Abstract
In patients with myelodysplastic syndromes (MDS), apoptosis in hematopoietic cells is up-regulated in low-grade disease, whereas advanced disease is characterized by apoptosis resistance. We have shown that marrow stroma–derived signals convey sensitivity to tumor-necrosis-factor alpha (TNF-α)–mediated apoptosis in otherwise-resistant KG1a myeloid cells and CD34+ cells from MDS marrow. Here, we used a PhosphoScan proteomic liquid chromatography–mass spectrometry method to identify signals relevant for this effect. The transcription factor DJ-1/PARK-7 (DJ-1) was highly phosphorylated in KG1a cells cultured without stroma but dephosphorylated after stroma coculture, whereas expression of p53 increased significantly, suggesting a stroma contact-dependent effect of DJ-1 on p53. In CD34+ marrow cells from advanced MDS, expression of DJ-1 was up-regulated, whereas p53 levels were low, resulting in significantly greater DJ-1/p53 ratios than in patients with low-grade MDS (P = .01). DJ-1 levels were correlated with increasing International Prognostic Scoring System scores (P = .006). Increasing DJ-1/p53 ratios were associated with an increased risk of mortality, although the correlation did not reach statistical significance (P = .18). These data suggest that DJ-1/p53 interactions contribute to apoptosis resistance in clonal myeloid cells and may serve as a prognostic marker in patients with MDS.
Introduction
The myelodysplastic syndromes (MDS) comprise a spectrum of clonal disorders of hematopoiesis, with hematopoietic precursors demonstrating high rates of programmed cell death (apoptosis) in early stages/low-grade disease but resistance to apoptosis in advanced disease.1,2 We have shown that apoptosis resistance is associated with up-regulation of nuclear factorκB and increased expression of the antiapoptotic adaptor molecule FLICE inhibitory protein.2 It is not clear, however, whether additional factors are involved, and there is growing evidence that not only cell-autonomous events but also extrinsic signals contribute. The bone marrow microenvironment has been shown to provide essential signals for the fate of normal hematopoiesis and for leukemic cells,3 and contact with stroma is thought to convey antiapoptotic signals to (clonal) leukemia cells.4-6 Several cytokines, including tumor necrosis factor alpha (TNF-α) and interleukin 32, are abnormally expressed in marrow stroma from patients with MDS,7 and preliminary data show aberrant methylation patterns of growth regulator and transcription factor gene promoters in MDS stroma.8
We observed that coculture with the human marrow stroma cell line HS5 unexpectedly rendered the apoptosis-resistant myeloid cell line KG1a sensitive to TNF-α–induced apoptosis. Sensitization was associated with up-regulation of PYCARD and p53 in KG1a cells.6 To further dissect the relevant signals leading to apoptosis, we chose to use a PhosphoScan Proteomics liquid chromatography–mass spectrometry (LC-MS) method to identify peptides (and consequently proteins) that were phosphorylated or dephosphorylated in myeloid cell lines, in response to stroma contact, hypothesizing that differences in phosphorylation with and without stroma contact identified proteins that were relevant for apoptosis. Those pathways would then be examined in primary cells from patients with MDS. In the absence of stroma, proteins associated with cell survival (eg, DJ-1/PARK-7, LGALS1, RPL38) were found to be phosphorylated. Conversely, with stroma contact, proapoptotic peptides/proteins showed prominent phosphorylation (eg, MCM4, EEF1A2, and PLCG1). We focused further studies on DJ-1/PARK-7 (DJ-1), a transcription factor with several functions that has been shown to interact with p539 and that was dephosphorylated in the presence of stroma, suggesting a loss of activity in the stroma coculture system.
Methods
Cell lines and primary cells
HS5 is an immortalized human marrow stroma cell line derived from the marrow aspirate of a healthy volunteer. Cells were immortalized by transduction with human papilloma virus E6/E7 constructs and characterized extensively.10-12 HS5 cells are a rich source of cytokines and support the growth of committed hematopoietic progenitor cells. The human myeloid leukemia cell lines, KG1a and the parent cell line KG1, were obtained from Dr D. Banker (Fred Hutchinson Cancer Research Center [FHCRC]). KG1a is a nondifferentiating myeloid subclone of the KG1 cell line, which was derived from a 59-year-old man with erythroleukemia who demonstrated megaloblastic and dysplastic morphology. We have shown previously that KG1a cells are apoptosis resistant to chemotherapeutic agents and proapoptotic ligands and express high levels of the antiapoptotic molecule termed FLICE inhibitory protein.13 However, the parent cell line KG1 is apoptosis sensitive.14 All cell lines were maintained in complete medium (RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum, 1% glutamine, and 1% sodium pyruvate) and propagated at 37°C in a humidified 5% CO2/air atmosphere.
Primary hematopoietic cells were derived from marrow aspirates from healthy volunteers and from patients with MDS, obtained at or near the initial examination date as part of the intake evaluation. All patients and healthy donors had given informed consent as required by the Institutional Review Board of FHCRC. Bone marrow mononuclear cells were isolated by Ficoll-Hypaque density gradient centrifugation. CD34+ cells were separated by magnetic-activated cell sorting (MACS) according to the manufacturer's protocol (Miltenyi Biotec).15,16 The purity of the cell preparation was verified by staining with fluorescently tagged antibody for CD34 expression and quantified by flow cytometric analysis on an LSR2 (BD Biosciences). RNA was extracted from CD34+ cells, and cDNA was generated for reverse-transcriptase polymerase chain reaction (RT-PCR) analysis.
Patient follow-up and data collection
Clinical follow-up of patients was approved by the FHCRC Institutional Review Board. Patient characteristics, treatment regimens, and clinical outcome data were collected prospectively and stored in the FHCRC database. We performed a retrospective analysis of results in 78 consecutive patients with a diagnosis of MDS. MDS cases were identified by established criteria and included the subtypes refractory anemia, refractory anemia with ringed sideroblasts, and refractory cytopenia with multilineage dysplasia (together referred to as early/low-grade MDS) and refractory anemia with excess blasts (RAEB) and refractory anemia with excess blasts in transformation (RAEB-t; referred to as late/advanced MDS). Of the 78 patients, 58 were previously untreated, and among these 13 subsequently underwent hematopoietic cell transplantation. Patient age ranged from 33 to 79 years; 30 were women and 28 were men.
Reagents
TNF-α (human recombinant) was purchased from PeproTech Inc; daunorubicin, etoposide, and pifithrin-α (PFT-α) were obtained from Sigma-Aldrich. All reagents were prepared as 1000× stocks in appropriate vehicles and diluted into cell culture medium to the desired working concentration.
Coculture model
Cocultures of KG1 and KG1a cells with HS5 stroma were carried out as described.6 In brief, KG1a cells (∼ 108) were cultured without HS5 stroma or in cocultures in contact with stroma (∼ 5 × 107 cells) in 100-mm plates in complete medium (RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum, 1% glutamine, and 1% sodium pyruvate). KG1a cells from cocultures, with or without additional manipulations, were harvested at 4 sequential time points (over a time course of 0.5 to 18 hours) and separated from CD45 negative stroma cells by MACS (Miltenyi Biotec).
Cell lysates
Cell lysates were prepared from KG1 and KG1a cells either cocultured with stroma or propagated in the absence of stroma. Sonicated cell lysates were cleared by centrifugation, and proteins were reduced with 0.5M dithiothreitol, stabilized, at a final concentration of 4.1mM and alkylated with iodoacetamide at 8.3mM. For digestion with trypsin, protein extracts were diluted in 20mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), pH 8.0, to a final concentration of 2M urea, and trypsin (Pierce Biotechnology Inc) was added at a ratio of 1 to 2.5 mL of beads (200 TAME units trypsin/mL) per 109 cells. Digestion was performed for 24 hours at 20 to 25°C.
