupus erythematosus (SLE). The aim of the present study is to establish the degree of the global DNA methylation in CD4+ T-cells in MDS patients and their potential dysfunction.
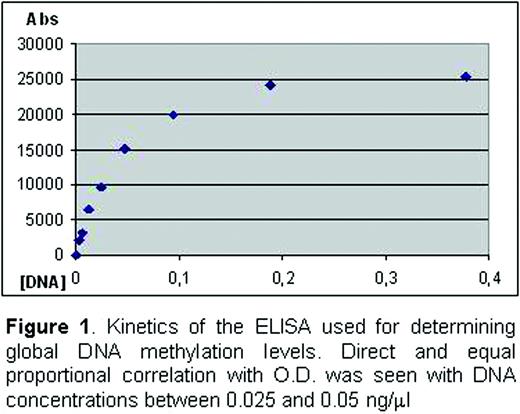
Eight MDS patients with low-risk, according to IPSS (between 0 and 1), diagnosed by cytology, cytogenetics and immunophenotype, and 4 healthy donors have been studied. Peripheral blood mononuclear cells were obtained by ficoll density gradient. Negative CD4+ selection followed by a positive selection were performed using the MACS system (Miltenyi). In patients with CD34+ cells expressing CD4+ antigen, lymphocyte isolation was done using a FACSAria sorter (BD Biosciences). Purity of CD4+ T-cells was higher than 90% in all cases. Cell DNA extraction was carried out with the QIAamp DNA blood mini kit (Qiagen). DNA concentration and 260:280 absorbance ratios were calculated with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Global DNA methylation content was measured by means of an ELISA using an anti-5-mC mAb (Calbiochem) 1/400 diluted. Before proceeding with patient samples, an optimal DNA concentration to evaluate global methylation levels was established. With this purpose, a reference DNA sample of purified CD4+ T cells (374 ng/ml) was twofold serially diluted in TE (starting at 1:1000 dilution) to observe the kinetics of our ELISA. The best DNA concentration range was 0.025–0.05 ng/ml (Figure 1). Methylation indices (MI) were calculated by getting the ratio between optical density (OD) and DNA concentration for each sample. To minimize experimental variability between plates a reference sample was included in each run. Control and patient MI were corrected by establishing the ratio with the reference MI.
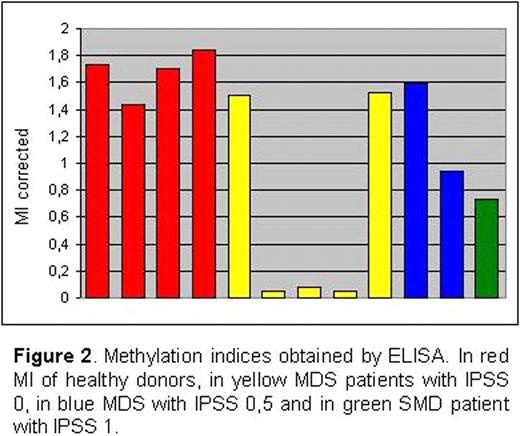
The global DNA methylation indices obtained by ELISA (Figure 2) displayed that most of MDS patients studied presented a global DNA hypomethylation in CD4+ T-cells, and 3 with IPSS 0 showed and important decrease.
These preliminary results showed that there are low-risk MDS patients with hypomethylation in CD4+ T-cells. This observation may suggest an autoimmune component in these malignancies as the one described in SLE. To address this hypothesis we are increasing the number of patients studied to all IPSS categories, to test if this phenomenon is highly represented in MDS. In the other hand, we are studying the presence of autoimmune-related factors, like the expression of integrins adhesive receptors such as LFA-1, in patients showed CD4 T-cells hypomethylation.
Guerra-Moreno:Celgene: Research Funding. Vallespi:Celgene: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.