Abstract
The transcription factor growth factor independence 1 (Gfi1) and the growth factor granulocyte colony-stimulating factor (G-CSF) are individually essential for neutrophil differentiation from myeloid progenitors. Here, we provide evidence that the functions of Gfi1 and G-CSF are linked in the regulation of granulopoiesis. We report that Gfi1 promotes the expression of Ras guanine nucleotide releasing protein 1 (RasGRP1), an exchange factor that activates Ras, and that RasGRP1 is required for G-CSF signaling through the Ras/mitogen–activated protein/extracellular signal-regulated kinase (MEK/Erk) pathway. Gfi1-null mice have reduced levels of RasGRP1 mRNA and protein in thymus, spleen, and bone marrow, and Gfi1 transduction in myeloid cells promotes RasGRP1 expression. When stimulated with G-CSF, Gfi1-null myeloid cells are selectively defective at activating Erk1/2, but not signal transducer and activator of transcription 1 (STAT1) or STAT3, and fail to differentiate into neutrophils. Expression of RasGRP1 in Gfi1-deficient cells rescues Erk1/2 activation by G-CSF and allows neutrophil maturation by G-CSF. These results uncover a previously unknown function of Gfi1 as a regulator of RasGRP1 and link Gfi1 transcriptional control to G-CSF signaling and regulation of granulopoiesis.
Introduction
Lineage-restricted transcription factors regulate differentiation of hematopoietic progenitors by promoting lineage-specific transcriptional programs and simultaneously repressing the transcriptional profile of alternative lineages. PU.1, CCAAT enhancing-binding protein (C/EBP) α and β are transcription factors that regulate neutrophil differentiation from progenitors.1 Deletion of either gene leads to the absence of neutrophils in conjunction with variable alterations of eosinophils, lymphocytes, and monocytes.
Growth factor independence 1 (Gfi1) is a zinc-finger transcription factor first identified as a gene frequently targeted for proviral integration contributing interleukin-2 growth independence in a rat lymphoma cell line and promoting T-cell lymphoma development.2-4 Gfi1 is expressed in thymus, spleen, testis, and the hematopoietic system.2,5 In hematopoietic stem cells, Gfi1 maintains quiescence.6,7 In myeloid cells, Gfi1 promotes differentiation.5,8,9 Gfi1-null mice lack normal neutrophils and accumulate a population of atypical Gr1+/CD11b+ cells that share characteristics of neutrophils and macrophages and can mature into macrophages but not neutrophils.5,8 The mice are small, die prematurely of bacterial infections, and have reduced T- and B-cell differentiation.5,8,10 Rare cases of severe congenital neutropenia have been linked to heterozygous Gfi1 mutations, which can act as dominant negative.9,11 The patients resemble Gfi1-null mice in displaying neutropenia, abnormal circulating myeloid precursors, and reduced B and T lymphocytes.9
Previous studies have concluded that Gfi1 regulates myeloid cell maturation by transcriptional repression of target genes, including suppressor of cytokine signaling 3 (SOCS3),12 neutrophil elastase,13 colony-stimulating factor1 (CSF1)/CSF1 receptor,11 and the microRNAs miR-21 and miR-196b.14 Derepression of these genes explains important aspects of defective neutrophil differentiation in Gfi1-null mice and patients with Gfi1 mutations. Gfi1 can bind consensus DNA target sequences by a C-terminal zinc-finger domain and repress transcription by its N-terminal SNAG domain,15 in part by recruiting corepressors.16-19 Besides acting as a transcriptional repressor, Gfi1 interacts with protein inhibitor of activated signal transducer and activator of transcription (STAT), which can bind to and inhibit STAT3 signaling.20 As a result, Gfi1 can relieve STAT3 from protein inhibitor of activated STAT3–induced inhibition, resulting in enhanced STAT3 signaling. Because STATs are activated by a variety of cytokines and growth factors, we hypothesized that Gfi1 could play a role as a modulator of cytokine and growth factor responses.
We focused on granulocyte (G)–CSF and its unique receptor (G-CSFR), which are critical to the generation of neutrophils from granulocyte/monocyte precursors and their release to the peripheral circulation. Homozygous deletion of G-CSFR or G-CSF causes severe neutropenia in mice,21,22 and heterozygous mutations of G-CSFR have been linked to sporadic cases of severe congenital neutropenia.23 As both Gfi1 transcriptional regulation and G-CSF/G-CSFR signaling are critical to neutrophil maturation, we have investigated whether their activities might be linked. We find that Gfi1 functions as a regulator of G-CSF/G-CSFR signaling in myeloid cells by promoting expression of RasGRP1 (Ras guanine nucleotide releasing protein 1), which is shown to be a critical modulator of G-CSF/G-CSFR–induced Ras activation.
Methods
Mice and cell isolation
Germline Gfi1−/− mice24 were bred and housed in the animal facilities at the National Cancer Institute. Genotyping was as described.24 All animal studies were approved by the National Cancer Institute Bethesda Animal Care and Use Committee and were carried out per protocol with mice 4 to 8 weeks of age. G-CSF–induced cell mobilization experiments were described.25 Bone marrow, thymic, and splenic cell suspensions were prepared from isolated organs by standard techniques. Peripheral blood cell counts and differentials were carried out by the National Institutes of Health Clinical Center with the use of an ADVIA 120 hematology system (Bayer).
Cell culture
Bone marrow mononuclear cells (MNCs) were obtained by flushing femurs and tibias, and Lin− cell bone marrow cell populations were derived with the Mouse Lineage Cell Depletion Kit (Miltenyi Biotec; 130-090-858) as described by the manufacturer. For signaling experiments, cells (1-2 × 106/mL) were incubated (0-180 minutes) in Iscove Dulbecco modification of Eagle medium (DMEM) with 0% to 10% heat-inactivated fetal bovine serum (FBS; Biosource) alone or with 25 ng/mL human G-CSF (PeproTech) or 10 ng/mL mouse IL-3 (PeproTech). For proliferation experiments, cells were cultured (1 × 105 cells/well in 96-well plate) for 3 days in Iscove modified Dulbecco medium with 10% FBS with or without G-CSF (25 ng/mL) or interleukin-3 (IL-3; 10 ng/mL). For differentiation experiments in liquid culture, cells were incubated (5 × 104 to 1 × 105) in Iscove DMEM with 10% FBS with or without G-CSF (25 ng/mL) with or without puromycin (2.5 mg/mL). Colony assays were performed with methocult M3234 (StemCell Technologies). Bone marrow cells (5 × 104; cultures supplemented with 10 ng/mL IL-3; PeproTech), 10 ng/mL IL-6 (R&D Systems), and 50 ng/mL stem cell factor (SCF; R&D Systems) or 4 × 105 cells (cultures supplemented with 25 ng/mL G-CSF) were plated into 35-mm2 petri dishes and incubated for 10 to 14 days. The murine 32Dcl3 cell line (32D; a gift of Dr A. Friedman, John Hopkins University) was maintained in DMEM supplemented with 10% FBS and 1 ng/mL murine IL-3. For signaling experiments, 32D cells were washed and preincubated in DMEM supplemented with 10% FBS medium for 4 hours; cells (1-2 × 106/mL) were harvested, washed, and activated with 25 ng/mL G-CSF or 10 ng/mL IL-3.
Cell proliferation
Proliferation was measured by 3H-thymidine deoxyribose incorporation (0.5 μCi/well [0.0185 Bq/well]; New England Nuclear) during the last 6 hours of culture, as described.25
Cell sorting, flow cytometry, and immunocytochemistry
Green fluorescent protein (GFP)–expressing cells and cells surface-stained with phycoerythrin (PE) rat anti–mouse Gr1 antibodies (BD PharMingen) were sorted with a FACSVantage SE (BD Biosciences). For analysis, cells were stained with specific antibodies: allophycocyanin (APC) rat anti–mouse CD45R/B220 APC rat anti–mouse CD11b, R (Red) PE–conjugated rat anti–mouse CD4, fluorescein isothiocyanate (FITC) and PE rat anti–mouse CD117 (cKit), FITC rat anti–mouse Ly-6A/E (Sca-1), FITC rat anti–mouse CD8a, PE/cyanine 5 (Cy5) rat anti–mouse CD45, PE/Cy5 anti–mouse CD45, PE/Cy5 anti–mouse CD48; PE/Cy5 anti–mouse CD150, PE rat anti–mouse CD16/CD32, and relevant isotype control antibodies (all from BD PharMmingen); PE rat anti–mouse CD34 (eBioscience); PE anti–mouse CD 244.2 (BioLegend); biotin-labeled Lin selection cocktail (StemCell Technologies); R-PE rat anti–mouse neutrophils:RPE (clone 7/4) and FITC rat anti–mouse F4/80 antigen (AbD Serotec); rabbit anti–phospho extracellular signal-related kinase 1/2 (Erk1/2) and total Erk1/2 monoclonal antibodies (Cell Signaling Technology) and PE-labeled goat anti–rabbit antibodies (Invitrogen); RasGRP1 monoclonal antibody m199 (Santa Cruz Biotechnology) or control mouse immunoglobulin G1 (IgG1; Invitrogen) followed by Alexa Fluor 488 goat anti–mouse IgG1 (Invitrogen). Surface G-CSFR was detected with PE-labeled G-CSF (R&D Systems) cross-linked with Bis(sulfosuccinimidyl)suberate (Pierce, Thermo Scientific).3
Staining for phospho and total Erk1/2 was carried out after fixation (10 minutes at room temperature in Iscove medium with 2% paraformaldehyde) and permeabilization in (60 minutes at 4°C in 90% methanol) with the use of specific primary antibodies followed by PE-labeled anti–rabbit secondary antibodies (BD PharMingen). Staining for RasGRP1 was carried out after cell fixation and permeabilization (Fix and Perm solution; BD PharMingen) with the use of RasGRP1 monoclonal antibody m199 (Santa Cruz Biotechnology) or control mouse IgG1 (Invitrogen) followed by Alexa Fluor 488 goat anti–mouse IgG1 (Invitrogen). For double-staining cells were first surface-stained and then fixed/permeabilized for RasGRP1 staining. Results were analyzed with CellQuest software (BD Biosciences) after acquisition of data from 104 cells. Cytospin preparations were immunostained for intracellular RasGRP1 as described for flow cytometry; nuclei were stained with DAPI (4,6 diamidino-2-phenylindole); cell morphology was evaluated by Giemsa staining. Fluorescence and bright-field images were acquired through a Nikon Eclipse E600 microscope equipped with Plan Apo 40×/0.95 DIC M, 60×/1.40 oil differential interference contrast (DIC) H, and 100×/1.40 oil DIC H lenses, and photographed with a digital camera (Retiga 1300; QImaging). Images obtained with IPLab for Windows software (Scanalytics) were imported into Adobe Photoshop.