Reversed-phase solid-phase extraction of digests
Trifluoroacetic acid (TFA) was added to protein digests for a final concentration of 1%, the precipitate was sedimented by centrifugation at 2000g for 5 minutes, loaded onto Sep-Pak C18 columns (Waters), and equilibrated with 0.1% TFA. A column volume of 0.7 to 1.0 mL was used per 2 × 108 cells. Columns were washed with 15 volumes of 0.1% TFA, followed by 4 volumes of 5% acetonitrile in 0.1% TFA. Peptide fraction I was obtained by eluting columns with 2 volumes each of 8%, 12%, and 15% acetonitrile in 0.1% TFA, and the eluates were combined. Fractions II and III were a combination of eluates after eluting columns with 18%, 22%, 25% acetonitrile in 0.1% TFA and with 30%, 35%, and 40% acetonitrile in 0.1% TFA, respectively. All peptide fractions were lyophilized.
Immunoaffinity purification of phosphopeptides
Peptides from each fraction, corresponding to 2 × 108 cells, were dissolved in 1 mL of inhibitor of apoptosis protein buffer (20mM Tris/HCl, pH 7.2; 10mM sodium phosphate; 50mM NaCl), and insoluble matter was removed by centrifugation. Immunoaffinity purification (IP) was performed on each peptide fraction separately. The phosphotyrosine monoclonal antibody P-Tyr-100 (Cell Signaling Technology Inc) was coupled noncovalently to protein G agarose (Roche) at 4 mg/mL beads by incubation overnight at 4°C with gentle shaking. After coupling, antibody resin was washed twice with phosphate-buffered saline and 3 times with IP buffer (5-10 bead volumes of buffer for each wash). Efficient coupling was verified by boiling an aliquot of antibody resin in sodium dodecyl sulfate–polyacrylamide gel electrophoresis sample buffer for 5 minutes and evaluating the yield of released antibody against a standard of purified antibody by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, followed by staining with Coomassie blue. Immobilized antibody was added as 1:1 slurry in IP buffer to 1 mL of each peptide fraction, and the mixture was incubated overnight. The immobilized antibody beads were washed 3 times with 1 mL of IP buffer and twice with 1 mL of water, all at 4°C. Peptides were eluted from beads by incubation with 75 mL of 0.1% TFA at 20°C to 25°C for 10 minutes.
Analysis by LC-MS/MS
Peptides in the IP eluate were concentrated and separated from eluted antibody by the use of 0.2 mL of StageTips. Peptides were eluted from the microcolumns with 1 mL of 40% acetonitrile, 0.1% TFA (fractions I and II), or 1 mL of 60% acetonitrile. Eluted peptide fractions were dried by speed vacuum and resuspended in 7 μL of water/0.1% formic acid. Samples were loaded onto a 21-cm 75-μm internal diameter (ID) PicoFrit capillary column (New Objective) packed with C18 reverse-phase resin and eluted with a linear acetonitrile/water gradient (2%-40% B in 40 minutes, A: 0.1% formic acid, B: acetonitrile/0.1% formic acid) at 400 nL/minute. Data were collected on a ThermoElectron LTQ ion trap mass spectrometer (Thermo Scientific) and coupled to a NanoLC 2Dimensional HPLC (Eksigent). The mass spectrometer was set to acquire a single MS scan followed by up to 5 data-dependent MS/MS scans (dynamic exclusion repeat count of 1, repeat duration of 30 seconds, exclusion duration of 90 seconds).
Assigning peptide sequences with the use of Sequest
MS/MS spectra were evaluated by use of the X!Tandem database searching engine.17 Data were searched against an International Protein Index human database (Version 3.41) with the following parameters. The specified enzyme was trypsin, and potential phosphorylation modifications at Ser, Thr, and Tyr residues were considered. Results were evaluated by the PeptideProphet statistical algorithm, where the PeptideProphet filter value was 0.9 or greater (with an error rate of 2%), and the IonPercent filter value was 0.15 or greater.
Quantitative RT-PCR analysis
Total RNA from cell lines and primary CD34+ marrow cells was isolated, and cDNA was synthesized by use of the μMACS One-Step cDNA Synthesis kit (Miltenyi Biotec) as instructed and as previously described.18 Quantitative PCR was carried out with the comparative Ct method as described by Livak and Schmittgen and as used previously.19 Quantitative RT-PCR (Applied Biosystems) was used to analyze expression of the human DJ-1, p53, and β-actin genes.
RT reactions of 1.5 μg of RNA used a Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science) in 20-μL volume with oligo dT primers following the protocol provided with the kit. The reaction mixes were incubated for 1 hour at 55°C followed by 5 minutes at 85°C to inactivate the reverse transcriptase enzyme. The cDNAs were brought to 300-μL volume in water (∼ 5 ng/μL of the original RNA) before analysis.
Analysis of DJ-1 and p53 by real-time PCR
β-actin (and as a second control β-glucuronidase) was used as a “housekeeping” gene for normalization of quantitative and qualitative RNA variation. Applied Biosystems Pre-Designed Gene Expression Assays (Applied Biosystems) containing both primers and fluorescent Taq-Man probes were used for all gene assessments. These assays are designed to span intronic sequences to avoid amplification of any contaminant DNA. ABI part numbers for the assays were as follows: DJ-1, Hs00697109_m1; p53, Hs01034249_m1; β-actin, Hs00607939. Each 20-μL reaction contained 2.0 μL of 10× PCR buffer without Mg2+, 2.8 μL of 25mM MgCl2 (3.5mM final concentration), 0.4 μL of ROX passive reference dye, 0.4 μL of 10mM dNTPs, 1.0 μL of ABI primer/probe, and 0.16 μL of (0.8 U) Fast Start Taq Polymerase (Roche); 8.24 μL of H2O; and 5 μL of the cDNA template. All reactions were carried out in triplicate in 384-well plates on an ABI7900HT (Applied Biosystems). For inclusion in the dataset, standard deviations of the triplicates had to be less than 0.15 CT. Additionally, we verified that the PCR efficiencies of the ABI assays were greater than 95% and that the slopes of the linear portion of the amplification curves varied by less than 5%.
RNA interference and transient transfection
The siRNA-expressing plasmid specific stealth RNA interference (RNAi) were obtained from Invitrogen. KG1a cells (106) were electroporated with 500 ng of siRNA by use of the Nucleofector Kit L (Amaxa Biosystems Inc). The sequences used for the scrambled siRNA were 5′-CGAAUCCUAAUGCUGCUCCCUACUU-3′ and 3′-AAGUAGGGAGGAGCAUUACCAUUCG-5′. For the specific knock-down of DJ-1, the sequences were as follows: 5′-GGUAAUCUGGGUGCACAGAAUUUAU-3′ and 3′-AUAAAUUCUGUGCACCCAGAUUACC-5′. For the specific knock-down of p53, we used a FlexiTube siRNA approach (SI02655170; QIAGEN) with the validated sequence AAGGAAATTTGCGTGTGGAGT.
