Abstract
Activating mutations in JAK1 have been reported in acute lymphoblastic leukemias (ALLs). In this study, we found a type I interferon (IFN) transcriptional signature in JAK1 mutation-positive human ALL samples. This signature was recapitulated in vitro by the expression of JAK1 mutants in BW5147 and BaF3 hematopoietic cell lines. Binding of JAK1 to the IFN receptor was essential because mutations in the FERM domain abrogated this effect. Beside the constitutive activation of the type I IFN signaling cascade, JAK1 mutations also strongly potentiated the response to IFN in vitro. Typically, the proliferation of cell lines expressing JAK1A634D was abrogated by type I IFNs. Interestingly, we found that different JAK1 mutations differentially potentiate responses to type I IFNs or to interleukin-9, another cytokine using JAK1 to mediate its effects. This suggests that the type of mutation influences the specificity of the effect on distinct cytokine receptor signaling. Finally, we also showed in an in vivo leukemia model that cells expressing JAK1A634D are hypersensitive to the antiproliferative and antitumorigenic effect of type I IFN, suggesting that type I IFNs should be considered as a potential therapy for ALL with JAK1-activating mutations.
Introduction
JAK1 is a member of the Janus kinase family comprising 4 nonreceptor tyrosine kinases (JAK2, JAK3, and TYK2) that associate to cytokine receptors without intrinsic kinase activity to mediate cytokine-induced signal transduction.1 Recently, we and others reported that JAK1 mutations occur in acute lymphoblastic leukemia (ALL) and are relatively common in adult patients with T-cell ALL.2-4 Some of these mutations, such as A634D and R724H, induce the autonomous growth of the cytokine-dependent BaF3 cell line, whereas the R897C and A634D mutants protect the murine ALL cell line BW5147 from dexamethasone-induced apoptosis, indicating that they represent gain-of-function mutations that participate in the leukemogenic process by inducing the constitutive activation of the JAK-STAT pathway.2
JAKs participate in the signal transduction of various cytokine receptor families, which regulate different cellular processes. JAK1-binding receptors include the common γ chain subfamily (interleukin-2 receptor [IL-2R], IL-4R, IL-7R, IL-9R, IL-15R, IL-21R), the gp130 subfamily (IL-6R, LIFR, IL-11R), and the type II cytokine receptor complexes (type I, II and III interferon [IFN] receptors, IL-10R). We showed that JAK1 mutants need to associate to cytokine receptors, such as IL-2R and IL-9R, to promote constitutive activation of the receptor complex and signal transduction via signal transducers and activators of transcription, STATs.5 A similar mechanism was shown for the JAK2 V617F mutant found in myeloproliferative neoplasms. JAK2 V617F needs to associate to homodimeric type I cytokine receptors, such as the erythropoietin or thrombopoietin receptor, to mediate the cytokine receptor JAK2-STAT5 constitutive activation responsible for the increased myeloproliferation.6,7
Type I interferons (IFN-α/β) are important cytokines involved in the defense against viral infection. The type I IFN receptor complex consists of 2 receptor subunits, IFNAR1 and IFNAR2, which are associated with TYK2 and JAK1, respectively.8 Ligand-induced stimulation of IFNAR results in cross-activation of TYK2 and JAK1, which in turn activate 2 members of the STAT family, STAT1 and STAT2.9 Activation of STAT1 and STAT2 leads to the formation of 2 transcriptional complexes. The first complex, IFN-α–activated factor, is a homodimer of activated STAT1. The second complex, IFN-stimulated gene factor 3 (ISGF3), consists of a complex of activated STAT2, STAT1, and IRF-9 (a member of interferon regulatory factors, IRFs). STAT1 activation is involved in signaling from various cytokine receptors,9 whereas activation of STAT2 and formation of the ISGF3 complex are specifically triggered by type I IFN receptor complex and in a small number of cell types by type III IFN receptor complex.10,11 In contrast to other JAK1-binding cytokine receptors, type I IFN receptors are ubiquitously expressed,8 and ligand-induced activation of type I IFN receptor complex was shown to induce inhibition of cell proliferation and apoptosis. Expression of antiproliferative and proapoptotic genes was shown to be mainly under the control of the ISGF3 complex, eg, STAT2 and IRF-9.12 The antiproliferative effect of INF-α is used in the clinic to treat patients with myeloproliferative neoplasms, melanoma, or renal cell carcinoma.13,14
In this study, we found that tumor cells from patients with JAK1 mutation-positive ALL exhibit a type I IFN signature. In vitro, we showed that several JAK1 mutants constitutively activate components of type I IFN receptor signaling cascade, such as TYK2, STAT2, and STAT1. Moreover, cell lines expressing JAK1 mutants were hypersensitive to the antiproliferative effect of type I IFNs, which in turn resulted in a better response to the antitumorigenic effect of IFN in an in vivo leukemia model.
Methods
Patients and microarray data analyses
Cohorts used for gene expression profiling included 3 JAK1 mutation-positive (S512L, R724H 2x) and 8 mutation-negative T-cell ALL (T-ALL) samples from patients enrolled in the adult clinical trial Gruppo Italiano Malattie Ematologiche dell'Adulto 0496 and 2000 as described previously.2 An independent cohort included 6 JAK1 mutation-positive (S512L, R724H/C/Q and Y652H 2x) and 11 mutation-negative T-ALL samples from the Group for Research on Adult Acute Lymphoblastic Leukemia study.15 Written informed consent for genetic analyses was obtained from all subjects according to the Declaration of Helsinki, and experimental procedures were approved by the institutional review board of all participating institutions. Raw Affymetrix microarray data were obtained as described2 and are accessible at the Gene Expression Omnibus public database (GSE18239). Raw data were normalized by global scaling on all probe sets, with a target signal of 100. Genes called absent in more than 8 samples were removed from the analysis, which was performed on the remaining 27 049 genes. For Gene Set Enrichment Analysis (GSEA), a supervised comparison of the JAK1 mutation–positive and –negative blast samples was performed with the Significance Analysis of Microarrays tool implemented in the TIGR MeV software (http://www.tm4.org/mev.html). Genes were classified from 1 (more expressed in the JAK1-mutated samples) to 27 049 (genes more expressed in the JAK1 wild-type samples) according to the Significance Analysis of Microarrays score. A preranked analysis was performed in the GSEA software (http://www.broad.mit.edu/cancer/software/gsea) with the curated gene sets.16 By this analysis, 20 gene sets were significantly (FWER P < .05) enriched in the JAK1 mutation–positive blast samples, and 20 gene sets were significantly enriched in the JAK1 mutation–negative blast samples. The clustering based on an independent list of IFN-stimulated genes (ISGs) was performed as follows: from the 39 genes induced by IFN in peripheral blood mononuclear cells (PBMCs),17 29 genes were called present in more than 3 ALL samples. These genes were represented by 80 probe sets. The analysis was performed according to these probe sets. A hierarchical clustering of the samples and of the genes, based on the Euclidian distance, was performed in the TIGR MeV software (Dana-Farber Cancer Institute).
Cell culture and cytokines
BaF3 mouse pro-B cells were cultured in Dulbecco modified Eagle medium with fetal bovine serum (10%) and IL-3 (100 U/mL). BW5147 mouse T-cell leukemia cells18 were grown in Iscove-Dulbecco medium supplemented with 10% fetal bovine serum, 0.55mM l-arginine, 0.24mM l-asparagine, and 1.25mM l-glutamine. Recombinant human IL-9 was produced and purified in our laboratory. Murine IFN-α6T, -α11, and -β supernatants were produced by transient transfection of HEK293 cells as described.19 Control supernatant was produced with the use of the corresponding empty vector. Typically, HEK293 cells produced supernatants that contained 250 000 U/mL IFNs.19 Recombinant murine IFN-β was purchased from PBL Interferon Source.
Plasmid construction, stable DNA transfections, and analysis of transfected cells
All JAK1 mutants (A634D, A634D/Y107A, R724H, R879C) were generated with the use of the QuickChange XL II site-directed mutagenesis kit (Stratagene) and subcloned into the pMX-GFP biscistronic retroviral vector as described.2,5,20 For stable transduction, 0.5 × 106 BW5147 or BaF3 cells were infected by retroviruses produced by BOSC packaging cells with the use of standardized protocol.21 Cells were subsequently sorted by fluorescence-activated cell sorting for equal level of green fluorescent protein.