Retroviral constructs, infection, and small interfering RNA
Gfi1-GFP-RV and GFP-RV26 and pBabe-RasGRP1 or pBabe27 were previously constructed. Retroviruses were packaged in the Phoenix packaging cell line, as described.28 Two-day culture supernatants were filtered, supplemented with Polybrene (4 mg/mL, final concentration; Sigma-Aldrich) and used for infection. Two days after infection with Gfi1-GFP-RV and GFP-RV retroviruses, GFP-expressing 32D cells were sorted; only cell populations at least 80% GFP+ were used for experiments. Two days after infection with pBabe-RasGRP1 or pBabe retroviruses, puromycin (1 mg/mL) was added to 32D cells for selection. For infection of primary mouse bone marrow cells, a similar method was applied except for the use of RetroNectin-coated plates (Takara) and cell preculture (18-72 hour) in culture medium supplemented with IL-3 (10 ng/mL), cKitL (25 ng/mL), SCF (25 ng/mL), and granulocyte-macrophage–CSF (5 ng/mL). Two days after retroviral infection, the GFP+ and GFP− cells were sorted, and the cells used for further experiments. RasGRP1 was silenced with small interfering RNAs (siRNAs) for mouse RasGRP1 (Ambion, Applied Biosystems); risc-free siRNA (Dharmacon) was used as control. siRNA was transfected into 32D cells with the Amaxa nucleofector system (Amaxa Biosystems) optimized for 32D cells (E-32), with Cell Line Nucleofector Solution V. Transfection efficiency was assessed by GFP positivity.
RNA isolation and semiquantitative and real-time polymerase chain reactions
We isolated total RNA with TRIzol reagent (Invitrogen). Semiquantitative polymerase chain reaction (PCR) was performed as described,25 using primers for amplification of mouse Gfi1,26 mouse RasGRP1 (5′-GGACCACGGTGATAGTGCTT-3′; 5′-TGCTCAGACTTTGCATGCTT-3′), and glyceraldehyde phosphate dehydrogenase.25 Real-time PCR was performed with the use of Assay-on-Demand TaqMan probes (Applied Biosystems) for mouse RasGRP1, Gfi1, G-CSFR, and glyceraldehyde phosphate dehydrogenase; rat Gfi1 and CSF1.
Immunoprecipitation and Western blotting
RasGRP1 was immunoprecipitated with rabbit anti-RasGRP1 antibody (H120; Santa Cruz Biotechnology) from cell extracts prepared in NP-40 lysis buffer supplemented with protease inhibitor cocktail set III (Calbiochem) supplemented with 50mM NaF and phenylmethyl sulfonyl fluoride. Precipitates were resolved through 8% Tris Glycine Gels (Invitrogen) and immunoblotted with mouse monoclonal anti-RasGRP1 antibody m199 or rabbit anti-RasGRP1(H120).29 Active Ras was immunoprecipitated with glutathione-S-transferase–Raf1 (residues 1-149; Upstate) following the manufacturer's instructions; precipitates were resolved through 10% NuPage Bis Tris gels (Invitrogen) and immunoblotted with anti-Ras antibody (05-516; Upstate). Protein extracts for phosphorylated proteins prepared in sodium dodecyl sulfate lysis buffer with protease inhibitor cocktail set III (Calbiochem), 50mM NaF, and 1mM sodium orthovanadate were resolved in 4% to 12% or 10% NuPAGE Bis Tris or 10% to 20% Tricine gels (Invitrogen), and nitrocellulose membranes were immunoblotted with the following specific antibodies: phospho-p44/42 mitogen-activated protein (MAP) kinase (Tyr202/Tyr204), total p44/42, phospho-MAP/Erk1/2 (MEK1/2) (Ser217/221), phospho–Stat-3 (Tyr705), total Stat-3 (BD Biosciences), phospho–Stat-1(Tyr701), phospho–Stat-5(Tyr694), phospho-Akt (Ser473), total Akt, phospho-Erk5 (Thr218/Tyr220), phospho-p38 MAP kinase (MAPK) (Thr180/Tyr182), phospho-Src (Tyr527 and Tyr416), non–phospho-Src (Tyr527), phospho-MAP phosphatase 1 (MKP1) (Ser359,125E2), phospho-KSR1 (Ser392), phospho–c-Raf (Ser259), SOCS3 (2923) (all from Cell Signaling Technology); SOCS3 (M-20 and H-103; Santa Cruz Biotechnology), SOCS3 (Zymed Laboratories), MKP1 (C-19; Santa Cruz Biotechnology), and phospho-cJun (Ser73; Upstate).
Statistical analysis
Group differences were evaluated by 2-tailed Student t test. P values less than .05 were considered significant.
Results
Altered G-CSF signaling in Gfi1-deficient hematopoietic cells
Consistent with previous studies, Gfi1-null mice from our colony have a reduction in circulating granulocytes (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and in the mobilization response to G-CSF administration in vivo (supplemental Table 2), deficiencies that could derive from defective G-CSF responses.
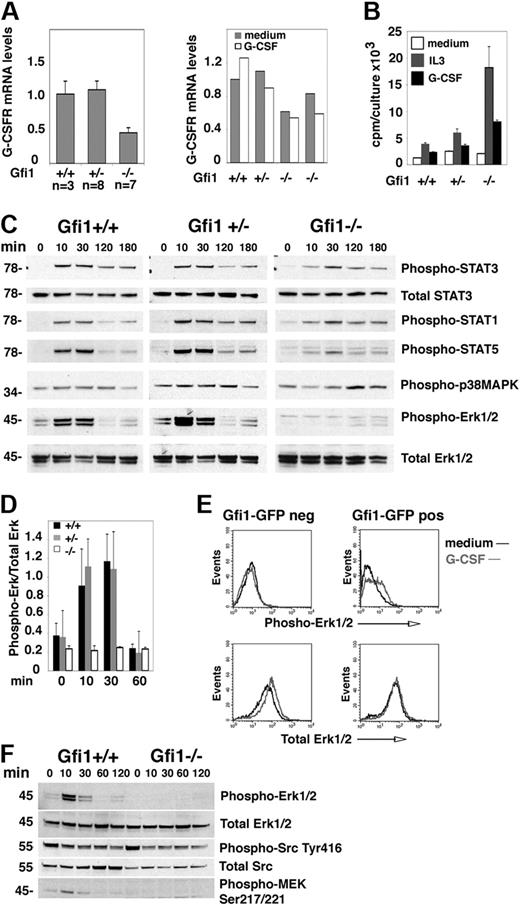
G-CSF signals through the G-CSFR, which is expressed exclusively from myeloid-restricted progenitor cells to mature neutrophils.30 We found G-CSFR expression levels to be reduced by approximately 50% in bone marrow MNCs from Gfi1-null mice in comparison to control wild-type and heterozygous mice, with or without G-CSF stimulation in vitro (Figure 1A). Because G-CSFR expression increases with myeloid cell differentiation,30 reduced G-CSFR expression in Gfi1-null bone marrow MNCs is likely due to the reduction in neutrophils.