Immunoblotting
Total lysates for KG1a cells were prepared as follows: Cells were isolated from cocultures by MACS (Miltenyi Biotec) by the use of anti-CD45 antibody and lysed with cell lysis buffer (Cell Signaling Technology Inc) containing 1% Triton X-100 and Proteinase Inhibitor Cocktail (EMD Biosciences). The lysates were sonicated for 2 minutes and cleared by centrifugation at 20 000g for 10 minutes. Protein concentrations were quantified by use of the bicinchoninic acid assay (Pierce Biotechnology Inc), and equal amounts of protein (30 μg) in each lysate were diluted in Laemmli sodium dodecyl sulfate sample buffer, resolved by gel electrophoresis on 4% to 12% Bis-Tris precast NuPage gels (Invitrogen) in running buffer (50mM 2-[N-morpholino] ethane sulfonic acid, 50mM Tris base, 0.1% sodium dodecyl sulfate, and 1mM EDTA [ethylenediaminetetraacetic acid]) as described by the manufacturer and transferred to polyvinylidene difluoride membrane. The membranes were blocked in 5% nonfat dry milk diluted in Tris-buffered saline containing 0.1% Tween-20 for 1 hour at room temperature and then incubated overnight at 4°C in 5% nonfat dry milk/0.1% Tween-20 containing either mouse anti-p53 total or anti-p53 ser46 antibody (1:1000, Cell Signaling Technology Inc) or rabbit anti–DJ-1 antibody (1:200; AbCam Inc). Secondary human anti–mouse or anti–rabbit antibodies (1:2000; Santa Cruz Biotechnology) conjugated to horseradish peroxidase were used for enhanced chemoluminescence (Pierce Biotechnology), and the membranes were exposed to film.
Nuclear lysates of KG1a cells were prepared as follows: CD45+ KG1a cells were isolated as described,20 and nuclear and cytoplasmic proteins were extracted by use of the nuclear extraction kit from Marligen Biosciences Inc. Total protein was determined by the use of a modified BCA procedure. The amount of nuclear and cytoplasmic DJ-1 protein in cell lysates was determined by Western blot with the use of a rabbit anti–DJ-1 antibody (1:200; AbCam Inc); each lane was loaded with 24 μg of total protein. Nuclear lysates were probed for Histone-H2A (Cell Signaling Technology Inc) for loading normalization purposes.
Immunoprecipitation
Immunoprecipitation assays were performed as described previously.21 In brief, 5 × 106 KG1a cells were centrifuged and resuspended in 1 mL of lysis buffer. The cells were placed on ice for 1 hour and shaken gently during the incubation, then sedimented at 16 000g for 10 minutes at 4°C. Next, the supernatant was transferred to a clean tube containing 100 μL of Protein G beads and incubated for 1 hour at 4°C for pretreatment. The Protein G beads were discarded after centrifugation. Antibody to goat anti–DJ-1 was then added at 4°C and left overnight. An isotype matched anti–STRO-1 antibody (R&D Systems) was added as negative control. Protein G, 100 μL, was then added at 4°C, and after 1 to 2 hours, the supernatant was discarded. The beads containing the aggregate immune-complexes were washed 5 times with lysis buffer, and the protein complexes associated with the beads were loaded with 2× loading buffer and dithiothreitol. The membrane was processed as described for Western blotting.
Statistical analysis
Each experimental treatment was performed in triplicate, and the experiments were repeated at least 3 times. For gene expression purposes all values were expressed as the mean plus or minus SEM. A Student t test or Mann-Whitney U test was used to compare continuous variables between 2 groups; 1-way analysis of variance (ANOVA) analysis was applied to compare continuous variables among 3 or more groups. The correlation between continuous variables (eg, International Prognostic Scoring System and DJ-1/p53 ratio) was assessed by linear regression, and the 2-sample t test was used to compare the mean values of a continuous measure between groups (eg, ratio between the group with low-grade vs advanced disease). Cox regression was used to assess the correlation between ratio and the hazard of overall mortality. Presence of transplantation was included in the regression model and was treated as a time-dependent covariate.
Results
Tyrosine phosphorylation profiles in TNF-α–resistant KG1a cells and the apoptosis-sensitive parent cell line KG1
We have shown that apoptosis-resistant KG1a cells and primary CD34+ cells from patients with MDS (but not from healthy donors) are rendered sensitive to TNF-α–triggered apoptosis when in contact with stroma.6 Stroma contact leads to up-regulation of several apoptosis-related factors, including p53.6 To further characterize potential signals that lead to TNF-α sensitivity in the context of stroma contact, we determined and compared protein phosphorylation patterns in KG1 and KG1a cells before and after stroma contact. The peptide “hits” for protein classes under both conditions (before and after stroma contact) are listed in supplemental Tables 1 and 2 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Phosphorylation of DJ-1/PARK-7 (DJ-1) in KG1a cells in the presence and absence of stroma contact
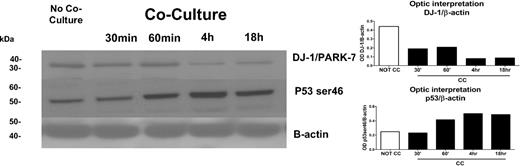
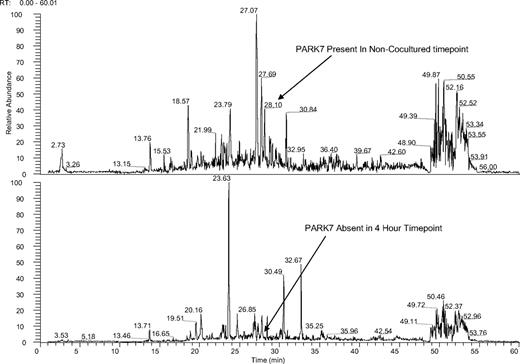
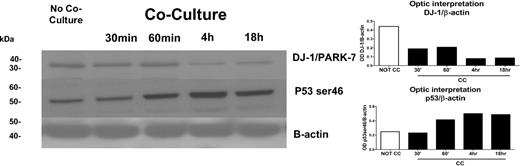
We first determined differences in phosphopeptide patterns between KG1a cells cultured with and without stroma contact. Cell lysates from KG1a cells with stroma contact were analyzed at 4 sequential time points (Figure 1). We focused on the role of DJ-1 because DJ-1 expression has been associated with repression of p53 and resistance to apoptosis22 and because our previous observations had shown up-regulation of p53 in KG1a cells in stroma cocultures.6 Results showed highly phosphorylated DJ-1 in KG1a cells and low levels of p53 (mRNA and protein) in the absence of stroma. When cocultured with stroma, levels of phosphorylated DJ-1 in KG1a cells were basically undetectable, and levels of p53 protein (phosphorylated at ser46) increased (Figure 2) over a time course of 0.5 to 18 hours. In KG1, the apoptosis-sensitive parent cell line of KG1a, DJ-1 phosphorylation was undetectable even in the absence of stroma, and p53 was expressed constitutively at greater levels than in KG1a cells, consistent with the observed stroma-independent activation of caspase-3 and induction of apoptosis in KG1 (but not in KG1a) cells.14
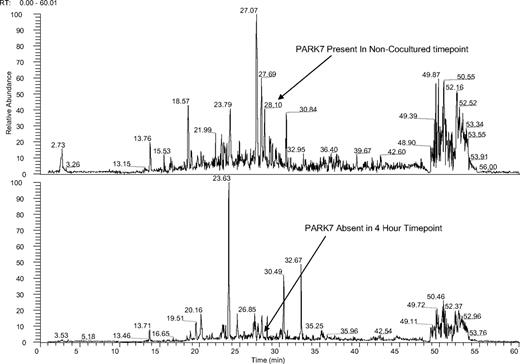
Ion chromatogram of KG1a cells before and after coculture with HS5 stroma. Mass spectra of noncocultured (top panel) and cocultured (bottom panel) KG1a cells at 4-hour time points both spectra filtered for PARK-7 peptide sequence ALVILA at a mass to charge ratio of 364.5. KG1a cell lysates were trypsin digested, solid-phase extraction purified, and exposed to phospho-Tyr-100 immunoaffinity beads as described in “Cell lysates.” The phosphopeptide-enriched samples were dried, resuspended, and analyzed by LC-MS. Tandem mass spectra were searched against a human International Protein Index database by use of the X!Tandem protein search engine and evaluated by the Bayesian statistical algorithm, PeptideProphet. Two unique peptides for PARK-7 were identified in the noncocultured assay; the peptide with sequence ALVILA (amino acid sequence) and mass to charge ratio of 364.5 eluted at 28.1 minutes in the filtered ion chromatogram (X). The ion peak for ALVILA was not observed in filtered ion chromatograms for the 30-minute, 1-hour, 4-hour, and 24-hour cocultured time points (X through X; shown is the 4-hour time point); the database search identified no PARK-7 peptides in these samples (see supplemental Figure 1).