Transient cell transfection and luciferase assay
Two reporter plasmids were used: pGL3-pap1-luc and pIRF7-luc. STAT3-mediated transcription was evaluated with pGL3-pap1-luc plasmid (obtained from Dr Jan Tavernier, Ghent University) containing the luciferase gene under the control of the STAT3-inducible rat Pap1 promoter.22 The pIRF7-luc reporter plasmid contains the murine IRF-7 promoter,23 which is able to bind ISGF3 complex. Another plasmid, pRL-TK, encoding Renilla luciferase, was cotransfected as an internal control. Briefly, 12 × 106 BW5147 cells stably transduced with the different JAK1 constructs were electroporated with 15 μg of luciferase reporter plasmid and 1 μg of pRL-TK plasmid and were seeded in 96-well plates at 5 × 105 cells/well. Cells were stimulated by mock/control or IFN-β HEK293 supernatants or hIL-9 (100 U/mL) for 2 hours before luciferase assays were performed with the use of the dual luciferase reporter kit (Promega).
Western blots
BW5147 or BaF3 cells (106) were starved for 4 hours, then collected and lysed in 250 μL of Laemmli buffer (Bio-Rad). Phosphorylation of signaling proteins was investigated by Western blot as described previously5 with the use of the following phospho-specific antibodies from Cell Signaling Technology: anti-pY1022/1023 JAK1 (no. 3331S), anti-pY1054/1055 TYK2 (no. 9321S), anti-pY701 STAT1 (no. 9171S) and from Upstate anti-pY689 STAT2 (no. 07-224). Blots were reprobed with anti-JAK1 (no. 3332), anti-STAT2 (no. 4597; Cell Signaling Technology), anti-STAT1 (no. S21120; Signal Transduction Laboratory) or anti–β-actin (Sigma-Aldrich) antibodies.
Proliferation assay
BW5147 or 3000 BaF3 stably transduced cells (4000) were seeded in 96-well plates and stimulated by murine IFN-α6T, -α11, -β, and mock/control HEK293 supernatants or recombinant murine IFN-β at different dilutions, starting from 1%. Alternatively, cells were treated by various concentrations of doxorubicin (Sigma-Aldrich) or cytosine arabinoside (ARA-C; Sigma-Aldrich). For all conditions, 150 U/mL IL-3 was added to the BaF3 cell cultures to ensure proliferation of the nonautonomous cells. After 72 hours, tritiated thymidine was added to the cells for 4 hours, and thymidine incorporation was measured with a Top Count microplate scintillation counter (Canberra-Packard).
Quantitative reverse transcription–polymerase chain reaction
Cells (106) were stimulated for 3 hours with IL-9 (100 U/mL) or with IFN-β (1% HEK293 SN) or left unstimulated. Total RNA was extracted with the use of the TriPure isolation reagent (Roche). Reverse transcription was performed on 1 μg of total RNA with an oligo-(dT) primer (Roche) and M-MLV RT (Invitrogen). Quantitative polymerase chain reactions (q-PCRs) were performed with the use of 50 μL of cDNA and primer sets corresponding to IRF-7, Oasl-2, Bcl-3, and β-actin with qPCR Mastermix for SYBR Green I (Eurogentec). The sequences of murine primers (final concentration, 300nM) were as follows: mIRF-7, 5′-CAGCGAGTGCTGTTTGGAGAC-3′ (forward) and 5′-AAGTTCGTACACCTTATGCGG-3′ (reverse); mOasl-2, 5′-GGATGCCTGGGAGAGAATCG-3′ (forward) and 5′-TCGCCTGCTCTTCGAAACTG-3′ (reverse); mBcl-3, 5′-TCCTGAGCGCAAGTACTCTGT-3′ (forward) and 5′-CTGATCCACATCTGCTGGAAG-3′(reverse); mβ-actin, 5′-GCTGGAAGGTGGACAGTGAG-3′ (forward) and 5′-CTCTGGCTCCTAGCACCATGAAG-3′ (reverse). The sequences of human primers were as follows: hIRF-7, 5′-AGAAGAGCCTGGTCCTGGTGAA-3′ (forward) and 5′-CGATGTCGTCATAGAGGCTGTTG-3′ (reverse); hMX1, 5′-GAGCTGCCAGGCTTTGTGAA-3′ (forward) and 5′-GGCGGATCAGCTTCTCACCT-3′ (reverse); hβ-actin, 5′-GTCATACTCCTGCTTGCTGA-3′ (forward) and 5′-ATTGCCGAC-AGGATGCAGAA-3′ (reverse) and 5′-TCAAGATCATTGCTCCTCCTGAGC-3′ (probe). Samples were first heated 2 minutes at 50°C then 10 minutes at 95°C. cDNA was amplified as follows: 40 cycles of a 2-step PCR program at 95°C for 15 seconds and 60°C for 1 minute. Melting point analysis was carried out by heating the amplicon from 60°C to 95°C. Results were analyzed by MyiQ software (Bio-Rad). Starting quantity of the transcripts was calculated with the use of using standard curves from a cDNA clone.
Mice
Female 6- to 9-week-old severe combined immunodeficient or RAG2−/− mice were obtained from the animal facility of the Université Catholique de Louvain, Belgium. Handling of mice and experimental procedures were conducted in accordance with national and institutional guidelines for animal care.
In vivo leukemia model
BW5147 is a T-leukemia cell line obtained from AKR mice.18 BW5147 cells or BW5147 cells expressing JAK1A634D were resuspended at the concentration of 106 cells in 200 μL of phosphate-buffered saline. Cells (106) were injected intraperitoneally into mice 2 days after the intramuscular electroinjection of murine IFN-α6T subcloned into pcDNA3 or the empty vector. Mice were examined daily for 3 months in a blind way for tumor growth. Results were represented by Kaplan-Meier curves, and statistics were performed with the Gehan-Breslow-Wilcoxon test in the package “survival” of the program R (http://www.R-project.org).
DNA electroinjection
Mice were anesthetized with 200μL of a mix of medetomidine hydrochloride (Dormitor; Pfizer) 100 μg/mL and ketamine (Anesketin; Eurovet) 500 μg/mL given intramuscularly. Before DNA injection, mice were shaved locally. Endotoxin-free plasmid DNA (10 μg) in 25 μL of phosphate-buffered saline was injected in the tibialis anterior muscles of the mice. Electric pulses (80 V per 4 mm, 8 pulses, 20 msec/pulse; pause, 480 milliseconds) were administered with the use of a Cliniporator system (Cliniporator; IGEA) with 6-mm electrode plates. Mice were awakened up by intramuscular injection of 250 μL of atipamezole 500 μg/mL (Antisedan; Pfizer).