Erk activation is selectively defective in G-CSF–stimulated bone marrow cells from Gfi1-null mice. (A) G-CSFR mRNA levels in fresh (left) bone marrow MNCs from untreated Gfi1+/+, Gfi1+/−, and Gfi1−/− mice and after 24-hour incubation in vitro with or without G-CSF (25 ng/mL; right) measured by quantitative real-time PCR. (Left) Gfi1+/+ (n = 3); Gfi1+/− (n = 8); Gfi1−/− (n = 7); the results reflect the means ± SD. (Right) Means (duplicate measurements) of G-CSFR mRNA levels in bone marrow MNCs from individual mice. (B) Proliferation of bone marrow MNCs from Gfi1+/+, Gfi1+/−, and Gfi1−/− mice after 3-day incubation in medium only, with IL-3 (10 ng/mL), or with G-CSF (25 ng/mL). The results reflect the mean 3H thymidine uptake (cpm) of triplicate cultures ± SD and are representative of 3 independent experiments. (C) Immunoblot analysis of G-CSF–induced signaling in bone marrow MNCs from Gfi1+/+, Gfi1+/−, and Gfi1−/− mice after derivation (time 0, no culture) and after incubation with G-CSF (25 ng/mL) for the indicated time intervals. All results reflect probing of single membranes with specific antibodies. The results are representative of 5 experiments. (D) Quantitative measurement of phopho-Erk1/2 activation by G-CSF (25 ng/mL) in bone marrow MNCs from Gfi1+/+, Gfi1−/−, and Gfi1−/− mice at the indicated time points. The results reflect the mean ± SD band intensity ratios of phospho-Erk1/2/total Erk1/2 (n = 5 mice/group evaluated at each time point). (E) G-CSF–induced Erk1/2 phosphorylation in bone marrow MNCs from Gfi1−/− mice after transduction in vitro with Gfi1-GFP retroviruses, cultured for 48 hours in medium (Iscove DMEM with 10% FCS) supplemented with 10 ng/mL IL-3, 25 ng/mL SCF, 25 ng/mL cKitL, and 5 ng/mL granulocyte-macrophage–CSF, cell sorting the GFP+ and GFP− cells, and starved (1 hour) in culture medium without growth factors. Phospho-Erk1/2 and total Erk was evaluated by fluorescence-activated cell sorting analysis after intracellular staining with specific antibodies. (F) G-CSF–induced signaling evaluated by immunoblotting with specific antibodies. Bone marrow MNCs from Gfi1+/+, Gfi1−/−, and Gfi1−/− mice were incubated with G-CSF (25 ng/mL) for the indicated time intervals. The results reflect reprobing of a single membrane. The results are representative of 3 experiments performed.
Erk activation is selectively defective in G-CSF–stimulated bone marrow cells from Gfi1-null mice. (A) G-CSFR mRNA levels in fresh (left) bone marrow MNCs from untreated Gfi1+/+, Gfi1+/−, and Gfi1−/− mice and after 24-hour incubation in vitro with or without G-CSF (25 ng/mL; right) measured by quantitative real-time PCR. (Left) Gfi1+/+ (n = 3); Gfi1+/− (n = 8); Gfi1−/− (n = 7); the results reflect the means ± SD. (Right) Means (duplicate measurements) of G-CSFR mRNA levels in bone marrow MNCs from individual mice. (B) Proliferation of bone marrow MNCs from Gfi1+/+, Gfi1+/−, and Gfi1−/− mice after 3-day incubation in medium only, with IL-3 (10 ng/mL), or with G-CSF (25 ng/mL). The results reflect the mean 3H thymidine uptake (cpm) of triplicate cultures ± SD and are representative of 3 independent experiments. (C) Immunoblot analysis of G-CSF–induced signaling in bone marrow MNCs from Gfi1+/+, Gfi1+/−, and Gfi1−/− mice after derivation (time 0, no culture) and after incubation with G-CSF (25 ng/mL) for the indicated time intervals. All results reflect probing of single membranes with specific antibodies. The results are representative of 5 experiments. (D) Quantitative measurement of phopho-Erk1/2 activation by G-CSF (25 ng/mL) in bone marrow MNCs from Gfi1+/+, Gfi1−/−, and Gfi1−/− mice at the indicated time points. The results reflect the mean ± SD band intensity ratios of phospho-Erk1/2/total Erk1/2 (n = 5 mice/group evaluated at each time point). (E) G-CSF–induced Erk1/2 phosphorylation in bone marrow MNCs from Gfi1−/− mice after transduction in vitro with Gfi1-GFP retroviruses, cultured for 48 hours in medium (Iscove DMEM with 10% FCS) supplemented with 10 ng/mL IL-3, 25 ng/mL SCF, 25 ng/mL cKitL, and 5 ng/mL granulocyte-macrophage–CSF, cell sorting the GFP+ and GFP− cells, and starved (1 hour) in culture medium without growth factors. Phospho-Erk1/2 and total Erk was evaluated by fluorescence-activated cell sorting analysis after intracellular staining with specific antibodies. (F) G-CSF–induced signaling evaluated by immunoblotting with specific antibodies. Bone marrow MNCs from Gfi1+/+, Gfi1−/−, and Gfi1−/− mice were incubated with G-CSF (25 ng/mL) for the indicated time intervals. The results reflect reprobing of a single membrane. The results are representative of 3 experiments performed.
To assess G-CSFR function, we measured G-CSF–induced cell proliferation, which excludes the nondividing neutrophils; as a control for G-CSF, we used IL-3. In all cases, IL-3 was a stronger inducer of proliferation than G-CSF, probably reflecting the wider distribution of IL-3 as opposed to G-CSF receptors in bone marrow cells. G-CSF induced proliferation in bone marrow MNC from all mice, including Gfi1−/− mice, indicating that the G-CSFR is functional in Gfi1-null bone marrow MNCs (Figure 1B). Note, G-CSF promoted greater proliferation in MNCs from Gfi1−/− mice than in Gfi1+/+ and Gfi1+/− controls, probably attributable to the absence of neutrophils in Gfi1−/− bone marrows and the relative enrichment of immature cells with greater proliferative potential (Figure 1B). However, when G-CSF–induced signaling was examined (Figure 1C-D), Erk1/2 phosphorylation was found reproducibly reduced in Gfi1−/− bone marrow MNCs, whereas STAT1, STAT3, and p38MAPK phosphorylation was similar. STAT5 phosphorylation was also somewhat lower in Gfi1−/− bone marrow MNCs than in controls. We tested whether expression of Gfi1 in Gfi1−/− bone marrow MNCs could reconstitute Erk1/2 activation by G-CSF. Using a retroviral vector for expression of GFP-Gfi1 and sorting the GFP+ and GFP− cell populations, we found that expression of Gfi1 in Gfi1−/− bone marrow MNCs reconstituted Erk1/2 phosphorylation in a proportion of cells stimulated with G-CSF (Figure 1E).
To identify signaling alterations underlying defective G-CSF activation of Erk in Gfi1-null bone marrow MNCs, we queried several pathways linked to G-CSFR activation or Erk1/2 activation (Figure 1E; data not shown). MEK1/2 phosphorylation (Ser217/221), which is upstream of Erk1/2 and contributes to G-CSF–induced myeloid cell differentiation,31 was not induced by G-CSF stimulation in bone marrow MNCs from Gfi1−/− mice but was induced in MNCs from Gfi1+/+ controls (Figure 1F). By contrast, we detected a similar degree of basal Src activity (phosphorylation of tyrosine [Tyr] 416) in bone marrow MNCs of Gfi1+/+ and Gfi1−/− mice, which was not modulated by G-CSF stimulation (Figure 1F). SOCS3, a negative regulator of G-CSF signaling that was reportedly expressed at higher levels in progenitor cells from Gfi1−/− mice,12 was detected at similarly low levels in bone marrow MNCs from Gfi1+/+ and Gfi1−/− mice (not shown). Phosphorylated (serine [Ser] 359) MKP1 and Akt (Ser473) were also detected at similarly low levels in bone marrow MNCs from Gfi1−/− and Gfi1+/+ mice, with or without G-CSF (not shown).
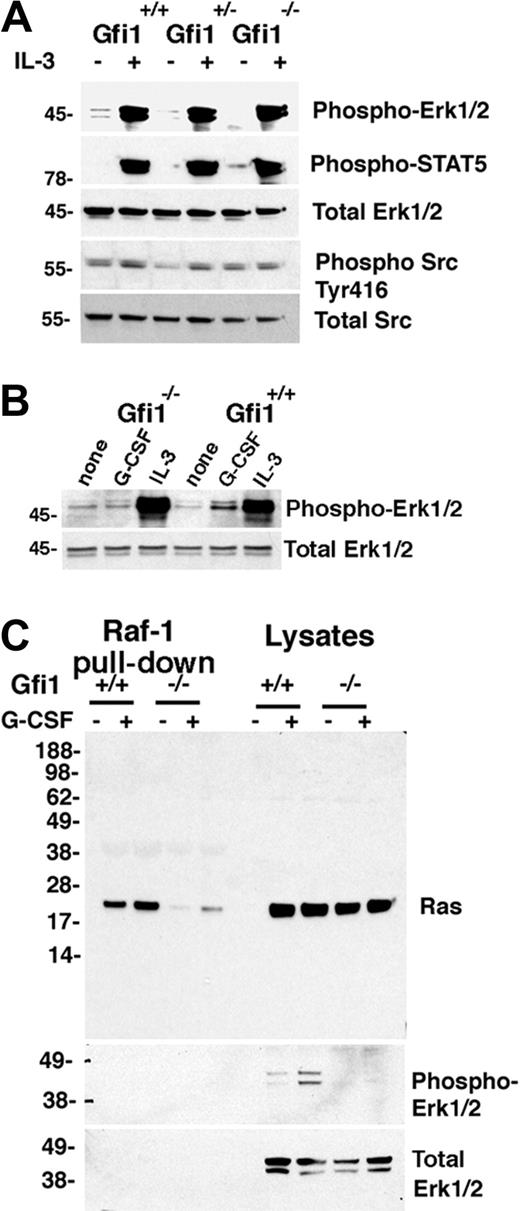
To explore whether there might be a global defect in Erk1/2 signaling, we stimulated bone marrow MNCs with IL-3, which activates the Erk1/2 pathway through the IL-3 receptor, a receptor that is functional in most nucleated hematopoietic cells.32 However, bone marrow MNCs from Gfi1+/+, Gfi1+/−, and Gfi1−/− mice similarly activated Erk1/2 phosphorylation with 10 ng/mL IL-3 (Figure 2A), which was a stronger activator of Erk1/2 than G-CSF (Figure 2B), presumably reflective of the wider expression of IL-3–responsive cells in bone marrow MNCs.