Ion chromatogram of KG1a cells before and after coculture with HS5 stroma. Mass spectra of noncocultured (top panel) and cocultured (bottom panel) KG1a cells at 4-hour time points both spectra filtered for PARK-7 peptide sequence ALVILA at a mass to charge ratio of 364.5. KG1a cell lysates were trypsin digested, solid-phase extraction purified, and exposed to phospho-Tyr-100 immunoaffinity beads as described in “Cell lysates.” The phosphopeptide-enriched samples were dried, resuspended, and analyzed by LC-MS. Tandem mass spectra were searched against a human International Protein Index database by use of the X!Tandem protein search engine and evaluated by the Bayesian statistical algorithm, PeptideProphet. Two unique peptides for PARK-7 were identified in the noncocultured assay; the peptide with sequence ALVILA (amino acid sequence) and mass to charge ratio of 364.5 eluted at 28.1 minutes in the filtered ion chromatogram (X). The ion peak for ALVILA was not observed in filtered ion chromatograms for the 30-minute, 1-hour, 4-hour, and 24-hour cocultured time points (X through X; shown is the 4-hour time point); the database search identified no PARK-7 peptides in these samples (see supplemental Figure 1).
Expression of DJ-1 and p53 proteins as determined by LC-MS/MS/PhosphoScan method. DJ-1/PARK-7 protein levels in KG1a cells with or without coculture with HS5 stroma at 0.5, 1, 4, and 18 hours were determined by Western blot (left) and confirmed by densitometry (right). There was a strong correlation between the decrease in DJ-1 over time in KG1a cells after coculture with HS5 stroma and an increase in protein levels of p53 as determined with anti–serine 46 phospho antibodies. The increase in p53 was associated with an increase in apoptosis (see Figure 5A). One of 3 identical experiments is shown.
Expression of DJ-1 and p53 proteins as determined by LC-MS/MS/PhosphoScan method. DJ-1/PARK-7 protein levels in KG1a cells with or without coculture with HS5 stroma at 0.5, 1, 4, and 18 hours were determined by Western blot (left) and confirmed by densitometry (right). There was a strong correlation between the decrease in DJ-1 over time in KG1a cells after coculture with HS5 stroma and an increase in protein levels of p53 as determined with anti–serine 46 phospho antibodies. The increase in p53 was associated with an increase in apoptosis (see Figure 5A). One of 3 identical experiments is shown.
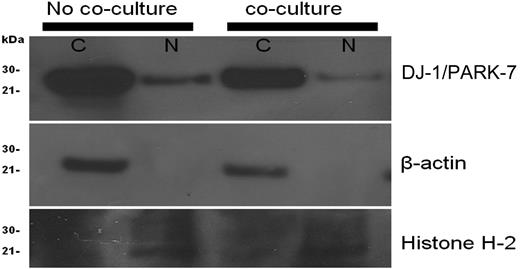
Subcellular localization of DJ-1 in KG1a cells is dependent upon stroma contact
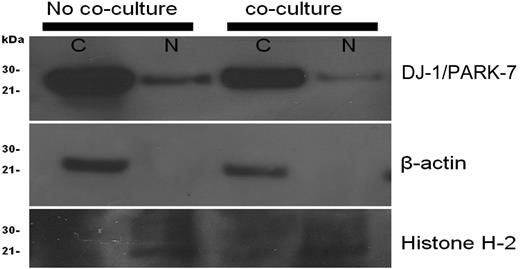
To assess subcellular localization of DJ-1 in KG1a leukemic cells, the cells were stained with appropriate antibody, and cytoplasm and nucleus were examined. Both compartments expressed DJ-1 before contact with stroma; however, after stroma contact, DJ-1 localization shifted from the nucleus to the cytoplasm, suggesting decreased nuclear-binding activity. Loading controls for cytoplasm (β-actin) and nucleus (histone H-2A) remained constant during the experiments (Figure 3).
Cellular localization of DJ-1/PARK-7 and p53 in KG1a cells. KG1a cells before coculture showed more prominent DJ-1 in the nucleus (N) than after coculture. Included were loading controls for cytoplasm (C; β-actin) and nucleus (N; Histone H-2A).
Cellular localization of DJ-1/PARK-7 and p53 in KG1a cells. KG1a cells before coculture showed more prominent DJ-1 in the nucleus (N) than after coculture. Included were loading controls for cytoplasm (C; β-actin) and nucleus (N; Histone H-2A).
Interactions of DJ-1 and p53 in cocultures of KG1a cells and stroma
To determine protein-protein interactions between DJ-1 and p53, which are likely to be relevant for the apoptotic function of p53, we tested p53 expression and its possible interaction with DJ-1. Protein extracts from KG1a cells were immunoprecipitated with anti–DJ-1 antibody and subjected to Western blot analysis with the use of the anti-p53 antibody (Figure 4). The results indicated that DJ-1 protein was associated with p53. DJ-1 and p53 also coprecipitated from KG1a cells after contact with stroma (Figure 4). These results, taken together with the subcellular localization of DJ-1 and p53, suggest that cellular shifts (coculture versus no coculture) accounted for increased levels of active p53 after coculture and enhanced apoptosis sensitivity (Figures 2–3). DJ-1 expression in the nucleus of KG1a cells was suppressed by contact with stroma, and a shift from the nucleus to the cytoplasm (Figure 3) could account for the “release” of p53 in the presence of stroma, which would then allow for the enhanced facilitation of apoptosis. That p53 was instrumental in facilitating apoptosis in KG1a cells in this model is further supported by the fact that addition of the p53 inhibitor PFT-α or inhibition of p53 by siRNA reduced the rate of apoptosis even in the presence of stroma contact (Figure 5D-E). Of note, simultaneous siRNA repression of both p53 and DJ-1 was also associated with inhibition of TNF-mediated apoptosis, unless TNF-α concentrations of 50 ng/mL or greater were used (Figure 5E).
Interaction of p53 and DJ-1 protein in KG1a cells. Whole-cell lysates from KG1a cells were immunoprecipitated with an anti–DJ-1 antibody and protein-A beads, or alternatively, with anti–TWIST-1 or Stro-1 antibodies (as positive [+] and negative [−] controls, respectively). The protein complexes brought down by anti–DJ-1 or anti-TWIST antibody contained p53, consistent with protein-protein interactions of p53 and DJ-1. One of 4 identical experiments is shown.
Interaction of p53 and DJ-1 protein in KG1a cells. Whole-cell lysates from KG1a cells were immunoprecipitated with an anti–DJ-1 antibody and protein-A beads, or alternatively, with anti–TWIST-1 or Stro-1 antibodies (as positive [+] and negative [−] controls, respectively). The protein complexes brought down by anti–DJ-1 or anti-TWIST antibody contained p53, consistent with protein-protein interactions of p53 and DJ-1. One of 4 identical experiments is shown.