Results
A type I IFN transcriptional signature characterizes JAK1 mutation–positive ALL
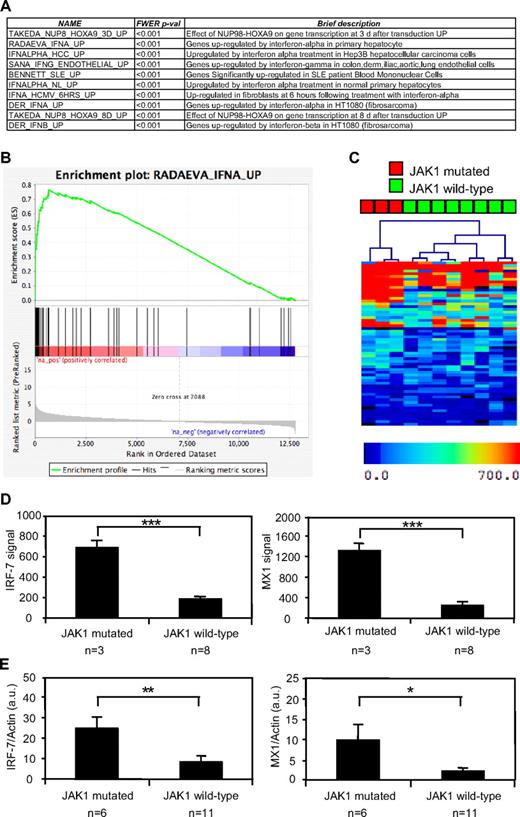
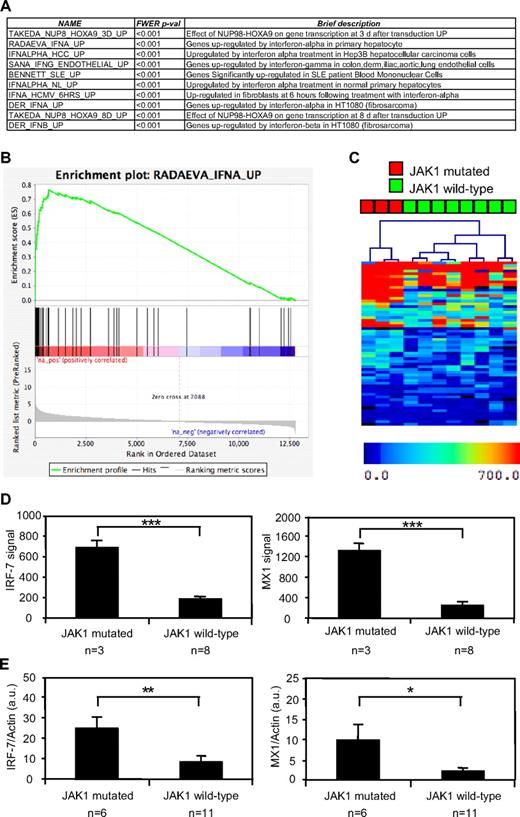
To identify the signaling pathways regulated by constitutively active mutations in JAK1, we performed a GSEA based on the genes differentially expressed by 3 JAK1 mutation–positive T-ALL samples compared with 8 JAK1 mutation–negative T-ALL samples. GSEA is a computational method for determining whether a rank-ordered list of genes for a particular comparison of interest (eg, samples with mutated versus wild-type JAK1) is enriched in genes derived from an independently generated gene set (eg, genes induced by IFN-α in primary hepatocytes). Among the 1892 predefined gene sets available from the curated Molecular Signature Database,24 the GSEA analysis showed that 20 gene sets were significantly overexpressed in the JAK1 mutation–positive cases compared with the samples without a mutated JAK1 allele. As shown in Figure 1A for the 10 most enriched gene sets, most of these gene sets contained either type I IFN-induced genes or were composed of genes found in situations in which type I IFN can be produced to activate IFN-target genes, such as in systemic lupus erythematosus. Figure 1B shows a typical GSEA enrichment plot illustrating the enrichment in IFN target genes among those that were up-regulated in JAK1 mutation–positive ALL blasts. These results suggested that JAK1 mutation–positive T-ALL cells were characterized by the activation of pathways leading to the expression of type I IFN target genes. On the basis of this observation, we hypothesized that clustering of our ALL samples based on another IFN target gene signature should clearly distinguish those with or without a JAK1 mutation. Focusing on genes induced by IFN in hematopoietic cells, we selected a list of genes shown to be induced by IFN in PBMCs.17 As shown in Figure 1C, hierarchical clustering based on this signature perfectly separated JAK1 mutation–positive ALL cases from those without a JAK1 mutation. To confirm these observations, we used an independent set of 6 JAK1 mutation–positive and 11 mutation-negative T-ALL samples15 to assess the expression of 2 IFN target genes, IRF-7 and MX1. These genes are good surrogates of the IFN signature and were overexpressed in the first cohort of JAK1-mutated samples based on the microarray data (Figure 1D). As shown in Figure 1E, both IFN target genes were significantly up-regulated in the 6 independent JAK1 mutation–positive compared with 11 mutation-negative T-ALL samples. Altogether, these data show that JAK1 mutation–positive T-ALL cells are characterized by a type I IFN transcriptional signature.
JAK1 mutation–positive T-ALL cells show a type I IFN transcriptional signature. The gene expression profile of human JAK1 mutation–positive and mutation–negative T-ALL cells was assessed with pangenomic microarrays. (A) The list of the 10 gene sets most enriched in cells with a mutated JAK1 allele, according to a GSEA analysis, is shown. (B) The figure represents the GSEA enrichment plot of one of the most enriched gene sets. The lower part of this plot shows the ranking of the genes relative to their enrichment in JAK1 mutation–positive cells. The medium part shows the ranking of the hits from the IFN-induced gene set and the top part shows the resulting enrichment profile. (C) Hierarchical clustering based on the genes induced by IFN-α in PBMCs. (D) The mean microarray signal for the IFN targets hIRF-7 (right) and hMX1 (left) is represented for 3 JAK1 mutation–positive compared with 8 mutation-negative T-ALL samples. The differences between JAK1 mutation–positive and mutation–negative samples were extremely significant (***P < .001, t test). Results are mean ± SEM. (E) Quantitative analysis hIRF-7 (left) and hMX1 (right) expression, based on an independent set of 6 JAK1 mutation–positive and 11 mutation–negative T-ALL samples. Transcripts were normalized to the level of the β-actin transcripts. The differences between JAK1 mutation–positive and mutation–negative ALL samples were significant (**P < .007, *P < .035, t test). Results are mean ± SEM.
JAK1 mutation–positive T-ALL cells show a type I IFN transcriptional signature. The gene expression profile of human JAK1 mutation–positive and mutation–negative T-ALL cells was assessed with pangenomic microarrays. (A) The list of the 10 gene sets most enriched in cells with a mutated JAK1 allele, according to a GSEA analysis, is shown. (B) The figure represents the GSEA enrichment plot of one of the most enriched gene sets. The lower part of this plot shows the ranking of the genes relative to their enrichment in JAK1 mutation–positive cells. The medium part shows the ranking of the hits from the IFN-induced gene set and the top part shows the resulting enrichment profile. (C) Hierarchical clustering based on the genes induced by IFN-α in PBMCs. (D) The mean microarray signal for the IFN targets hIRF-7 (right) and hMX1 (left) is represented for 3 JAK1 mutation–positive compared with 8 mutation-negative T-ALL samples. The differences between JAK1 mutation–positive and mutation–negative samples were extremely significant (***P < .001, t test). Results are mean ± SEM. (E) Quantitative analysis hIRF-7 (left) and hMX1 (right) expression, based on an independent set of 6 JAK1 mutation–positive and 11 mutation–negative T-ALL samples. Transcripts were normalized to the level of the β-actin transcripts. The differences between JAK1 mutation–positive and mutation–negative ALL samples were significant (**P < .007, *P < .035, t test). Results are mean ± SEM.