G-CSF– but not IL-3–induced Erk1/2 activation in Gfi1-null bone marrow cells is impaired, correlating with defective Ras activation. (A) Immunoblot analysis of IL-3–induced signaling responses in bone marrow MNCs from Gfi1+/+, Gfi1+/−, and Gfi1−/− mice. Freshly obtained cells were incubated with for 10 minutes in either medium only or with IL-3 (10 ng/mL). The results reflect reprobing of a single membrane. The results are representative of 7 experiments. (B) Phospho-Erk1/2 activation by IL-3 or G-CSF in Gfi1+/+ and Gfi1−/− bone marrow MNCs evaluated by immunoblotting with specific antibodies. Cells were incubated for 10 minutes in medium only or with IL-3 (10 ng/mL) or G-CSF (25 ng/mL). The results are representative of 5 experiments. (C) Active Ras was pulled down from bone marrow cell lysates with agarose-bound Raf-1 protein (residues 1-149 corresponding to the binding domain for Ras-GTP) and immunoblotted with Ras-specific antibody. Bone marrow MNCs from 2 Gfi1+/+ and 2 Gfi1−/− mice were incubated in medium only or with G-CSF (25 ng/mL) for 10 minutes; the cell lysates were either immunoblotted directly or after Ras-GTP pull down. The membrane was reprobed with specific antibodies to phospho-Erk1/2 or total Erk. The results are representative of 3 experiments performed.
G-CSF– but not IL-3–induced Erk1/2 activation in Gfi1-null bone marrow cells is impaired, correlating with defective Ras activation. (A) Immunoblot analysis of IL-3–induced signaling responses in bone marrow MNCs from Gfi1+/+, Gfi1+/−, and Gfi1−/− mice. Freshly obtained cells were incubated with for 10 minutes in either medium only or with IL-3 (10 ng/mL). The results reflect reprobing of a single membrane. The results are representative of 7 experiments. (B) Phospho-Erk1/2 activation by IL-3 or G-CSF in Gfi1+/+ and Gfi1−/− bone marrow MNCs evaluated by immunoblotting with specific antibodies. Cells were incubated for 10 minutes in medium only or with IL-3 (10 ng/mL) or G-CSF (25 ng/mL). The results are representative of 5 experiments. (C) Active Ras was pulled down from bone marrow cell lysates with agarose-bound Raf-1 protein (residues 1-149 corresponding to the binding domain for Ras-GTP) and immunoblotted with Ras-specific antibody. Bone marrow MNCs from 2 Gfi1+/+ and 2 Gfi1−/− mice were incubated in medium only or with G-CSF (25 ng/mL) for 10 minutes; the cell lysates were either immunoblotted directly or after Ras-GTP pull down. The membrane was reprobed with specific antibodies to phospho-Erk1/2 or total Erk. The results are representative of 3 experiments performed.
The defect in MEK1/2 and Erk1/2 phosphorylation after G-CSF stimulation of bone marrow MNCs from Gfi1−/− mice suggested that Ras activation might be impaired in these cells. Consistent with this possibility, a pull-down assay of active Ras (using agarose-bound glutathione-S-transferase–Raf1 residues 1-149, corresponding to the Raf1 domain that binds to Ras–guanosine triphosphate [GTP]) detected significantly greater amounts of active Ras (constitutive and G-CSF–induced) in bone marrow cell lysates from Gfi1+/+ in comparison to Gfi1−/− mice (Figure 2C representative results), differences (both constitutive and G-CSF-induced) that were not attributable to a Ras protein deficiency in the bone marrow cell lysates from Gfi1−/− mice (Figure 2C). In addition, consistent with active Ras serving as an inducer of the Raf/MEK/Erk pathway, reduced Ras-GTP was accompanied by reduced phospho-Erk1/2 in Gfi1−/− cells (Figure 2C).
RasGRP1 deficiency in Gfi1-deficient mice
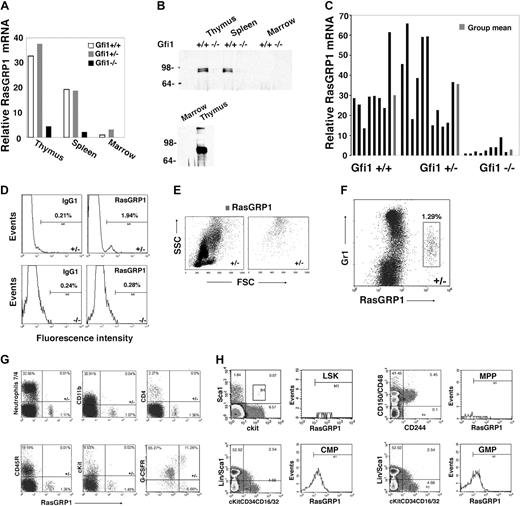
To identify genes whose expression might underlie the defective Ras activation in Gfi1-null mice, we used microarray analysis to compare patterns of gene expression in bone marrow MNCs from Gfi1−/− and Gfi1+/+ mice. Among the genes whose expression differed by at least 5-fold, we found that RasGRP1 mRNA was approximately 8-fold less abundant in bone marrow MNCs from Gfi1−/− mice than in Gfi1+/+ controls. RasGRPs are Ras guanine nucleotide-exchange factors (Ras GEFs) that positively regulate Ras activity by catalyzing the release of guanosine diphosphate, allowing Ras binding to cellular GTP.33 By real-time PCR, we confirmed that levels of RasGRP1 mRNA were reduced in the bone marrow MNCs of Gfi1−/− mice compared with Gfi1+/+ and Gfi1+/− mice (Figure 3A). In the thymus and spleen, where RasGRP1 mRNA is substantially more abundant than in bone marrow, levels of RasGRP1 mRNA were also reduced in Gfi1−/− mice compared with controls (Figure 3A). This difference in RasGRP1 mRNA was confirmed at the protein level in the thymus and spleen by immunoprecipitation/immunoblotting (Figure 3B). Consistent with the low level of mRNA in bone marrow (Figure 3A), RasGRP1 protein was not clearly detected in bone marrow cell lysates even from wild-type mice (Figure 3B). Nonetheless, RasGRP1 mRNA was consistently detected in bone marrow MNCs from Gfi1+/+ and Gfi1+/− mice and to a significantly (P < .05) lower degree in Gfi1−/− mice (Figure 3C).
Reduced RasGRP1-expressing cells in Gfi1-null mice. (A) Levels of RasGRP1 mRNA were measured by quantitative real-time PCR in thymus, spleen, and bone marrow extracted from individual Gf1+/+, Gfi1+/−, and Gfi1−/− mice. The results are representative of at least 5 mice/group. (B) RasGRP1 was immunoprecipitated from lysates of thymus (300 μg, top; 800 μg, bottom), spleen (300 μg), and bone marrow MNCs (300 μg, top; 3 mg bottom) from individual Gfi1+/+ and Gfi1−/− mice. The immunoprecipitates were immunoblotted with RasGRP1-specific antibody. The bottom panel relates to tissues from a Gfi1+/+ mouse (representative of 3 experiments). (C) RasGRP1 mRNA levels measured by quantitative real-time PCR in individual bone marrows from Gfi1+/+ (n = 8), Gfi1+/− (n = 11), and Gfi1−/− (n = 9) mice. Results from individual mice, ■; and group means,  . (D) Flow cytometric detection of RasGRP1 in a population of bone marrow MNCs from a Gfi1+/− but not Gfi1−/− mouse. After fixation and permeabilization, the cells were immunostained with a mouse monoclonal antibody to RasGRP1 or isotype control antibody (mouse IgG1) followed by a goat anti–mouse IgG Alexa Fluor 488–conjugated antibody. The results reflect the percentage of RasGRP1-positive cells and are representative of 5 independent experiments. (E) Forward scattering counter (FSC) and SSC of bone marrow MNCs from a Gfi1+/− mouse after RasGRP1 immunostaining (as described in panel D). Left panel shows total; right panel shows RasGRP1 only. The results are representative of Gfi1+/+ (n = 4) and Gfi1+/− (n = 3) mice. (F) Flow cytometric analysis of Gr1 and RasGRP1 expression within bone marrow MNCs from a Gfi1+/− mouse, showing that RasGRP1-positive cells display low-level surface Gr1. The MNCs were first immunostained for surface Gr1 (PE-labeled) and after fixation/permeabilization immunostained for RasGRP1 (as described in panel D). Representative of 5 independent experiments is shown. The percentage of cells in the box is shown. (G) Flow cytometric detection of the surface markers neutrophils 7/4, CD11b, CD4, CD45R/B220, cKit, G-CSFR, and intracellular RasGRP1in bone marrow MNCs from a Gfi1+/− mouse. G-CSFR was detected by PE-labeled G-CSF cross-linked to the cell surface with Bis(sulfosuccinimidyl)suberate3 . The results are representative of 3 experiments. The percentage of cells in each quadrant is shown. (H) Flow cytometric analysis of RasGRP1-positive cells within LSK (lin−c-Kit+Sca+), MPP (CD150−CD48−CD244+), CMP (lin−Sca−c-Kit+CD34+CD16/32mid), and GMP (lin−Sca−c-Kit+CD34+CD16/32hi) bone marrow cells populations from Gfi1+/+ MNCs. Representative of 3 experiments is shown. The values reflect percentage of cells in each quadrant.
