Abstract
Abstract 677
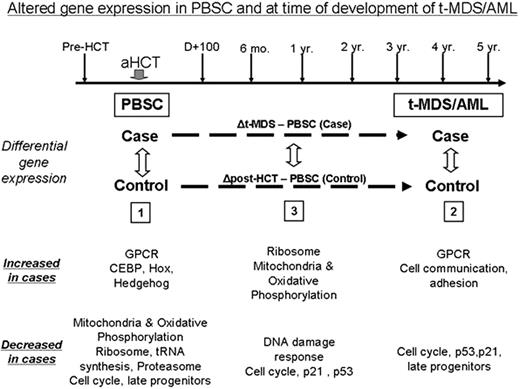
t-MDS/AML is a lethal complication of aHCT for HL and NHL. The pathogenesis of t-MDS/AML is unclear. We are addressing this gap by conducting a prospective study of patients undergoing aHCT for HL/ NHL at City of Hope, with serial collection of biospecimens from pre-aHCT to 5 years after aHCT (Figure). In this report we describe alterations in gene expression (analyzed using Affymetrix HG-U133 plus 2.0 arrays) in CD34+ hematopoietic stem and progenitor cells (HSPC) associated with development of t-MDS/AML. Patients who developed t-MDS/AML after aHCT (”cases”) were compared with patients who did not develop t-MDS/AML after aHCT (“controls”: matched for primary diagnosis, age, race/ethnicity, and time since aHCT). CD34+ cells were selected from specimens obtained at the following time points: 1) pre-aHCT: peripheral blood stem cell (PBSC) product; and 2) at time of t-MDS/AML for cases and comparable time for controls: bone marrow (BM). Conditional logistic model was used to identify differences in gene expression between cases and controls in: 1) PBSC samples (18 patients who subsequently developed t-MDS/AML [cases]; 37 who did not [controls]); 2) BM samples at time of t-MDS/AML (12 cases; 21 controls); and 3) changes in gene expression from PBSC to t-MDS/AML (Figure). Gene Set Enrichment Analysis (GSEA) showed that PBSC from patients that later developed t-MDS/AML showed significant downregulation of gene sets related to mitochondria and oxidative phosphorylation, ribosomes, tRNA synthesis, proteasome, late progenitors and cell cycle (FDR<10-5for each) and upregulation of genes related to G-protein coupled receptors (GPCR, FDR=0.003) and hematopoietic regulation (CEBP, HOX, Hedgehog, FDR=0.003, 0.04, 0.06 respectively). These pathway alterations were confirmed by Ingenuity and Gene Ontology analysis. BM CD34+ cells obtained at t-MDS/AML showed downregulation of gene sets for cell cycle, p21 and p53 signaling and late progenitors (FDR<10-5 for each) and upregulation of gene sets for GPCR and cell communication/adhesion (FDR<10-5). Analysis of changes in gene expression from PBSC to t-MDS/AML revealed that compared to the controls, the cases demonstrated increased expression of mitochondrial and ribosomal gene sets and reduced expression of DNA damage response and cell cycle regulation gene sets. These results show that specific abnormalities in gene expression associated with t-MDS/AML are present in HSPC long before the development of clinical disease, and that other gene expression abnormalities occur later in the course of disease development. Indeed 22 of the top 50 upregulated and 20 of the top 50 downregulated gene sets at t-MDS/AML were represented within the top 50 up- and down-regulated sets prior to aHCT in the PBSC sample. A gene set comprised of genes differentially expressed between cases and controls at t-MDS/AML was significantly enriched amongst genes differentially expressed in PBSC (NES= -1.74, P = 0.003, FDR=0.024), further demonstrating that gene expression abnormalities associated with t-MDS/AML were present at the time of PBSC collection. To explore the hypothesis that reduced mitochondrial oxidative phosphorylation may lead to enhanced generation of reactive oxygen species (ROS) and oxidative damage, we measured ROS levels in PBSC in untreated cells and after VP16 exposure. ROS level were increased at baseline in PBSC CD34+ cells from t-MDS/AML cases compared to controls (P=0.003) and ROS elimination after VP16 exposure was reduced (P=0.05, 2 hours after VP-16 treatment). The observed gene expression changes support a model for t-MDS/AML development in which 1) reduced mitochondrial oxidative phosphorylation leads to increased ROS levels and increased damage to HSPC following therapeutic exposures, and 2) subsequent loss of DNA damage response and cell cycle regulatory mechanisms in damaged HSPC leads to the emergence of t-MDS/AML.
Disclosures:
No relevant conflicts of interest to declare.
Author notes
*
Asterisk with author names denotes non-ASH members.
© 2009 by The American Society of Hematology
2009