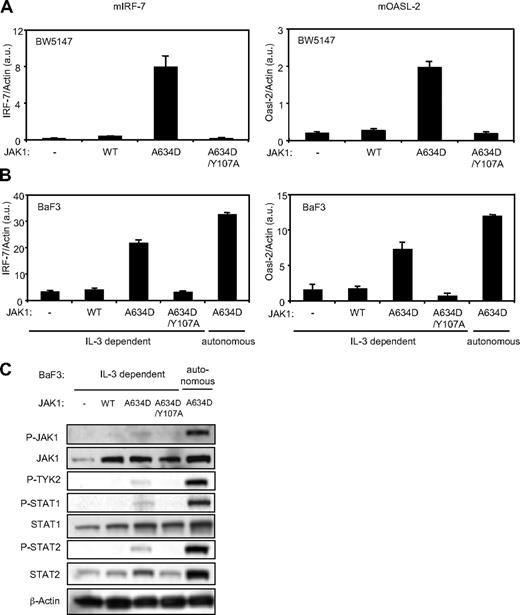
The JAK1A634D mutant, but not JAK1A634D/Y107A, mediates constitutive expression of type I IFN–regulated genes and promotes constitutive phosphorylation of the components of the type I IFN signal transduction pathway in BW5147 and BaF3 cells
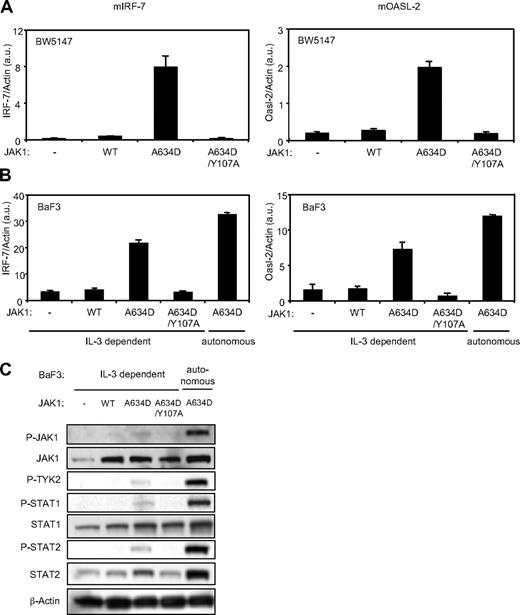
On the basis of the interferon signature observed in JAK1 mutation–positive ALL samples, we hypothesized that ALL-associated JAK1 mutants bind to type I IFN receptor complex and constitutively activate type I IFN receptor signal transduction. To address this hypothesis, we stably expressed JAK1A634D, one of the ALL-associated JAK1 mutants, in BW5147, a murine T-ALL cell line, and BaF3, a murine pro-B cell line by retroviral infection. The level of expression of IRF-7 and Oasl-2, 2 specific type I IFN–regulated genes, was assessed by quantitative reverse transcription (RT)–PCR in cells transduced by JAK1WT and JAK1A634D, with or without the FERM domain mutation Y107A, which is known to abolish JAK1 binding to receptors. As shown in Figure 2A and B, expression of the JAK1A634D mutant promoted constitutive expression of IRF-7 and Oasl-2. As expected, the FERM domain mutation abolished this effect, supporting the idea that JAK1 binding to the receptor is required for the activating effect of the JAK1 mutation on signaling.5 Expression of the JAK1A634D mutant in BaF3 allows for selection of IL-3–independent autonomous cells, which was associated with constitutive STAT5 activation.2 As shown in Figure 2, these autonomously growing cells expressed even higher levels of IRF-7 and Oasl-2 than JAK1A634D-expressing BaF3 nonautonomous cells proliferating in a medium containing IL-3, suggesting a stronger activation of type I IFN signal transduction in the former. To dissect the mechanism of constitutive expression of type I IFN–regulated genes in JAK1A634D-expressing BaF3 cells, we studied the activation status of proteins involved in the signal transduction of type I IFN receptor complex. We observed that JAK1A634D was constitutively phosphorylated in BaF3 cells. Interestingly, phosphorylation of TYK2 paralleled the JAK1 phosphorylation pattern (Figure 2C). Because TYK2 and JAK1 are, respectively associated with IFNAR1 and IFNAR2, this suggested that TYK2 represents a partner of JAK1 in mediating the type I IFN–stimulated signal and is cross-phosphorylated and activated by JAK1A634D. Constitutive phosphorylation of STAT1 and of STAT2, which is a specific feature of the type I IFN receptor complex activation, was also detected in BaF3 cells expressing the JAK1 mutant, confirming constitutive activation of the pathway. Phosphorylation and activation of all these signaling molecules was increased after autonomous selection. Of note, total STAT2 protein level was also increased in cells expressing JAK1A634D (Figure 2C), in line with previous observations showing that STAT2 expression is up-regulated on IFN stimulation.25 Constitutive phosphorylation of JAK1, STAT1, and STAT2 was also observed in JAK1A634D-expressing BW5147 cells (data not shown).
The JAK1A634D mutant, but not JAK1A634D/Y107A, mediates constitutive expression of type I IFN–regulated genes and promotes constitutive phosphorylation of the components of the type I IFN signal transduction pathway in BW5147 and BaF3 cells. Total RNA was extracted from 106 BW5147 (A) and BaF3 (B) cells stably transduced by JAK1WT or JAKA634D with intact or mutated Y107A FERM domain (JAK1A634D/Y107A). To quantify basal expression of type I IFN–induced genes, such as mIRF-7 (left) and mOasl-2 (right), quantitative RT-PCR was performed. Transcripts were normalized to the level of the murine β-actin transcripts. Results are mean ± SD of biologic triplicates. Similar results were obtained in 3 independent experiments. (C) BaF3 cells (106) stably transduced with JAK1WT or JAKA634D, with intact or mutated Y107A FERM domain (JAK1A634D/Y107A), were starved for 4 hours, lysed, and subjected to Western blot analysis. Basal phosphorylation of JAK1, TYK2, STAT1, and STAT2 was detected with specific anti-pY1022/1023 JAK1, anti-pY1054/1055 TYK2, anti-pY701 STAT1, and anti-pY689 STAT2 antibodies. Membranes were reprobed with anti-JAK1, anti-STAT1, anti-STAT2, and anti–β-actin antibodies as control. Similar results were obtained in 3 independent experiments for the BaF3 and BW5147 cell lines. WT indicates wild-type.
The JAK1A634D mutant, but not JAK1A634D/Y107A, mediates constitutive expression of type I IFN–regulated genes and promotes constitutive phosphorylation of the components of the type I IFN signal transduction pathway in BW5147 and BaF3 cells. Total RNA was extracted from 106 BW5147 (A) and BaF3 (B) cells stably transduced by JAK1WT or JAKA634D with intact or mutated Y107A FERM domain (JAK1A634D/Y107A). To quantify basal expression of type I IFN–induced genes, such as mIRF-7 (left) and mOasl-2 (right), quantitative RT-PCR was performed. Transcripts were normalized to the level of the murine β-actin transcripts. Results are mean ± SD of biologic triplicates. Similar results were obtained in 3 independent experiments. (C) BaF3 cells (106) stably transduced with JAK1WT or JAKA634D, with intact or mutated Y107A FERM domain (JAK1A634D/Y107A), were starved for 4 hours, lysed, and subjected to Western blot analysis. Basal phosphorylation of JAK1, TYK2, STAT1, and STAT2 was detected with specific anti-pY1022/1023 JAK1, anti-pY1054/1055 TYK2, anti-pY701 STAT1, and anti-pY689 STAT2 antibodies. Membranes were reprobed with anti-JAK1, anti-STAT1, anti-STAT2, and anti–β-actin antibodies as control. Similar results were obtained in 3 independent experiments for the BaF3 and BW5147 cell lines. WT indicates wild-type.