. (D) Flow cytometric detection of RasGRP1 in a population of bone marrow MNCs from a Gfi1+/− but not Gfi1−/− mouse. After fixation and permeabilization, the cells were immunostained with a mouse monoclonal antibody to RasGRP1 or isotype control antibody (mouse IgG1) followed by a goat anti–mouse IgG Alexa Fluor 488–conjugated antibody. The results reflect the percentage of RasGRP1-positive cells and are representative of 5 independent experiments. (E) Forward scattering counter (FSC) and SSC of bone marrow MNCs from a Gfi1+/− mouse after RasGRP1 immunostaining (as described in panel D). Left panel shows total; right panel shows RasGRP1 only. The results are representative of Gfi1+/+ (n = 4) and Gfi1+/− (n = 3) mice. (F) Flow cytometric analysis of Gr1 and RasGRP1 expression within bone marrow MNCs from a Gfi1+/− mouse, showing that RasGRP1-positive cells display low-level surface Gr1. The MNCs were first immunostained for surface Gr1 (PE-labeled) and after fixation/permeabilization immunostained for RasGRP1 (as described in panel D). Representative of 5 independent experiments is shown. The percentage of cells in the box is shown. (G) Flow cytometric detection of the surface markers neutrophils 7/4, CD11b, CD4, CD45R/B220, cKit, G-CSFR, and intracellular RasGRP1in bone marrow MNCs from a Gfi1+/− mouse. G-CSFR was detected by PE-labeled G-CSF cross-linked to the cell surface with Bis(sulfosuccinimidyl)suberate3 . The results are representative of 3 experiments. The percentage of cells in each quadrant is shown. (H) Flow cytometric analysis of RasGRP1-positive cells within LSK (lin−c-Kit+Sca+), MPP (CD150−CD48−CD244+), CMP (lin−Sca−c-Kit+CD34+CD16/32mid), and GMP (lin−Sca−c-Kit+CD34+CD16/32hi) bone marrow cells populations from Gfi1+/+ MNCs. Representative of 3 experiments is shown. The values reflect percentage of cells in each quadrant.
Reduced RasGRP1-expressing cells in Gfi1-null mice. (A) Levels of RasGRP1 mRNA were measured by quantitative real-time PCR in thymus, spleen, and bone marrow extracted from individual Gf1+/+, Gfi1+/−, and Gfi1−/− mice. The results are representative of at least 5 mice/group. (B) RasGRP1 was immunoprecipitated from lysates of thymus (300 μg, top; 800 μg, bottom), spleen (300 μg), and bone marrow MNCs (300 μg, top; 3 mg bottom) from individual Gfi1+/+ and Gfi1−/− mice. The immunoprecipitates were immunoblotted with RasGRP1-specific antibody. The bottom panel relates to tissues from a Gfi1+/+ mouse (representative of 3 experiments). (C) RasGRP1 mRNA levels measured by quantitative real-time PCR in individual bone marrows from Gfi1+/+ (n = 8), Gfi1+/− (n = 11), and Gfi1−/− (n = 9) mice. Results from individual mice, ■; and group means,  . (D) Flow cytometric detection of RasGRP1 in a population of bone marrow MNCs from a Gfi1+/− but not Gfi1−/− mouse. After fixation and permeabilization, the cells were immunostained with a mouse monoclonal antibody to RasGRP1 or isotype control antibody (mouse IgG1) followed by a goat anti–mouse IgG Alexa Fluor 488–conjugated antibody. The results reflect the percentage of RasGRP1-positive cells and are representative of 5 independent experiments. (E) Forward scattering counter (FSC) and SSC of bone marrow MNCs from a Gfi1+/− mouse after RasGRP1 immunostaining (as described in panel D). Left panel shows total; right panel shows RasGRP1 only. The results are representative of Gfi1+/+ (n = 4) and Gfi1+/− (n = 3) mice. (F) Flow cytometric analysis of Gr1 and RasGRP1 expression within bone marrow MNCs from a Gfi1+/− mouse, showing that RasGRP1-positive cells display low-level surface Gr1. The MNCs were first immunostained for surface Gr1 (PE-labeled) and after fixation/permeabilization immunostained for RasGRP1 (as described in panel D). Representative of 5 independent experiments is shown. The percentage of cells in the box is shown. (G) Flow cytometric detection of the surface markers neutrophils 7/4, CD11b, CD4, CD45R/B220, cKit, G-CSFR, and intracellular RasGRP1in bone marrow MNCs from a Gfi1+/− mouse. G-CSFR was detected by PE-labeled G-CSF cross-linked to the cell surface with Bis(sulfosuccinimidyl)suberate3 . The results are representative of 3 experiments. The percentage of cells in each quadrant is shown. (H) Flow cytometric analysis of RasGRP1-positive cells within LSK (lin−c-Kit+Sca+), MPP (CD150−CD48−CD244+), CMP (lin−Sca−c-Kit+CD34+CD16/32mid), and GMP (lin−Sca−c-Kit+CD34+CD16/32hi) bone marrow cells populations from Gfi1+/+ MNCs. Representative of 3 experiments is shown. The values reflect percentage of cells in each quadrant.
. (D) Flow cytometric detection of RasGRP1 in a population of bone marrow MNCs from a Gfi1+/− but not Gfi1−/− mouse. After fixation and permeabilization, the cells were immunostained with a mouse monoclonal antibody to RasGRP1 or isotype control antibody (mouse IgG1) followed by a goat anti–mouse IgG Alexa Fluor 488–conjugated antibody. The results reflect the percentage of RasGRP1-positive cells and are representative of 5 independent experiments. (E) Forward scattering counter (FSC) and SSC of bone marrow MNCs from a Gfi1+/− mouse after RasGRP1 immunostaining (as described in panel D). Left panel shows total; right panel shows RasGRP1 only. The results are representative of Gfi1+/+ (n = 4) and Gfi1+/− (n = 3) mice. (F) Flow cytometric analysis of Gr1 and RasGRP1 expression within bone marrow MNCs from a Gfi1+/− mouse, showing that RasGRP1-positive cells display low-level surface Gr1. The MNCs were first immunostained for surface Gr1 (PE-labeled) and after fixation/permeabilization immunostained for RasGRP1 (as described in panel D). Representative of 5 independent experiments is shown. The percentage of cells in the box is shown. (G) Flow cytometric detection of the surface markers neutrophils 7/4, CD11b, CD4, CD45R/B220, cKit, G-CSFR, and intracellular RasGRP1in bone marrow MNCs from a Gfi1+/− mouse. G-CSFR was detected by PE-labeled G-CSF cross-linked to the cell surface with Bis(sulfosuccinimidyl)suberate3 . The results are representative of 3 experiments. The percentage of cells in each quadrant is shown. (H) Flow cytometric analysis of RasGRP1-positive cells within LSK (lin−c-Kit+Sca+), MPP (CD150−CD48−CD244+), CMP (lin−Sca−c-Kit+CD34+CD16/32mid), and GMP (lin−Sca−c-Kit+CD34+CD16/32hi) bone marrow cells populations from Gfi1+/+ MNCs. Representative of 3 experiments is shown. The values reflect percentage of cells in each quadrant.
By flow cytometry, we identified in the bone marrow MNCs of control mice a small (0.5%-3.0%) population of cells with intracellular RasGRP1, which was not found in the Gfi1-null mice (representative results in Figure 3D). These cells expressing RasGRP1 displayed a high side scattering counter (SSC; Figure 3E) and low surface Gr1 expression (Figure 3F), characteristics compatible with immature granulocytic precursors.34 By double staining, the RasGRP1-expressing cells did not stain for the neutrophil marker 7/4, the monocyte/macrophage marker CD11b, the T-cell marker CD4, the pan-B cell marker CD45R/B220, or the hematopoietic cell progenitor marker cKit; most of the cells expressed the G-CSFR, confirming their probable myeloid lineage derivation (Figure 3G). Further analysis showed that the RasGRP1-expressing cells were found within the CMP (lin−Sca−c-Kit+CD34+CD16/32mid) and GMP ((lin−Sca−c-Kit+CD34+CD16/32hi) cell populations (Figure 3H).