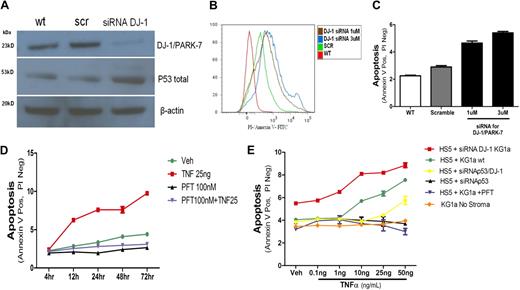
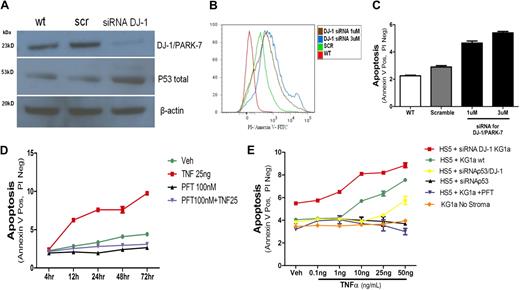
p53 and apoptosis induction in KG1a cells. (A) DJ-1 and p53 protein levels in KG1a cells cultured without stroma. The expression of DJ-1 was analyzed by Western blot (wt). KG1a cells were transfected either with a scrambled siRNA (scr) or a DJ-1–specific sequence (siRNA DJ-1). There was an inverse correlation between decrease in protein expression of DJ-1 and levels of p53. (B-C) Inhibition of DJ-1 by siRNA and TNF-α–triggered apoptosis in KG1a cells cultured without stroma. Apoptosis (determined by flow cytometry) in control cultures containing unmodified KG1a cells (WT) and in cocultures with HS5 cells transfected either with a scrambled siRNA sequence (scramble) or siRNA specific for DJ-1 (at 1μM and 3μM). (B) Histogram shows intensity of annexin V staining in KG1a CD45+ cells. (C) Proportion of apoptotic cells; only CD45+ (KG1a) cells were considered. (D) Inhibition of p53 in coculture prevents TNF-α–induced apoptosis. KG1a cells were analyzed by flow cytometry. The y-axis shows proportion of APC-CD45+ KG1a cells positive for fluorescein isothiocyanate/annexin V and negative for propidium iodide (early apoptosis). Shown are untreated cells (veh), cells treated with TNF-α at 10 or 25 ng/mL, with PFT-α at 100nM (in dose-response experiments inhibition of apoptosis was seen at PFT-α concentrations > 1nM), or a combination of TNF-α (at 25 ng/mL) plus PFT-α (at 100nM), over a time course of 4 to 72 hours. (E) Dose-response to TNF-α under different conditions. TNF-α achieved measurable apoptosis at concentrations greater than 1 ng/mL in cocultures of HS5 cells with unmodified (wt) KG1a cells. Apoptosis was enhanced if DJ-1 on KG1a cells was repressed by siRNA. Apoptosis was prevented by PFT or siRNA inhibition of p53. Apoptosis was also prevented by simultaneous repression of DJ-1 and p53 by specific siRNAs; only at very high TNF concentrations was a low rate of apoptosis observed. Data are represented as mean percentage of early apoptotic cells ± SEM (annexin V positive but negative with propidium iodine). Results summarize data from at least 3 experiments, each conducted in triplicate.
p53 and apoptosis induction in KG1a cells. (A) DJ-1 and p53 protein levels in KG1a cells cultured without stroma. The expression of DJ-1 was analyzed by Western blot (wt). KG1a cells were transfected either with a scrambled siRNA (scr) or a DJ-1–specific sequence (siRNA DJ-1). There was an inverse correlation between decrease in protein expression of DJ-1 and levels of p53. (B-C) Inhibition of DJ-1 by siRNA and TNF-α–triggered apoptosis in KG1a cells cultured without stroma. Apoptosis (determined by flow cytometry) in control cultures containing unmodified KG1a cells (WT) and in cocultures with HS5 cells transfected either with a scrambled siRNA sequence (scramble) or siRNA specific for DJ-1 (at 1μM and 3μM). (B) Histogram shows intensity of annexin V staining in KG1a CD45+ cells. (C) Proportion of apoptotic cells; only CD45+ (KG1a) cells were considered. (D) Inhibition of p53 in coculture prevents TNF-α–induced apoptosis. KG1a cells were analyzed by flow cytometry. The y-axis shows proportion of APC-CD45+ KG1a cells positive for fluorescein isothiocyanate/annexin V and negative for propidium iodide (early apoptosis). Shown are untreated cells (veh), cells treated with TNF-α at 10 or 25 ng/mL, with PFT-α at 100nM (in dose-response experiments inhibition of apoptosis was seen at PFT-α concentrations > 1nM), or a combination of TNF-α (at 25 ng/mL) plus PFT-α (at 100nM), over a time course of 4 to 72 hours. (E) Dose-response to TNF-α under different conditions. TNF-α achieved measurable apoptosis at concentrations greater than 1 ng/mL in cocultures of HS5 cells with unmodified (wt) KG1a cells. Apoptosis was enhanced if DJ-1 on KG1a cells was repressed by siRNA. Apoptosis was prevented by PFT or siRNA inhibition of p53. Apoptosis was also prevented by simultaneous repression of DJ-1 and p53 by specific siRNAs; only at very high TNF concentrations was a low rate of apoptosis observed. Data are represented as mean percentage of early apoptotic cells ± SEM (annexin V positive but negative with propidium iodine). Results summarize data from at least 3 experiments, each conducted in triplicate.
DJ-1–specific siRNA interference results in up-regulation of p53 and increased apoptosis in KG1a cells
KG1a cells transfected with a DJ-1–specific construct (siRNA DJ-1) showed decreased DJ-1 protein expression, whereas levels of p53 increased (Figure 5A). As shown in Figure 5B and C, in parallel to increased expression of p53 the rate of apoptosis in KG1a in response to TNF-α increased after inhibition of DJ-1. Those findings were progressive with time in coculture (4-72 hours; Figure 5D).
To determine whether DJ-1 knockdown would also affect apoptosis induced by conventional chemotherapeutic agents, KG1a cells were treated with siRNA (DJ-1 siRNA or scrambled sequence siRNA), exposed to daunorubicin or etoposide, and analyzed after annexin V–fluorescein isothiocyanate/propidium iodide staining. Exposure to daunorubicin or etoposide resulted in significantly increased apoptosis (P < .001 and P < .001, respectively) in cells transfected with DJ-1–specific siRNA in comparison with cells treated with scrambled siRNA (supplemental Figure 2A-B).
DJ-1 expression in primary CD34+ marrow cells
The insights derived from studies with KG1 and KG1a cells strongly suggested that expression of DJ-1 (and interaction with p53) was relevant for apoptosis control in clonal hematopoietic cells. Therefore, we determined DJ-1 expression in primary CD34+ marrow cells derived from patients with MDS and healthy donors. Quantitation of the transcript levels for DJ-1 in highly purified CD34+ cells by quantitative RT-PCR revealed significantly greater levels of DJ-1 in MDS patients than in healthy donors (DJ-1/β2M ratio 1.2 in MDS patients vs healthy donors, P < .001; Table 1; Figure 6A). DJ-1 expression levels correlated with International Prognostic Scoring System classification (R2 = 0.14; P = .006) and disease stage as determined by marrow blast count (low-grade vs advanced; Figure 6B); however, values were significantly different from healthy controls only in patients with advanced MDS (mean value, 1.21; P < .019).