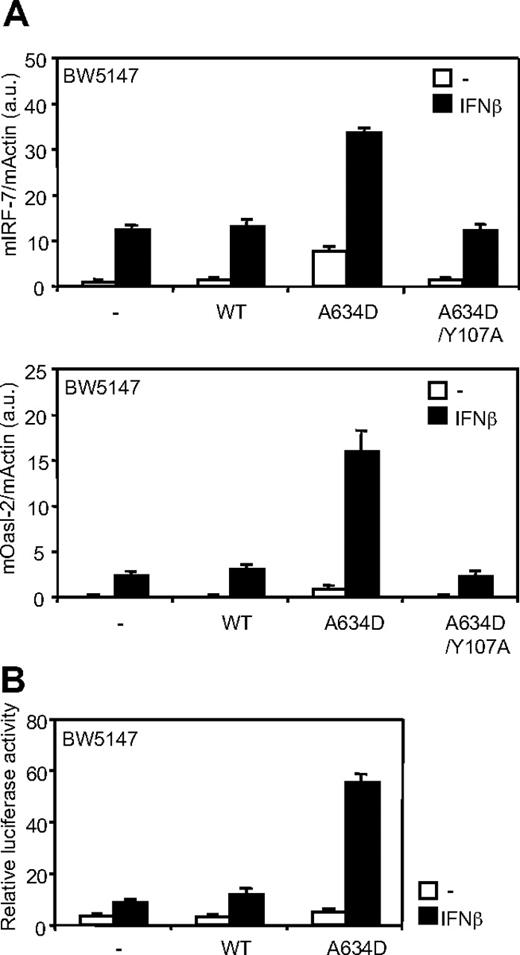
JAK1A634D up-regulates type I IFN responsiveness
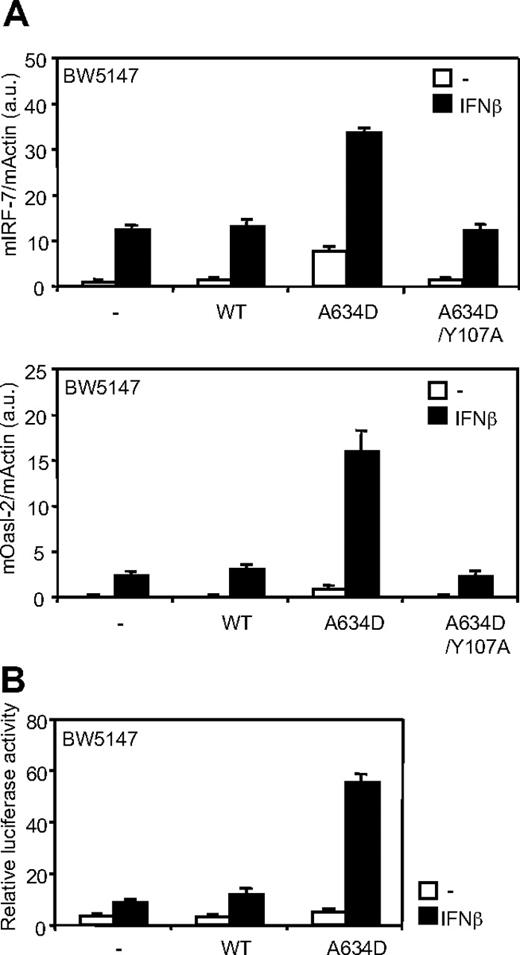
The V678F amino acid substitution in TYK2, the IFNAR1-associated kinase, has been previously shown to constitutively activate STAT3 without affecting ligand-induced signaling through the type I IFN receptor.26 We investigated whether the JAK1A634D mutant could enhance type I IFN responsiveness. Figure 3A shows that expression of JAK1A634D not only promoted constitutive transcription of type I IFN–regulated genes (IRF-7 and Oasl-2) but also significantly up-regulated IFN-β–dependent induction of these genes. To confirm that the JAK1 mutant up-regulates ligand-induced signaling through the type I IFN receptor, we took advantage of the luciferase reporter construct, pIRF7-luc, which uses the ISGF3-responsive IRF-7 promoter to control luciferase transcription. As shown on Figure 3B, reporter transactivation was increased in all IFN-β–stimulated BW5147 cells, but this induction was 3- to 4-fold higher in cells expressing JAK1A634D. Consistent results were obtained in BaF3 cells (data not shown). Overall, these data strongly suggest that, in contrast to the TYK2V678F mutant, the ALL-associated JAK1A634D mutant potentiates ligand-induced signaling through the type I IFN receptor complex.
JAK1A634D up-regulates ligand-induced signaling through the type I IFN receptor. (A) BW5147 cells (106) stably transduced with JAK1WT or JAKA634D, with intact or mutated Y107A FERM domain (JAK1A634D/Y107A), were stimulated for 3 hours with 1% IFN-β or mock/control HEK293 supernatant, lysed, and used for total RNA extraction. To quantify ligand-induced expression of type I IFN–induced genes, quantitative RT-PCR was performed with primers for mIRF-7 and mOasl-2 genes. Transcripts were normalized to the level of the murine β-actin transcripts. Results are mean ± SD of biologic triplicates. Similar results were obtained in 3 independent experiments in BW5147 and BaF3 cell lines. (B) BW5147 cells (12 × 106) stably transduced by JAK1WT or JAK1A634D were electroporated with 15 μg of pIRF7-luc luciferase reporter plasmid, which contains the promoter of the murine IRF-7, and 1 μg of internal control pRL-TK plasmid. Cells were seeded in 96-well plates at 5 × 105 cells/well and stimulated with 1% IFN-β or mock/control HEK293 supernatants for 2 hours before luciferase assay was performed. Results are mean ± SD of biologic triplicates. Similar results were obtained in 3 independent experiments. WT indicates wild-type.
JAK1A634D up-regulates ligand-induced signaling through the type I IFN receptor. (A) BW5147 cells (106) stably transduced with JAK1WT or JAKA634D, with intact or mutated Y107A FERM domain (JAK1A634D/Y107A), were stimulated for 3 hours with 1% IFN-β or mock/control HEK293 supernatant, lysed, and used for total RNA extraction. To quantify ligand-induced expression of type I IFN–induced genes, quantitative RT-PCR was performed with primers for mIRF-7 and mOasl-2 genes. Transcripts were normalized to the level of the murine β-actin transcripts. Results are mean ± SD of biologic triplicates. Similar results were obtained in 3 independent experiments in BW5147 and BaF3 cell lines. (B) BW5147 cells (12 × 106) stably transduced by JAK1WT or JAK1A634D were electroporated with 15 μg of pIRF7-luc luciferase reporter plasmid, which contains the promoter of the murine IRF-7, and 1 μg of internal control pRL-TK plasmid. Cells were seeded in 96-well plates at 5 × 105 cells/well and stimulated with 1% IFN-β or mock/control HEK293 supernatants for 2 hours before luciferase assay was performed. Results are mean ± SD of biologic triplicates. Similar results were obtained in 3 independent experiments. WT indicates wild-type.
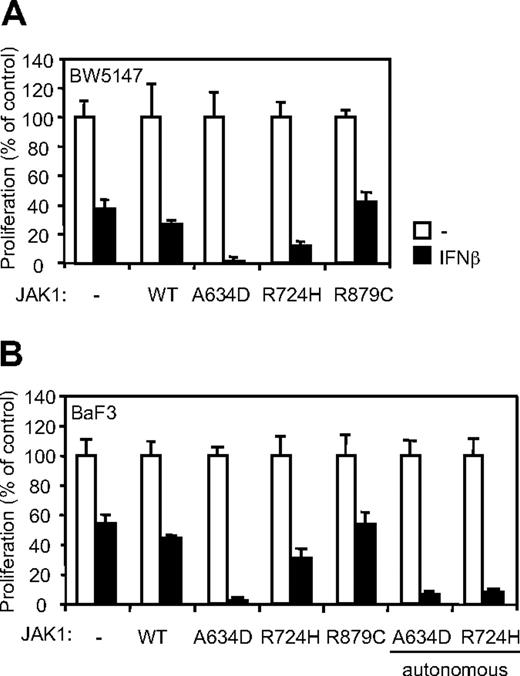
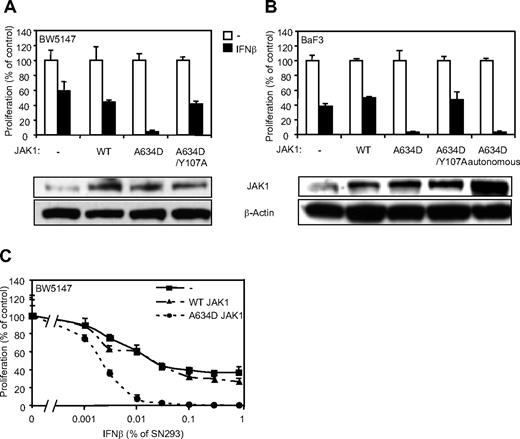
JAK1A634D potentiates type I IFN–induced cell growth inhibition
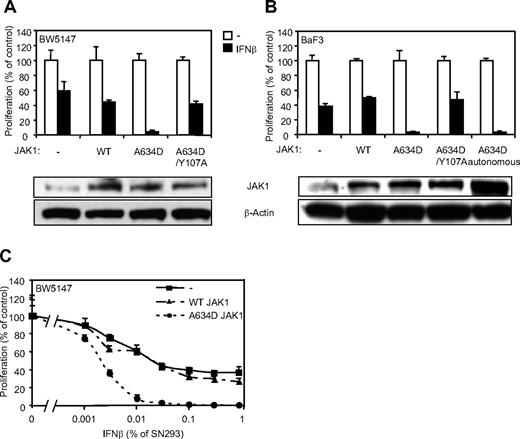
Type I IFNs are known to exert antiproliferative activity on various cell types. Our observations suggest that JAK1A634D-expressing cells might be particularly sensitive to this activity. To address this hypothesis, we analyzed proliferation of cells expressing a wild-type or mutated JAK1 in the presence of IFN-β. Although IFN-β reduced the proliferation of untransduced and JAK1WT-expressing BW5147 cells after 3 days (Figure 4A,C), it completely inhibited proliferation of cells expressing the JAK1A634D mutant. In line with our previous observations, the effect of the A634D mutation on cell proliferation in the presence of IFN-β was dependent on receptor binding, because the expression of JAK1A634D/Y107A abolished cell hypersensitivity to IFN-β. Similar experiments were performed in BaF3 cells, in which IFN-β treatment resulted in complete inhibition of proliferation of JAK1A634D-expressing cells, which was not observed in cells expressing JAK1WT or JAK1A634D/Y107A (Figure 4B). Interestingly, autonomous and IL-3–dependent BaF3 cells expressing JAK1A634D exhibited a similar hypersensitivity to IFN-β, indicating that even cells that depend on the JAK1 mutant for their autonomous proliferation retain hypersensitivity to IFNs. Because all previous experiments were performed with the use of IFN-β supernatant produced by HEK293 cells, we used purified mouse recombinant IFN-β to confirm the specificity of IFN-β–related inhibition of proliferation. As expected, mouse recombinant IFN-β completely inhibited proliferation of JAK1A634D-expressing cells but not that of parental or JAK1WT-expressing BW5147 cells (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
JAK1A634D confers hypersensitivity to IFN-β in BW5147 and BaF3 cells. (A) BW5147 (4000) or (B) BaF3 (3000) cells stably transduced by JAK1WT or JAK1A634D, with intact or mutated Y107A FERM domain (JAK1A634D/Y107A), were seeded in 96-well plates and stimulated by 1% IFN-β or mock/control HEK293 supernatants. After 72 hours, tritiated thymidine was added to the cells for 4 hours, and thymidine incorporation was measured. In parallel 106 BW5147 or BaF3 cells were starved for 4 hours, lysed, and subjected to Western blot analysis. Total JAK1 and β-actin levels were assessed with anti-JAK1 and anti–β-actin antibodies. (C) BW5147 cells (4000) stably transduced by JAK1WT or JAK1A634D, with intact or mutated Y107A FERM domain (JAK1A634D/Y107A), were seeded in 96-well plates and stimulated by IFN-β HEK293 supernatants at different dilutions, starting from 1%. After 72 hours, tritiated thymidine was added to the cells for 4 hours, and thymidine incorporation was measured. Results are mean ± SD of biologic triplicates. Similar results were obtained in 3 independent experiments in BW5147 and BaF3 cell lines. WT indicates wild-type.