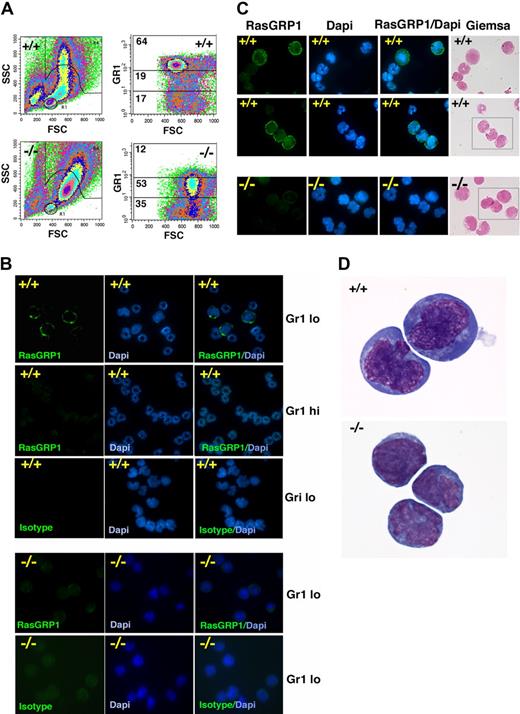
We sorted bone marrow MNCs that exhibited high SSC and low- or high-level Gr1 expression. Consistent with previous observations,5 forward scattering counter (FSC) and SSC of bone marrow MNCs from Gfi1−/− mice differed from that of Gfi1+/+ mice in showing a reduction of cells with high SSC and intermediate forward scattering counter (Figure 4A). Consistent with their defective neutrophil maturation, Gfi1-null mice showed a reduction in the Gr1hi cell population and an expansion of Gr1lo cell population (Figure 4A right). Within the Gr1lo cell population from Gfi1+/+ mice, we identified cells that specifically stained brightly with RasGRP1 mAb (Figure 4B). However, no RasGRP1-staining cells were identified within the Gr1lo cell population from Gfi1−/− mice (Figure 4B). RasGRP1 staining was mostly confined to the plasma membrane/cytoplasm, and the cells that stained for RasGRP1 displayed the nuclear morphology of immature cells (Figure 4B-D). No RasGRP1-brightly positive cells were identified within the more mature myeloid cells, based on nuclear morphology (DAPI and Giemsa staining) and Gr1hi expression (Figure 4B-C).
Characterization of RasGRP1-expressing cells in the bone marrow. (A) FSC and SSC of bone marrow cells from a representative Gf11+/+ and Gfi1−/− mouse showing the different distribution pattern (left). Distribution of bone marrow cells expressing high (hi), intermediate, and low (lo) levels of surface Gr1 showing a reduction of Gr1hi and an increase in Gr-1lo in a representative Gfi1−/− mouse compared with control Gfi1+/+ mouse. The percentage of cells within each gate is displayed (representative results from 5 experiments). (B) Cytospin preparations of sorted bone marrow Gr1hi and Gr1lo cell populations from a control Gfi1+/+ mouse showing that RasGRP1-stained cells have the nuclear morphology of immature myeloid cells and are confined to the Gr1lo cell populations. Mouse IgG1 isotype antibody was used as a negative control; DAPI shows nuclear morphology. RasGRP1 is undetectable in bone marrow Gr1lo cell populations from Gfi1−/− mice (representative results from 5 experiments). (C) Giemsa and DAPI staining of Gr1lo-sorted bone marrow populations after immunostaining for intracellular RasGRP1 showing cellular morphology. RasGRP1 is detected in Gr1lo bone marrow cell populations from a Gfi1+/+ mouse but not from a Gfi1−/− mouse. Representative images are shown. (D) Enlarged image from Giemsa stained Gr1lo-sorted bone marrow populations showing the morphology of cells expressing RasGRP1 from the control Gfi1+/+ mouse (top). The morphology of Gr1lo cell populations from the Gfi1−/− mouse that fail to display RasGRP1 immunostaining is shown (bottom) in representative images.
Characterization of RasGRP1-expressing cells in the bone marrow. (A) FSC and SSC of bone marrow cells from a representative Gf11+/+ and Gfi1−/− mouse showing the different distribution pattern (left). Distribution of bone marrow cells expressing high (hi), intermediate, and low (lo) levels of surface Gr1 showing a reduction of Gr1hi and an increase in Gr-1lo in a representative Gfi1−/− mouse compared with control Gfi1+/+ mouse. The percentage of cells within each gate is displayed (representative results from 5 experiments). (B) Cytospin preparations of sorted bone marrow Gr1hi and Gr1lo cell populations from a control Gfi1+/+ mouse showing that RasGRP1-stained cells have the nuclear morphology of immature myeloid cells and are confined to the Gr1lo cell populations. Mouse IgG1 isotype antibody was used as a negative control; DAPI shows nuclear morphology. RasGRP1 is undetectable in bone marrow Gr1lo cell populations from Gfi1−/− mice (representative results from 5 experiments). (C) Giemsa and DAPI staining of Gr1lo-sorted bone marrow populations after immunostaining for intracellular RasGRP1 showing cellular morphology. RasGRP1 is detected in Gr1lo bone marrow cell populations from a Gfi1+/+ mouse but not from a Gfi1−/− mouse. Representative images are shown. (D) Enlarged image from Giemsa stained Gr1lo-sorted bone marrow populations showing the morphology of cells expressing RasGRP1 from the control Gfi1+/+ mouse (top). The morphology of Gr1lo cell populations from the Gfi1−/− mouse that fail to display RasGRP1 immunostaining is shown (bottom) in representative images.
Gfi1 regulates RasGRP1 expression and signaling
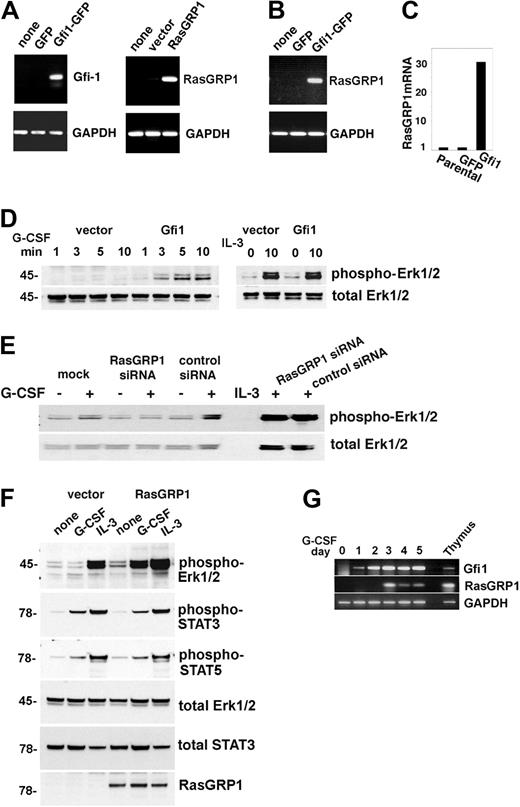
To test for the possibility that Gfi1 may induce RasGRP1 expression and that RasGRP1 deficiency may contribute to defective G-CSF signaling and function in Gfi1−/− mice, we stably transduced the murine myeloid 32D cells with rat Gfi1 or rat RasGRP1 retroviral vectors, because these cells do not constitutively express detectable levels of Gfi1 or RasGRP1 (Figure 5A). Forced expression of Gfi1 in these cells was accompanied by increased expression of endogenous RasGRP1 mRNA (Figure 5B-C), but not G-CSFR (not shown), and increased phosphorylation of Erk1/2 with G-CSF, but not IL-3 (Figure 5D). Conversely, silencing RasGRP1 in Gfi1-transduced 32D cells resulted in decreased phosphorylation of Erk1/2 with G-CSF, but not IL-3 (Figure 5E). In addition, overexpression of RasGRP1 in 32D cells resulted in increased phosphorylation of Erk1/2 on stimulation with G-CSF, but less clearly with IL-3. By contrast, RasGRP1 expression in 32D cells was accompanied by minimal change in G-CSF–induced activation of STAT3 and STAT5 (Figure 5F) and expression of G-CSFR (not shown).