DJ-1 mRNA expression in cells from healthy donors (n = 13), and patients with low-grade or advanced MDS (n = 58). (A) There was a statistically significant difference between the mRNA from healthy donors and patients with untreated MDS (P < .001) *Statistical significance as determined by Student t test compared with healthy control patients. (B) In comparison, patients with advanced MDS showed increased levels of DJ-1 (P < .019). The difference between low-grade MDS (refractory anemia, refractory anemia with ringed sideroblasts, and refractory cytopenia with multilineage dysplasia) and healthy controls was not significant. **Statistical significance as determined by ANOVA, comparison of healthy versus advanced MDS and healthy controls versus low-grade MDS. (C) Expression of p53 and DJ-1 as determined by RT-PCR in CD34+ cells from healthy donors (normal controls [NC]) and patients with MDS CD34 cells. In CD34+ cells from healthy donors (NCD34+; left), p53 and DJ-1 levels did not differ with and without stroma-contact. In clonal cells (MDSCD34+), the expression of p53 after stroma contact (CC) showed prominent p53 up-regulation; the inverse correlation was found for levels of DJ-1, which declined. (D) DJ-1 and p53 mRNA expression in cells for healthy donors (n = 13), and patients with low-grade (n = 30) and advanced MDS (n = 28). The ratio of DJ-1 and p53 expression was assessed by subgroup. A ratio greater than 1.01 would indicate inhibition of p53 expression. *Statistical significance as determined by ANOVA, comparison of DJ-1/p53 ratios of healthy versus advanced MDS healthy controls versus low-grade MDS and low-grade versus advanced MDS (P < .014). (E) DJ-1/p53 mRNA ratio in patients with MDS. There was a statistically significant difference between DJ-1/p53 ratios in low-grade (mean = 1.029; variance = 0.060064) and advanced (mean = 1.176468, variance = 0.137022) MDS patients (P = .014). *Statistical significance as determined by ANOVA compared low-grade with advanced MDS (P < .014).
DJ-1 mRNA expression in cells from healthy donors (n = 13), and patients with low-grade or advanced MDS (n = 58). (A) There was a statistically significant difference between the mRNA from healthy donors and patients with untreated MDS (P < .001) *Statistical significance as determined by Student t test compared with healthy control patients. (B) In comparison, patients with advanced MDS showed increased levels of DJ-1 (P < .019). The difference between low-grade MDS (refractory anemia, refractory anemia with ringed sideroblasts, and refractory cytopenia with multilineage dysplasia) and healthy controls was not significant. **Statistical significance as determined by ANOVA, comparison of healthy versus advanced MDS and healthy controls versus low-grade MDS. (C) Expression of p53 and DJ-1 as determined by RT-PCR in CD34+ cells from healthy donors (normal controls [NC]) and patients with MDS CD34 cells. In CD34+ cells from healthy donors (NCD34+; left), p53 and DJ-1 levels did not differ with and without stroma-contact. In clonal cells (MDSCD34+), the expression of p53 after stroma contact (CC) showed prominent p53 up-regulation; the inverse correlation was found for levels of DJ-1, which declined. (D) DJ-1 and p53 mRNA expression in cells for healthy donors (n = 13), and patients with low-grade (n = 30) and advanced MDS (n = 28). The ratio of DJ-1 and p53 expression was assessed by subgroup. A ratio greater than 1.01 would indicate inhibition of p53 expression. *Statistical significance as determined by ANOVA, comparison of DJ-1/p53 ratios of healthy versus advanced MDS healthy controls versus low-grade MDS and low-grade versus advanced MDS (P < .014). (E) DJ-1/p53 mRNA ratio in patients with MDS. There was a statistically significant difference between DJ-1/p53 ratios in low-grade (mean = 1.029; variance = 0.060064) and advanced (mean = 1.176468, variance = 0.137022) MDS patients (P = .014). *Statistical significance as determined by ANOVA compared low-grade with advanced MDS (P < .014).
Correlation of DJ-1 and p53 mRNA levels in primary CD34+ marrow cells and clinical course
We next characterized the expression of p53 and DJ-1 in primary marrow cells. There was a significantly greater DJ-1 mRNA to p53 mRNA ratio in advanced MDS (RAEB-1 and 2, ie, the population in whom apoptosis resistance was observed) than in healthy controls (P = .019), consistent with a DJ-1–mediated inhibitory effect on p53-mediated apoptosis (Figure 6C).
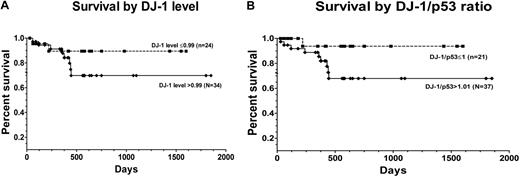
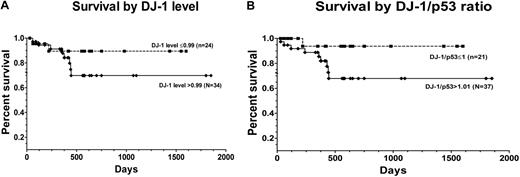
Stroma coculture resulted in decreased levels of DJ-1 and up-regulation of levels of p53 in marrow cells from patients with MDS. In contrast, no appreciable expression changes occurred in marrow cells from healthy subjects. The ratio of DJ-1 to p53 was greater in patients with advanced disease (mean value, 1.21) compared with the ratio in patients with low-grade disease (mean value, 0.99; P = .019; Figure 6D-E). Increasing ratios of DJ-1 and p53 were associated with an increased risk of overall mortality (after adjusting for patients who were transplanted), although the difference was not statistically significant (P = .18). Figure 7 shows the unadjusted Kaplan-Meier curves of overall survival by DJ-1 values (DJ-1 expression > 0.99 or ≤ 0.99) and by DJ-1/p53 ratios (> 1.01 vs ≤ 1.01).
Patient survival after diagnosis of MDS by DJ-1 expression. (A) Survival among patients with DJ-1 levels ≤ 0.99 (broken line; n = 24) and among patients with DJ-1 levels greater than 0.99 (solid line; n = 34; P = .10) log-rank (Mantel-Cox). (B) Survival was superior in patients with DJ-1/p53 ratios 1.01 or less (broken line; n = 21) than among patients with ratios greater than 1.01 (solid line; n = 37; P = .08) log-rank (Mantel-Cox) test (7 patients with DJ-1/p53 ratios 1.01 or less, and 6 patients with DJ-1/p53 ratios greater than 1.01 were censored at the time they underwent hematopoietic cell transplantation).
Patient survival after diagnosis of MDS by DJ-1 expression. (A) Survival among patients with DJ-1 levels ≤ 0.99 (broken line; n = 24) and among patients with DJ-1 levels greater than 0.99 (solid line; n = 34; P = .10) log-rank (Mantel-Cox). (B) Survival was superior in patients with DJ-1/p53 ratios 1.01 or less (broken line; n = 21) than among patients with ratios greater than 1.01 (solid line; n = 37; P = .08) log-rank (Mantel-Cox) test (7 patients with DJ-1/p53 ratios 1.01 or less, and 6 patients with DJ-1/p53 ratios greater than 1.01 were censored at the time they underwent hematopoietic cell transplantation).