JAK1A634D confers hypersensitivity to IFN-β in BW5147 and BaF3 cells. (A) BW5147 (4000) or (B) BaF3 (3000) cells stably transduced by JAK1WT or JAK1A634D, with intact or mutated Y107A FERM domain (JAK1A634D/Y107A), were seeded in 96-well plates and stimulated by 1% IFN-β or mock/control HEK293 supernatants. After 72 hours, tritiated thymidine was added to the cells for 4 hours, and thymidine incorporation was measured. In parallel 106 BW5147 or BaF3 cells were starved for 4 hours, lysed, and subjected to Western blot analysis. Total JAK1 and β-actin levels were assessed with anti-JAK1 and anti–β-actin antibodies. (C) BW5147 cells (4000) stably transduced by JAK1WT or JAK1A634D, with intact or mutated Y107A FERM domain (JAK1A634D/Y107A), were seeded in 96-well plates and stimulated by IFN-β HEK293 supernatants at different dilutions, starting from 1%. After 72 hours, tritiated thymidine was added to the cells for 4 hours, and thymidine incorporation was measured. Results are mean ± SD of biologic triplicates. Similar results were obtained in 3 independent experiments in BW5147 and BaF3 cell lines. WT indicates wild-type.
It is well known that a higher cell proliferation rate can result in a higher sensitivity to all kind of drugs. To rule out the possibility that expression of JAK1A634D would induce a nonspecific increase in drug sensitivity, we studied the activity of doxorubicin and ARA-C, 2 standard chemotherapy used for ALL, on the proliferation of BW5147 cells expressing JAK1WT or JAK1A634D. As shown in supplemental Figure 1B and C, the hypersensitivity of the JAK1A634D-expressing cells was specific for IFN-β, because the antiproliferative activity of ARA-C or doxorubicin was not affected by the expression of JAK1A634D.
To check whether JAK1A634D also confers hypersensitivity to other type I IFNs, we compared the proliferation of BW5147 cells expressing JAK1WT or JAK1A634D in the presence of murine IFN-α6T and IFN-α11. Previous studies on the murine B6 melanoma cell line documented that IFN-α11 displays an antiproliferative effect that is comparable with that exerted by IFN-β, whereas IFN-α6T shows a lower antiproliferative activity.19 As shown in supplemental Figure 2, JAK1A634D-expressing BW5147 cells, but not untransduced or JAK1WT-expressing BW5147 cells, were hypersensitive to all tested type I IFNs.
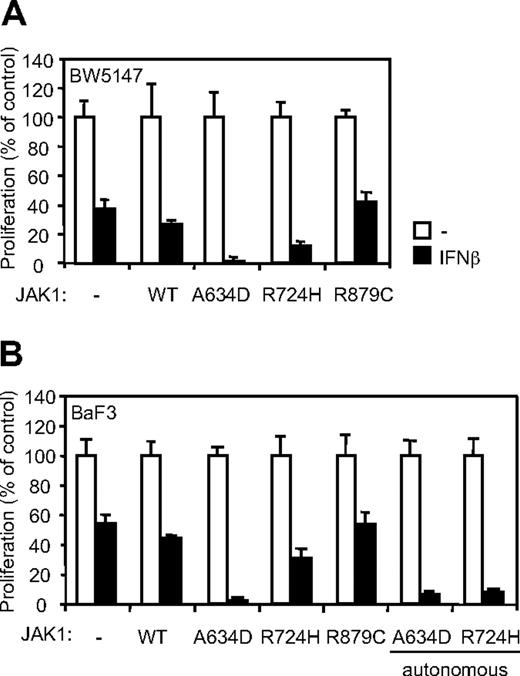
The R724H but not the R879C mutation in JAK1 confers IFN-β hypersensitivity in BW5147 and BaF3 cells
We showed previously that, beside JAK1A634D, the ALL-associated JAK1R724H mutant induced autonomous growth of the cytokine-dependent BaF3 cell line, whereas a third ALL-associated mutant, JAK1R897C, protected the BW5147 cell line from dexamethasone-induced apoptosis.2 These observations supported a gain-of-function role for these mutations. It also suggested that such a differential biologic outcome might be dependent on a specific effect of JAK1 mutants on signaling mediated by distinct endogenously expressed cytokine receptors. We therefore examined whether the JAK1R724H and JAK1R879C mutants confer hypersensitivity to IFNs in BW5147 and BaF3 cells. Remarkably, expression of the JAK1R724H mutant in both cell lines slightly potentiated the inhibitory effect of IFN-β on cell proliferation, whereas expression of JAK1R879C at a similar level did not affect the IFN response in both cell lines (Figure 5). Of note, the JAK1R724H-mediated inhibition of proliferation appeared to be more pronounced in BaF3 cells selected for autonomous growth. These results show a differential effect of ALL-associated JAK1 mutations in potentiating IFN response, with the A634D mutation having the strongest activity.
The JAK1R724H and the JAK1A634D mutants, but not JAK1R879C, confer IFN-β hypersensitivity in BW5147 and BaF3 cells. (A) BW5147 (4000) or (B) BaF3 (3000) cells stably transduced by JAK1 wild-type (WT) or different JAK1 mutants were seeded in 96-well plates and stimulated by 1% IFN-β or mock/control HEK293 supernatants. After 72 hours, tritiated thymidine was added to the cells for 4 hours, and thymidine incorporation was measured. Results are mean ± SD of biologic triplicates. Similar results were obtained in 3 independent experiments in BW5147 and BaF3 cell lines.
The JAK1R724H and the JAK1A634D mutants, but not JAK1R879C, confer IFN-β hypersensitivity in BW5147 and BaF3 cells. (A) BW5147 (4000) or (B) BaF3 (3000) cells stably transduced by JAK1 wild-type (WT) or different JAK1 mutants were seeded in 96-well plates and stimulated by 1% IFN-β or mock/control HEK293 supernatants. After 72 hours, tritiated thymidine was added to the cells for 4 hours, and thymidine incorporation was measured. Results are mean ± SD of biologic triplicates. Similar results were obtained in 3 independent experiments in BW5147 and BaF3 cell lines.