Gfi1 regulates expression of RasGRP1, which is critical for Erk activation by G-CSF. (A) Stable expression of GFP, Gfi1-GFP (left); vector and RasGRP1 (right) in 32D cells assessed by reverse transcription (RT)–PCR with specific primers. Glyceraldehyde phosphate dehydrogenase (GAPDH) expression was used as a control. Representative results from 3 experiments are shown. (B) Expression of Gfi1 in 32D cells specifically promotes expression of endogenous RasGRP1 as measured by RT-PCR. Representative results from 3 experiments are shown. (C) Relative levels of RasGRP1 mRNA expression in parental 32D cells and in 32D cells stably transduced with GFP or GFP-Gfi1 vectors as measured by real-time PCR. Representative results from 3 experiments are shown. (D) Stable expression of Gfi1 in 32D promotes increased Erk1/2 activation by G-CSF (left) but not by IL-3 (right) as assessed by immunoblotting with specific antibodies to phosphorylated Erk1/2. The membrane was reprobed for total Erk1/2. Representative results from 6 experiments are shown. (E) Effect of RasGRP1 silencing in 32D cells stably transduced with RasGRP1. Erk1/2 activation by G-CSF or IL-3 is assessed by immunoblotting. The membranes were reprobed for total Erk1/2. The reduction of RasGRP1 expression in this experiment was 78%, as assessed by real-time PCR. Representative results from 3 experiments are shown. (F) Erk1/2 activation by G-CSF or IL-3 in 32D cells stably expressing RasGRP1 or control cells as assessed by immunoblotting. The membranes were reprobed with the indicated antibodies to assess loading and activation of STAT3 and STAT5. Representative results from 3 experiments are shown. (G) Time-dependent activation of endogenous RasGRP1 in 32D cells cultured with G-CSF as assessed by semiquantitative PCR. Thymus-derived RNA from a Gfi1+/+ mouse was used as a positive control. Representative results from 5 experiments are shown.
Gfi1 regulates expression of RasGRP1, which is critical for Erk activation by G-CSF. (A) Stable expression of GFP, Gfi1-GFP (left); vector and RasGRP1 (right) in 32D cells assessed by reverse transcription (RT)–PCR with specific primers. Glyceraldehyde phosphate dehydrogenase (GAPDH) expression was used as a control. Representative results from 3 experiments are shown. (B) Expression of Gfi1 in 32D cells specifically promotes expression of endogenous RasGRP1 as measured by RT-PCR. Representative results from 3 experiments are shown. (C) Relative levels of RasGRP1 mRNA expression in parental 32D cells and in 32D cells stably transduced with GFP or GFP-Gfi1 vectors as measured by real-time PCR. Representative results from 3 experiments are shown. (D) Stable expression of Gfi1 in 32D promotes increased Erk1/2 activation by G-CSF (left) but not by IL-3 (right) as assessed by immunoblotting with specific antibodies to phosphorylated Erk1/2. The membrane was reprobed for total Erk1/2. Representative results from 6 experiments are shown. (E) Effect of RasGRP1 silencing in 32D cells stably transduced with RasGRP1. Erk1/2 activation by G-CSF or IL-3 is assessed by immunoblotting. The membranes were reprobed for total Erk1/2. The reduction of RasGRP1 expression in this experiment was 78%, as assessed by real-time PCR. Representative results from 3 experiments are shown. (F) Erk1/2 activation by G-CSF or IL-3 in 32D cells stably expressing RasGRP1 or control cells as assessed by immunoblotting. The membranes were reprobed with the indicated antibodies to assess loading and activation of STAT3 and STAT5. Representative results from 3 experiments are shown. (G) Time-dependent activation of endogenous RasGRP1 in 32D cells cultured with G-CSF as assessed by semiquantitative PCR. Thymus-derived RNA from a Gfi1+/+ mouse was used as a positive control. Representative results from 5 experiments are shown.
G-CSF can stimulate Gfi1 expression in 32D cells under culture conditions that promote their terminal differentiation into neutrophils.6,25 We now found that G-CSF additionally stimulates expression of endogenous RasGRP1 in 32D cells after the induction of Gfi1 (Figure 5G). Thus, Gfi1 promotes expression of RasGRP1 in 32D cells, which specifically enhances Erk1/2 phosphorylation in response to G-CSF.
RasGRP1 promotes G-CSF–dependent neutrophil differentiation from hematopoietic progenitor cells
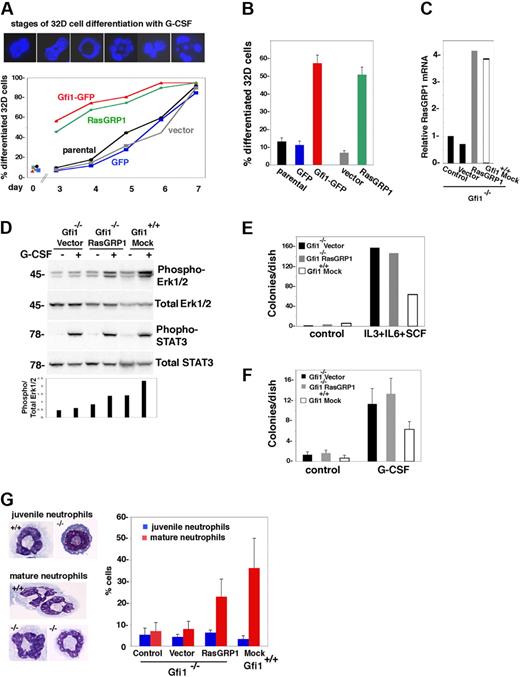
On withdrawal of IL-3 and the addition of G-CSF, 32D cells differentiate from myeloblasts into more mature granulocytic cells over a period of days. We examined whether forced RasGRP1 expression led to increased 32D cell differentiation with G-CSF. Staining with DAPI distinguishes the nuclei of myeloblasts from those of more mature myeloid cells, based on the absence of cavitation, lobulation, or fragmentation (Figure 6A). In kinetic experiments, we found that RasGRP1-transduced 32D cells differentiated more rapidly than control cells (Figure 6A). Gfi1-transduced 32D cells showed differentiation kinetics similar to those of RasGRP1-transduced cells (Figure 6A). After a 4-day incubation with G-CSF, granulocytic differentiation was consistently greater in Gfi1- or RasGRP1-transduced 32D cells than in controls (P < .05; Figure 6B). Thus, functionally, Gfi1 and RasGRP1 similarly support G-CSF–induced granulocytic differentiation of 32D cells.
RasGRP1 restores G-CSF–dependent myeloid cell differentiation from Gfi1-null bone marrow progenitors. (A) Retrovirus-mediated expression of RasGRP1 or Gfi1 in 32D cells accelerates G-CSF–induced differentiation. The top panel shows representative nuclear morphology from DAPI staining of 32D cells during G-CSF incubation; for quantification, stages 1 and 2 were considered undifferentiated nuclei; stages 3 to 6 were considered differentiated nuclei. The bottom panel reflects the kinetics of G-CSF–induced differentiation of control 32D cells (parental, GFP, vector), and those transduced with RasGRP1 or Gfi1. Representative results from 3 experiments are shown. (B) Enhanced differentiation of Gfi1 or RasGRP1-transduced 32D cells after incubation with G-CSF for 3 days (5 experiments; the error bars reflect SDs). Cell populations are described in panel A. (C) Relative RasGRP1 expression in primary Gfi1−/− and Gfi1+/+ bone marrow MNCs 48 hours after retroviral transduction (RasGRP1 or vector only), no transduction (control) or mock transduction (Gfi1 mock) as assessed by real-time PCR. The results are representative of 5 independent transductions. (D) Effects of RasGRP1 transduction on G-CSF–induced activation of Erk1/2 in primary Gfi1-null bone marrow MNCs as assessed by immunoblotting with specific antibodies. After transduction, bone marrow MNCs were cultured for 72 hours in medium (Iscove DMEM with 10% FCS) supplemented with 10 ng/mL IL-3, 25 ng/mL SCF, and 5 ng/mL granulocyte-macrophage–CSF. Before G-CSF activation, the transduced cells were cytokine-starved by incubation for 2 hours in medium only. The bar graph reflects quantitative analysis of band intensities (phospho-Erk/total Erk) as measured by National Institutes of Health ImageJ software. Representative results are shown (of 3 performed). (E) Colony formation in semisolid methylcellulose medium supplemented with IL-3, IL-6, and SCF. Bone marrow MNCs transduced 72 hours earlier (vector only, RasGRP1 retrovirus, or mock-transduced) were cultured in methylcellulose for 14 days; the results reflect the number of colonies/dish (representative experiment of 4 performed). (F) Colony formation in semisolid methylcellulose medium supplemented with G-CSF. The cell populations (described in panel E) were transduced 72 hours before culture in methylcellulose for 12 days. The results reflect the mean number of colonies/dish (± SD) from 3 independent experiments. (G) G-CSF–induced myeloid cell differentiation in liquid cultures. The indicated cell populations were seeded 48 hours after transduction, mock transduction, or no transduction. After 4 days of incubation, cytospin preparations were stained with Giemsa. The left panel shows typical juvenile and mature neutrophils from G-CSF–stimulated primary bone marrow cells from Gfi1+/+ vector-transduced and Gfi1−/− cells transduced with RasGRP1. The right panel reflects the percentage of cells with juvenile and more mature granulocytic morphology as assessed microscopically by an independent observer. The results reflect the means ± SD from 3 independent experiments.