Discussion
The pathophysiology of MDS is incompletely understood. In particular, the transition from a disease state with high rates of apoptosis (early/low-grade MDS) to apoptosis resistance (advanced MDS) is not clear. Numerous studies3,4,23 have documented an essential role of the marrow microenvironment and its interactions with hematopoietic cells in the regulation of normal hematopoiesis and myeloid malignancies. We showed previously that coculture of clonal myeloid KG1a cells with stroma modified the response of KG1a cells to proapoptotic signals.24 The acquisition of apoptosis sensitivity by KG1a cells was associated with up-regulation of p53. Here, we used our in vitro coculture model to further characterize in KG1a cells signals that interfered with or were permissive for apoptosis. We applied a global approach using phospho-proteomics to identify potentially relevant peptides/proteins, which were modified in coculture and, therefore, might be relevant for the coculture dependent change in apoptosis sensitivity.6
Among the proteins that showed significant losses of phosphorylation, we selected DJ-1 for further study. DJ-1 had been shown by others to affect apoptosis, possibly by interacting with p53, which we showed to be up-regulated in KG1a cells in coculture.6 Those studies supported the hypothesis that lower levels of expression of p53 (in the absence of stroma contact) contributed to apoptosis resistance, possibly by overexpression of DJ-1 and its interference with p53. The current results are consistent with such a model, and this hypothesis is further supported by the fact that chemical inhibition of p53 by PFT-α or knockdown by specific siRNA prevented TNF-α–induced apoptosis in the coculture model. Although p53 has been associated primarily with stress-induced apoptosis,25 these observations suggest a role in TNF receptor-initiated apoptosis as well.
Inhibition of p53 and interference with apoptosis has been described in other models. For example, human double-minute 2, an oncoprotein classified among the “miniature proteins,” interacts with p53 in an indirect way, apparently facilitated by histone deacetylase, which provides corepressor activity for p53.26,27 Fan et al9 showed that DJ-1 binds to the C terminus region of p53, thereby interfering with the binding of p53 to promoter DNA. Their experiments also demonstrated that DJ-1 inhibition was associated with increased apoptosis sensitivity. Our data are in agreement in so far as they show that functional interference with DJ-1 by siRNA in KG1a cells enhanced apoptosis in response to apoptotic stimuli beyond levels achieved in unmodified cocultures. Combined inhibition of DJ-1 and p53 prevented TNF-α–induced apoptosis. Only at very high concentrations of TNF-α was low-grade apoptosis noted, possibly related to activation of additional pathways or nonspecific toxic effects. These results are supportive of a model in which down-regulation of DJ-1 in coculture would lead to p53-facilitated apoptosis. The fact that apoptosis in coculture in response to TNF-α occurred in clonal hematopoietic cells but not in marrow cells from healthy donors (which did not express DJ-1) is consistent with the concept that DJ-1 is important for apoptosis resistance in transformed or clonal cells but not in normal hematopoietic precursors. Liu et al28 also showed up-regulation of DJ-1 in leukemic cell lines and in patients with acute myeloid leukemia, in agreement with the present findings in patients with MDS, particularly as our data show progressively increasing levels of DJ-1 with more advanced MDS.
Studies in neuronal tissue29,30 or prostate and lung cancer cells31 indicated that DJ-1 has oncogenic potential, presumably related to its antiapoptotic function. Overexpression of DJ-1 in prostatic benign hyperplasia (BPH-1) cell lines caused resistance to apoptosis induced by cytotoxic agents, whereas knockdown of DJ-1 enhanced apoptosis in PC-3 (prostatic cancer) cells by the same cytotoxic reagents.31 The mechanism responsible for the antiapoptotic function of DJ-1 involves sumoylation of the K130 residue of DJ-1. For the antiapoptotic function to occur, DJ-1 needs to transit from cytoplasm to the nucleus. Such a pattern does fit with the present results, which show a loss of DJ-1 from the nucleus of KG1a cells in coculture, which was associated with acquisition of apoptosis sensitivity.
DJ-1 expression also has been shown to decrease levels of Bax protein and inhibit caspase activation.32 The effect on Bax is of particular interest in view of the recently reported direct (nontranscriptional) interaction of p53 with Bax.9 A study in zebrafish33 showed that morpholino knock-down of DJ-1 was associated with increased transcription of p53 and increased levels of Bax, leading to increased neuronal cell death in response to hydrogen peroxide.
Another recent report by Vasseur et al34 suggested that DJ-1 may protect cells against apoptosis via yet another pathway by functioning as an upstream activator of hypoxemia-inducible factor 1. The bone marrow microenvironment is characterized by regions of fluctuating or chronic hypoxia and nutrient depletion. The report showed that under hypoxic conditions, the metabolic sensor AMP-activated protein kinase was rapidly activated in DJ-1–expressing cells but not in DJ-1 knockdown cells. Those findings suggest that DJ-1 is part of the complex cellular response to hypoxia that involves AMP-activated protein kinase and hypoxemia-inducible factor 1 and that the effector functions of these molecules are highly dependent on DJ-1 expression.
Although we showed previously that up-regulation of p53 and PYCARD6 was associated with stroma-dependent facilitation of apoptosis in cocultured hematopoietic cells, it was not clear why stroma contact should result in differential responses in cells from patients with MDS (presumably clonal) and in marrow cells from healthy subjects. One possibility is that the internal milieu in clonal MDS cells (eg, DNA damage, change in redox status) rendered these cells dependent upon p53-mediated apoptosis, which became possible upon inhibition of DJ-1. We did not determine whether p53 in MDS cells was mutated, which could have affected DJ-1/p53 interactions. However, published data suggest that only approximately 15% of MDS patients harbor p53 mutations.35-39 Thus, it is unlikely that p53 mutations affected the DJ-1/p53 interactions in the majority of patients, in whom there should be no interference with binding. In fact, high levels of expression of DJ-1 should effectively bind p53, although it is not clear why DJ-1 is overexpressed.
In summary, the present data suggest an important role of DJ-1 in the control of apoptosis in clonal hematopoietic cells. Findings in primary marrow cells suggest that dysregulation of DJ-1 expression in MDS is involved in the pathophysiology of MDS and could possibly serve as a molecular target for MDS treatment. The results of our study also indicated that an immunoaffinity strategy, when merged with qualitative mass spectrometry methods, is a useful approach to the identification of protein phosphorylation patterns that arise in response to specific treatment or experimental conditions. The technique should have substantial power in identifying activated tyrosine kinases and phosphorylated substrates in cancer cells, thereby elucidating oncogenic pathways and identifying targets for therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Helen Crawford and Bonnie Larson for help with manuscript preparation, and Chris Kemp and Derek Stirewalt for a critical reading of the manuscript.
This study was supported in part by National Heart, Lung and Blood Institute (National Institutes of Health) grant nos. HL036444 and HL082941. A.M.M. was also supported by the J. P. McCarthy Fund from the Community Foundation of Southeast Michigan.
National Institutes of Health
Authorship
Contribution: A.M.M. conceived of the study, conducted most of the experiments, and wrote the manuscript; X.L. carried out experiments; T.G. provided statistical expertise; B.M. contirbuted expertise on the application of phosphoscan proteomics and the analysis of results; and H.J.D. conceived of the study, provided critique, and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: A. Mario Marcondes, MD, Fred Hutchinson Cancer Research Center, D1-100, 1100 Fairview Ave North, Seattle, WA 98109; e-mail: mmarcond@fhcrc.org.