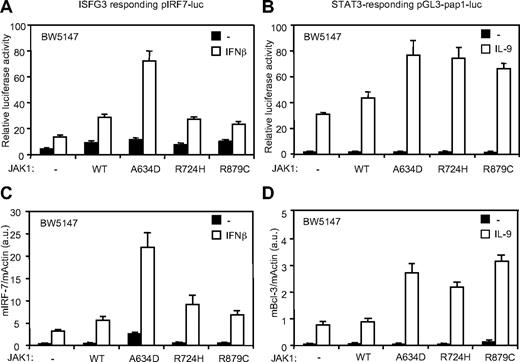
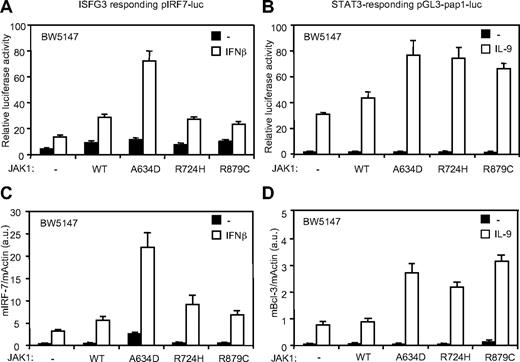
ALL-associated JAK1 mutants differentially potentiate IFN-β and IL-9 response
These observations support the hypothesis that different activating JAK1 mutations might have distinct activities because they potentiate signaling by distinct cytokine receptor complexes. We took advantage of the BW5147 cell line, which endogenously express 2 JAK1-binding receptors (IL-9Rα and IFNAR2) to compare the activity of different JAK1 mutants on distinct signaling pathways. The STAT3-responsive pGL3-pap1-luc reporter construct and quantitative RT-PCR for the STAT3-induced Bcl-3 gene were used as read-out for the IL-9 receptor signaling pathway.23,27 For the type I IFN pathway, we used the ISGF3 pIRF7-luc reporter plasmid and a quantitative RT-PCR for IRF-7. As shown in Figure 6, the effect of IL-9 was increased by the 3 JAK1 mutants, as documented by luciferase assay and Bcl-3 expression. By contrast, only JAK1A634D significantly increased IFN response, in line with the proliferation assays described above. Taken together, our observations suggest that ALL-associated JAK1 mutants have overlapping but also distinct intrinsic specificity to dysregulate signaling elicited by cytokine receptors, explaining the observed differential cytokine hypersensitivity.
JAK1A634D, JAK1R724H, and JAK1R879C differentially favor IFN-β and IL-9 signaling. (A-B) BW5147 cells (12 × 106) stably transduced by JAK1WT or JAK1A634D were electroporated with 15 μg of the ISGF-3 responsive pIRF7-luc (A) or the STAT3 responsive pGL3-pap1-luc (B) luciferase reporter plasmid and 1 μg of internal control pRL-TK plasmid. Cells were seeded in 96-well plates at 5 × 105 cells/well and stimulated with 1% IFN-β or mock/control HEK293 supernatants (for pIRF7-luc) and by 100 U/mL hIL-9 (for pGL3-pap1) for 2 hours before luciferase assay was performed. Results are mean ± SD of biologic triplicates. Similar results were obtained in 3 independent experiments. (C-D) BW5147 cells (106) stably transduced by JAK1 wild-type (WT) or different JAK1 mutants were stimulated with 1% IFN-β or mock/control HEK293 supernatants (C) and stimulated with 100 U/mL hIL-9 (D) for 3 hours, lysed, and used for total RNA extraction. Real-time PCR was performed to quantify transcripts of mIRF-7 and mBcl-3 genes. Gene expressions were normalized to the level of the murine β-actin transcripts. Results are mean ± SD of biologic triplicates. Similar results were obtained in 3 independent experiments.
JAK1A634D, JAK1R724H, and JAK1R879C differentially favor IFN-β and IL-9 signaling. (A-B) BW5147 cells (12 × 106) stably transduced by JAK1WT or JAK1A634D were electroporated with 15 μg of the ISGF-3 responsive pIRF7-luc (A) or the STAT3 responsive pGL3-pap1-luc (B) luciferase reporter plasmid and 1 μg of internal control pRL-TK plasmid. Cells were seeded in 96-well plates at 5 × 105 cells/well and stimulated with 1% IFN-β or mock/control HEK293 supernatants (for pIRF7-luc) and by 100 U/mL hIL-9 (for pGL3-pap1) for 2 hours before luciferase assay was performed. Results are mean ± SD of biologic triplicates. Similar results were obtained in 3 independent experiments. (C-D) BW5147 cells (106) stably transduced by JAK1 wild-type (WT) or different JAK1 mutants were stimulated with 1% IFN-β or mock/control HEK293 supernatants (C) and stimulated with 100 U/mL hIL-9 (D) for 3 hours, lysed, and used for total RNA extraction. Real-time PCR was performed to quantify transcripts of mIRF-7 and mBcl-3 genes. Gene expressions were normalized to the level of the murine β-actin transcripts. Results are mean ± SD of biologic triplicates. Similar results were obtained in 3 independent experiments.
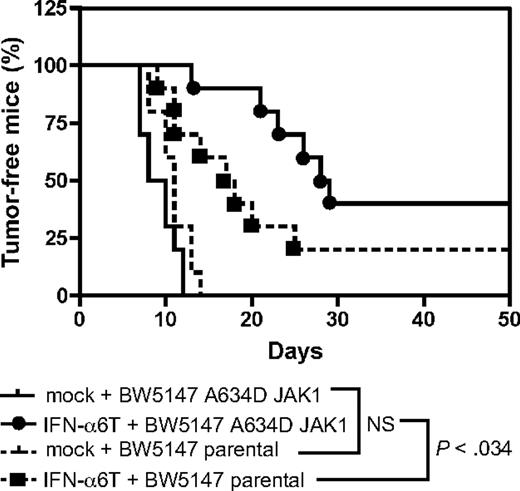
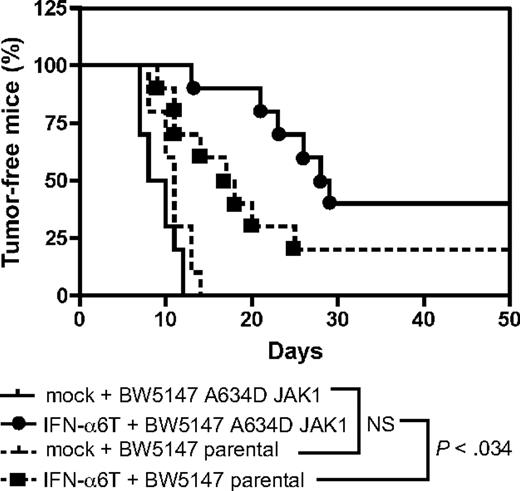
JAK1A634D-expressing leukemic cells are more sensitive to the antitumorigenic effect of type I IFN in vivo
Type I IFN are used in the clinic to treat several malignancies. Our in vitro data suggest that leukemic cells harboring a mutation in JAK1 are more sensitive to the antiproliferative effect of type I IFN. To check the relevance of this observation in vivo, we tested the effect of IFN-α in a murine model of leukemia, based on the intraperitoneal injection of the murine T-cell leukemia BW5147 cell line in immunocompromised mice. Treatment with IFN-α was achieved by intramuscular electroinjection of a murine IFN-α construct, a procedure that generates endogenous production of IFN-α with systemic effects lasting for 3 to 4 months.11 As shown in Figure 7, control mice receiving BW5147 cells showed tumor development after 1 week, and all mice developed a tumor after 2 weeks. This was similar for mice receiving JAK1A634D-expressing BW5147 cells. Treatment with IFN-α delayed the tumor formation of BW5147 cells, but this effect was more pronounced for JAK1A634D-expressing BW5147 cells. Thus, compared with control cells, JAK1A634D-expressing cells were significantly (P = .034) more sensitive to the antitumorigenic effect of IFN-α in vivo.
JAK1A634D potentiates the antitumoral effect of type I IFN in vivo. Rag2−/− mice received an intramuscular injection of an IFN-α6T construct or of an empty vector. Two days later, 106 BW5147 leukemia cells expressing or not JAK1A634D were injected intraperitoneally (10 mice/group). Kaplan-Meier curves of tumor development are shown. A Gehan-Breslow-Wilcoxon test shows that JAK1A634D expression significantly delayed tumor formation compared with control cells (P < .034). These results were reproduced in 3 experiments with either Rag2−/− or severe combined immunodeficient mice.