![Figure 4. Interaction of p53 and DJ-1 protein in KG1a cells. Whole-cell lysates from KG1a cells were immunoprecipitated with an anti–DJ-1 antibody and protein-A beads, or alternatively, with anti–TWIST-1 or Stro-1 antibodies (as positive [+] and negative [−] controls, respectively). The protein complexes brought down by anti–DJ-1 or anti-TWIST antibody contained p53, consistent with protein-protein interactions of p53 and DJ-1. One of 4 identical experiments is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/10/10.1182_blood-2009-08-236992/4/m_zh89991049420004.jpeg?Expires=1765995183&Signature=Z1Ye4cNDeCmvhdKrnYEQqfv3Yeda~YtvZjx-izcW5c7Fd622YECvPp8j4VUk6kzkl8o1GLS1N-VCeGgX9POx5n3Rzj6YE6UxauWYVZB-pj4-RerfzcKeZHMQ6CgNiIkH~oRjpM9cFUZv7kFbU3G-5I-SQPviZSD2Kc337I2UoTFhzoMjLC~YmrysQdpNq-tIjdDx2G3yHXTpjShZ2PqKTWRTMJGO~RG8QcTOH43mFXuOPAZAd9u0zAmphKwfxpXSakNZLMGqFnui3mxSJpbf7g6DE7woGevQ6cQUPoLtm4r4e6BrebAlx0DkbobQTN15AVeFDhOliUif5fv2o8xHfQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. DJ-1 mRNA expression in cells from healthy donors (n = 13), and patients with low-grade or advanced MDS (n = 58). (A) There was a statistically significant difference between the mRNA from healthy donors and patients with untreated MDS (P < .001) *Statistical significance as determined by Student t test compared with healthy control patients. (B) In comparison, patients with advanced MDS showed increased levels of DJ-1 (P < .019). The difference between low-grade MDS (refractory anemia, refractory anemia with ringed sideroblasts, and refractory cytopenia with multilineage dysplasia) and healthy controls was not significant. **Statistical significance as determined by ANOVA, comparison of healthy versus advanced MDS and healthy controls versus low-grade MDS. (C) Expression of p53 and DJ-1 as determined by RT-PCR in CD34+ cells from healthy donors (normal controls [NC]) and patients with MDS CD34 cells. In CD34+ cells from healthy donors (NCD34+; left), p53 and DJ-1 levels did not differ with and without stroma-contact. In clonal cells (MDSCD34+), the expression of p53 after stroma contact (CC) showed prominent p53 up-regulation; the inverse correlation was found for levels of DJ-1, which declined. (D) DJ-1 and p53 mRNA expression in cells for healthy donors (n = 13), and patients with low-grade (n = 30) and advanced MDS (n = 28). The ratio of DJ-1 and p53 expression was assessed by subgroup. A ratio greater than 1.01 would indicate inhibition of p53 expression. *Statistical significance as determined by ANOVA, comparison of DJ-1/p53 ratios of healthy versus advanced MDS healthy controls versus low-grade MDS and low-grade versus advanced MDS (P < .014). (E) DJ-1/p53 mRNA ratio in patients with MDS. There was a statistically significant difference between DJ-1/p53 ratios in low-grade (mean = 1.029; variance = 0.060064) and advanced (mean = 1.176468, variance = 0.137022) MDS patients (P = .014). *Statistical significance as determined by ANOVA compared low-grade with advanced MDS (P < .014).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/10/10.1182_blood-2009-08-236992/4/m_zh89991049420006.jpeg?Expires=1765995183&Signature=1AdTB3dgsetHYkGxxq-8mFVT~dKzvZT3sLlRFGDbf~INbI~CN5550weyDF1EbHVRTFN~AOAbX2nxDuZshz1p6mRT1QsxK2kLSimrxZyvHI71~YaVf-IgWN9G3YkmUGJwbOhmX7MQBqdPp7UaQDMgbrsZm51PeI6OwJd-LHPXyZuIxijicU0C-ul5n7cL0cbO8hi2UG~U8C5O3JeYLnEuScbcu8SLF6P6zyVt-cVCRM5-p9pRXlad~26VIjzT84s3r4Tz3~Rq13eGNUHBZUHaOwTCPtZHdP3D5cKW4SC5SilJteNIK-~256aXwSNv9NA6CIBmNTn3T1EIJh8ib9c3bQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 4. Interaction of p53 and DJ-1 protein in KG1a cells. Whole-cell lysates from KG1a cells were immunoprecipitated with an anti–DJ-1 antibody and protein-A beads, or alternatively, with anti–TWIST-1 or Stro-1 antibodies (as positive [+] and negative [−] controls, respectively). The protein complexes brought down by anti–DJ-1 or anti-TWIST antibody contained p53, consistent with protein-protein interactions of p53 and DJ-1. One of 4 identical experiments is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/10/10.1182_blood-2009-08-236992/4/m_zh89991049420004.jpeg?Expires=1766263428&Signature=T4p6ra8GJQgCp6QBNLxGAwf7SAikRJJzF9DhHpg67Y8R4-kEcradivtWc51zLnfiBSu4FdyDYrDf1oGQAEQ8owPu-K-PNd9Za~QVEzsFk1LBD1UZqU1Ekg-vyG3zqs96DDo14wyE4xCSW68HdL~7pAIVO7ddo3sOWH9j064U8JEz7glf3rcFaUtyS5OsQvTaDJuoUJKpxLaGJqcQAx0jGWVC345-5FeT1cgLLlLZhsOu8vG~3yblt8xaJBPaKfavX4wdURmFqUHPM1qUNavCbK4rXRgIEOM5mNzKhHVfjpaccK6SPmK0AYJEFxa5OIm~JbNDbL3YvOArpbFBLr8Wxw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. DJ-1 mRNA expression in cells from healthy donors (n = 13), and patients with low-grade or advanced MDS (n = 58). (A) There was a statistically significant difference between the mRNA from healthy donors and patients with untreated MDS (P < .001) *Statistical significance as determined by Student t test compared with healthy control patients. (B) In comparison, patients with advanced MDS showed increased levels of DJ-1 (P < .019). The difference between low-grade MDS (refractory anemia, refractory anemia with ringed sideroblasts, and refractory cytopenia with multilineage dysplasia) and healthy controls was not significant. **Statistical significance as determined by ANOVA, comparison of healthy versus advanced MDS and healthy controls versus low-grade MDS. (C) Expression of p53 and DJ-1 as determined by RT-PCR in CD34+ cells from healthy donors (normal controls [NC]) and patients with MDS CD34 cells. In CD34+ cells from healthy donors (NCD34+; left), p53 and DJ-1 levels did not differ with and without stroma-contact. In clonal cells (MDSCD34+), the expression of p53 after stroma contact (CC) showed prominent p53 up-regulation; the inverse correlation was found for levels of DJ-1, which declined. (D) DJ-1 and p53 mRNA expression in cells for healthy donors (n = 13), and patients with low-grade (n = 30) and advanced MDS (n = 28). The ratio of DJ-1 and p53 expression was assessed by subgroup. A ratio greater than 1.01 would indicate inhibition of p53 expression. *Statistical significance as determined by ANOVA, comparison of DJ-1/p53 ratios of healthy versus advanced MDS healthy controls versus low-grade MDS and low-grade versus advanced MDS (P < .014). (E) DJ-1/p53 mRNA ratio in patients with MDS. There was a statistically significant difference between DJ-1/p53 ratios in low-grade (mean = 1.029; variance = 0.060064) and advanced (mean = 1.176468, variance = 0.137022) MDS patients (P = .014). *Statistical significance as determined by ANOVA compared low-grade with advanced MDS (P < .014).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/10/10.1182_blood-2009-08-236992/4/m_zh89991049420006.jpeg?Expires=1766263428&Signature=UqNXUZkHPCaFUSMDKJXYFM1HZZxmKfqTDGmw6wDLcLl5OsIG3zK7v7ZbU~qzalMRL5P1PpPuAKBmG-XSGnbZDaXSVJk~sQrPHm2zt2WK1PDmWJvMBx0BjtED5ru8Oj-BsdItDW08sbP-g3jThsRLznKUitlL9WmrPduUuk34JvkMb1znsoiefLPuwLDsCix0A~-qU-0JeZGCF7JHaRzxTzyV0hQQ8EQiw1RcpJ~T1nTzUFtdjPzZYJ1CsZ2NKaURyeu8OIop0HaPW3QPtzvtZLsD0hLki-FGGjVxxEaFsN06BJXAqnZWY76XmCM-E60w3ACi7UdwoiGiYIx7vrabgw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)