JAK1A634D potentiates the antitumoral effect of type I IFN in vivo. Rag2−/− mice received an intramuscular injection of an IFN-α6T construct or of an empty vector. Two days later, 106 BW5147 leukemia cells expressing or not JAK1A634D were injected intraperitoneally (10 mice/group). Kaplan-Meier curves of tumor development are shown. A Gehan-Breslow-Wilcoxon test shows that JAK1A634D expression significantly delayed tumor formation compared with control cells (P < .034). These results were reproduced in 3 experiments with either Rag2−/− or severe combined immunodeficient mice.
Discussion
In this study, we made 3 important observations. First, we identified a type I IFN transcriptional signature in human ALL cells harboring activating mutations in JAK1. This signature can be explained by the constitutive activation of the IFN receptor–signaling complex by the mutated JAK1 protein. Second, we showed that different JAK1 mutants activate distinct cytokine-receptor signaling complexes, showing specificity in signaling pathway activation based on the type of mutation in JAK1. Finally, we demonstrated that the JAK1A634D mutant confers hypersensitivity to the antitumorigenic effect of type I IFN in vivo, suggesting that IFN-α can be used as adjuvant treatment for JAK1 mutation–positive malignancies.
Recently, we and others reported that acquired gain-of-function mutations in JAK1 contribute to leukemogenesis.2-4 These mutations appear to represent a relatively common event in adult T-cell ALL, whereas they are more rare in adult B-cell ALL and in childhood malignancies.2,4 We also showed that JAK1 mutants need to associate to cytokine receptors, such as IL-2R and IL-9R, to promote constitutive activation of the receptor complex and signal transduction by STAT3 and STAT5.5 In this model, the receptor has a dual role to play: it serves as a scaffold for cross-activation of JAKs and as a docking site for STATs. Activation of different STAT transcription factors and aberrant expression of the related STAT-regulated genes might therefore reflect the activation of distinct cytokine receptor complexes. In this work, we observed that human JAK1 mutation–positive T-ALL samples were characterized by an overexpression of type I ISGs, suggesting constitutive activation of the type I IFN signaling pathway. This observation is best explained by the activation of the type I IFN receptor complex by the activated JAK1 protein. Several arguments favor this hypothesis: (1) IFNAR2, which is one of the chain of the type-I IFN receptor, binds JAK1; (2) the Y107A mutation, which abolishes JAK1 capacity to bind to receptors, also abolishes the constitutive activation of the IFN pathway; (3) TYK2, the kinase associated to IFNAR1, which is the other chain of the receptor, is constitutively activated in cell lines expressing the mutated form of JAK1, suggesting cross-activation of TYK2 by hyperactive JAK1; (4) STAT2, which is specifically activated through the type I IFN receptor complex, is also constitutively activated in cell lines expressing the ALL-associated mutated JAK1 allele.
The other JAK required for type I IFN signaling is TYK2, which is appended to IFNAR1. An activating mutation in TYK2 (V678F, homologous to JAK2V617F),20 constitutively activated STAT3, but not the canonical IFN-signaling pathway, and did not lead to activation of JAK1.26 Furthermore, in the model of 11.1 mutant cells that lack TYK2, mutant TYK2 did not affect IFN-induced signaling.26 By contrast, in BW5147 and BaF3 cells, expression of JAK1 mutants, namely A634D and R724H, potentiated the activation of type I IFN receptor signal transduction by IFN. Most importantly, the increased ligand responsiveness resulted into the hypersensitivity of JAK1A634D mutant–expressing cells to type I IFN–mediated antiproliferative effect in vitro and in almost complete inhibition of proliferation of these cells by several type I IFN ligands.
The fact that JAK1A634D mutant–expressing tumor cells, which are hypersensitive to type 1 IFN, express a constitutive IFN signature, but yet proliferate in vivo might seem paradoxical. However, it must be stressed that the IFN signature that we observe in JAK1A634D mutant–expressing cells remains below the gene expression level reached on IFN-β stimulation, even if JAK1 is not mutated, as shown in Figure 3. Thus, JAK1 mutation results in a low level of IFN signaling, below the antiproliferative threshold, and renders cells hyperresponsive to further IFN stimulation.
Among the 3 functionally characterized ALL-associated JAK1 mutants, JAKA634D was observed to be the strongest potentiator of the IFN response, whereas JAK1R724H weakly potentiate cell response to IFN and JAK1R879C had no effect. However, in the same cell line endogenously expressing IL-9R, all 3 JAK1 mutants conferred increased IL-9 responsiveness to a similar extend. We also previously showed that, beside the A634D mutant, the R724H JAK1 mutant induced autonomous growth of the IL-3–dependent BaF3 cell line, whereas the R897C mutant did not. In contrast, this JAK1 R897C protected the BW5147 cell line from dexamethasone-induced apoptosis.2 These findings are best explained by the idea that the capability of each JAK1 mutant to promote intracellular signaling is strictly dependent on the receptor-dependent context and by the differential expression of a set of cytokine receptors. Altogether these data document a level of specificity based on the type of mutation in JAK1 and strongly suggest that the different ALL-associated JAK1 mutants do not have a homogeneous dysregulating effect on intracellular signaling. This specificity could be explained by either a mutation-specific conformational change of the kinase resulting in a different binding affinity for the diverse cytokine receptors, a different capacity to cross-activate other JAK bound to the second chain of the receptor complex, or a difference in substrate specificity. Of note, this hypothesis could explain why the V617F mutation in JAK2 is so frequent in myeloproliferative neoplasms. On the basis of the homology of JAK1 and JAK2, we can predict that mutations in JAK2 that are homologous to the mutations in JAK1 will activate JAK2. However, the V617F mutation is nearly the only one observed in patients. This mutation might specifically potentiate signaling pathways that would not be activated by other mutants, thereby representing the “best hit” to mediate myeloproliferation.
Type I IFNs are used in the clinic as adjuvant to treat patients with renal cell carcinoma and melanoma.13 The way INF exerts its antitumoral effect in these diseases is not perfectly understood, but it is considered as an immunotherapy that potentiate the immune response against the tumor. The treatment with IFN is safe and induces few side effects compared with chemotherapies that are usually used in these malignancies. The pegylated form of IFN allows for less frequent injection (once weekly) and easier administration. IFN-α is also used to treat patients with myeloproliferative neoplasms, such as essential thrombocythemia or polycythemia vera. In these diseases, IFN is thought to exert an antiproliferative effect. Recent data suggest that IFN is the only medication able to induce molecular remission in these diseases, as shown by the disappearance of the JAK2 V617F mutation in a substantial number of patients.14 IFN is not used to treat ALL. Our data show that different JAK1 mutants specifically potentiate the antiproliferative effect of IFN. Moreover, our in vivo leukemia model shows that treatment with IFN decreases tumor formation of cells expressing JAKA634D. These data suggest that IFN-α might represent a candidate as adjuvant-targeted therapy for patients with ALL harboring JAK1 mutations, which are present in up to 18% of adult subjects with T-ALL.2
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Pr Stefan N. Constantinescu for critical reading of the manuscript, Pr Thomas Michiels for the IFN constructs, and Pr André Tonon for flow cytometry expertise.
This work was supported in part by the Belgian Program on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming; the Actions de Recherche Concertées of the Communauté Française de Belgique; the Foundation Salus Sanguinis; the opération Télévie and AIRC IG 2009 8803 grant (M.T.). L.K. is an MD fellow of the Fonds National de la Recherche Scientifique (FNRS), Belgium.
Authorship
Contribution: T.H., L.K., and J.-C.R. designed the research and wrote the paper; S.C., R.F., and M.T. designed and performed microarray experiments; R.B.A. and V.A. provided cDNA from patient samples; T.H., L.K., M.M.L., and S.C. carried out the experiments and analyzed the data. All authors critically reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean-Christophe Renauld, Ludwig Institute for Cancer Research and de Duve Insitute, Université Catholique de Louvain, Avenue Hippocrate, 74, B-1200 Brussels, Belgium; e-mail: renauld@bru.licr.org.
References
Author notes
J.-C.R. and L.K. contributed equally to this study.