RasGRP1 restores G-CSF–dependent myeloid cell differentiation from Gfi1-null bone marrow progenitors. (A) Retrovirus-mediated expression of RasGRP1 or Gfi1 in 32D cells accelerates G-CSF–induced differentiation. The top panel shows representative nuclear morphology from DAPI staining of 32D cells during G-CSF incubation; for quantification, stages 1 and 2 were considered undifferentiated nuclei; stages 3 to 6 were considered differentiated nuclei. The bottom panel reflects the kinetics of G-CSF–induced differentiation of control 32D cells (parental, GFP, vector), and those transduced with RasGRP1 or Gfi1. Representative results from 3 experiments are shown. (B) Enhanced differentiation of Gfi1 or RasGRP1-transduced 32D cells after incubation with G-CSF for 3 days (5 experiments; the error bars reflect SDs). Cell populations are described in panel A. (C) Relative RasGRP1 expression in primary Gfi1−/− and Gfi1+/+ bone marrow MNCs 48 hours after retroviral transduction (RasGRP1 or vector only), no transduction (control) or mock transduction (Gfi1 mock) as assessed by real-time PCR. The results are representative of 5 independent transductions. (D) Effects of RasGRP1 transduction on G-CSF–induced activation of Erk1/2 in primary Gfi1-null bone marrow MNCs as assessed by immunoblotting with specific antibodies. After transduction, bone marrow MNCs were cultured for 72 hours in medium (Iscove DMEM with 10% FCS) supplemented with 10 ng/mL IL-3, 25 ng/mL SCF, and 5 ng/mL granulocyte-macrophage–CSF. Before G-CSF activation, the transduced cells were cytokine-starved by incubation for 2 hours in medium only. The bar graph reflects quantitative analysis of band intensities (phospho-Erk/total Erk) as measured by National Institutes of Health ImageJ software. Representative results are shown (of 3 performed). (E) Colony formation in semisolid methylcellulose medium supplemented with IL-3, IL-6, and SCF. Bone marrow MNCs transduced 72 hours earlier (vector only, RasGRP1 retrovirus, or mock-transduced) were cultured in methylcellulose for 14 days; the results reflect the number of colonies/dish (representative experiment of 4 performed). (F) Colony formation in semisolid methylcellulose medium supplemented with G-CSF. The cell populations (described in panel E) were transduced 72 hours before culture in methylcellulose for 12 days. The results reflect the mean number of colonies/dish (± SD) from 3 independent experiments. (G) G-CSF–induced myeloid cell differentiation in liquid cultures. The indicated cell populations were seeded 48 hours after transduction, mock transduction, or no transduction. After 4 days of incubation, cytospin preparations were stained with Giemsa. The left panel shows typical juvenile and mature neutrophils from G-CSF–stimulated primary bone marrow cells from Gfi1+/+ vector-transduced and Gfi1−/− cells transduced with RasGRP1. The right panel reflects the percentage of cells with juvenile and more mature granulocytic morphology as assessed microscopically by an independent observer. The results reflect the means ± SD from 3 independent experiments.
Because the transient expression of Gfi1 in Gfi1-null granulocyte progenitors partially corrected the defect in neutrophil differentiation and promoted generation of mature neutrophils,5 we tested the effects of RasGRP1 expression in these assays. With the use of a retroviral vector for expression of RasGRP1 and puromycin selection, we increased relative RasGRP1 expression in Gfi1−/− bone marrow MNCs by 3- to 4-fold (Figure 6C). When transduced with RasGRP1 and stimulated with G-CSF, Gfi1−/− bone marrow cells activated Erk1/2, albeit to a somewhat lower degree than control Gfi1+/+ cells, whereas Gfi1−/− bone marrow cells transduced with the empty retroviral vector essentially failed to do so (Figure 6D).
Colony assays in semisolid medium supplemented with IL-3, IL-6, and SCF showed an increase in progenitors responsive to these cytokines from Gfi1−/− bone marrow cells compared with Gfi1+/+ control, a difference that persisted when the cells were transduced with RasGRP1 (Figure 6E). Most (> 90%) of the colonies from the Gfi1−/− control and RasGRP1-transduced cells were megakaryocyte colony-forming unit (CFU-M), and the remainder (< 10%) were granulocyte-macrophage CFU (CFU-GM). By contrast, the colonies from the Gfi1+/+ precursors were 50% granulocyte CFU, 39% CFU-M, and the remaining 11% were CFU-GM. Colony assays in semisolid medium supplemented with G-CSF also showed an increase in progenitors responsive to this cytokine from Gfi1-null bone marrow cells compared with Gfi1+/+ control (Figure 6F). Virtually all the colonies from Gfi1+/+ precursors were granulocyte CFU, whereas virtually all the colonies from Gfi1−/− vector-transduced precursors were CFU-M. Instead, 30% to 50% of colonies from Gfi1−/− cells transduced with RasGRP1 were CFU-G, and the remainder were CFU-M. Importantly, microscopic analysis of G-CSF–induced colonies (cytospin preparations) from Gfi1-null cells transduced with RasGRP1 showed the presence of clusters of cells at different stages of granulocyte differentiation, including mature neutrophils, which were generally missing from control Gfi1-null bone marrow cells (not shown). To confirm that introduction of RasGRP1 in Gfi1−/− bone marrow cells promoted neutrophil differentiation, we used liquid cultures supplemented with G-CSF. After a 4-day incubation, we counted mature neutrophils in cytospin preparations. In 3 experiments, we found that expression of RasGRP1 in Gfi1−/− bone marrow cells promoted substantial neutrophil differentiation in the presence of G-CSF (P < .05 vector vs RasGRP1), which was largely missing from control cultures (Figure 6G). Thus, introduction of RasGRP1 corrects, at least in part, the defect in neutrophil differentiation resulting from the loss of Gfi1.
Discussion
The interplay between transcription factors and cytokines/growth factors that regulate differentiation of myeloid cells from hematopoietic precursors remains largely undefined. Individually, the transcription factor Gfi1 and the cytokine G-CSF are critical regulators of neutrophil maturation from granulocyte/monocyte progenitors. Here, we show that the transcription factor Gfi1 regulates G-CSF signaling through the Ras/MEK/Erk pathway, by stimulating the expression of RasGRP1, a critical regulator of Ras activation. Thus, the current results uncover a previously unknown link between Gfi1 transcriptional control and G-CSF signaling in the regulation of granulopoiesis.
RasGRP1 is a GEF that activates the small GTPase Ras.27 Levels of active Ras are regulated by the equilibrium between inactive guanosine diphosphate–bound Ras and active GTP-bound Ras. This equilibrium is regulated by Ras GEFs, which activate Ras, and Ras GTPase-activating proteins (GAPs), which inactivate Ras. RasGRP1 has a limited tissue distribution, being detected predominantly in thymocytes, T cells, and various T-cell lines and to a lower degree in B cells.33 Interestingly, RasGRP1 mRNA was detected in the promyelocytic leukemia cell line HL60 (Gene Atlas U133A, gcrma; 205590) and bone marrow from patients with acute myelogenous leukemia.35 Here, we show that RasGRP1 is expressed in the normal bone marrow but not in the bone marrow of Gfi1-null mice. We detect RasGRP1 in 1% to 3% of normal bone marrow MNCs; these RasGRP1+ cells display an immature myeloid cell phenotype, including myeloblasts and promyelocytes. Because promyelocytes comprise 3.7% (± 0.5%) of bone marrow MNCs from 4- to 6-week-old C57BL/6 mice,5 a considerable proportion of myeloid cells at this stage may express RasGRP1. The failure to detect RasGRP1 in Gfi1-null bone marrow cannot be attributed to a putative reduction of granulocyte progenitors, because such cells are increased in Gfi1-null bone marrow.5,6 Because we observed that RasGRP1 is expressed at abnormally low levels in thymus and spleen of Gfi1-null mice, it is possible that RasGRP1 deficiency may also contribute to the T- and B-cell deficiencies in Gfi1-deficient mice and patients.5,8,10
An important question is how RasGRP1 is biochemically linked to G-CSFR. In lymphocytes, immune receptor signaling leads to phospholipid hydrolysis. The resulting diacylglycerol positively regulates RasGRP1 both directly, through membrane recruitment, and indirectly, through protein kinase C–mediated phosphorylation.33 However, evidence that G-CSFR couples to phospholipid hydrolysis is lacking. G-CSFR signaling to Ras is thought to involve phosphorylation of one or more C-terminal tyrosines, which are presumed to serve as docking sites for adaptor proteins Grb2 and SHC, thereby facilitating membrane recruitment of the Ras GEF Sos.36 However, other studies have implicated a distinct membrane-proximal region of the cytoplasmic domain in Ras-Erk signaling.36 This proximal region may signal directly or indirectly, through protein kinases or adaptor proteins, to RasGRP1. The interplay of the 2 Ras GEFs in myeloid cells might allow a variety of intracellular and kinetic patterns of Ras-Erk signaling, resulting in distinct cellular outputs.
Another question is how RasGRP1 deficiency alone could account for defective Erk1/2 activation by G-CSF, given that only a small fraction of myeloid cells in the bone marrow express RasGRP1. It is possible that RasGRP1 is more widely expressed among G-CSF–responsive cell populations than we conservatively estimated, based on brightly positive immunofluorescence staining. It is also possible that other factors, besides RasGRP1, contribute to Erk activation by G-CSF. Consistent with the latter possibility, transduction of Gfi1 or RasGRP1 in Gfi1-null bone marrow cells only partially rescued Erk activation by G-CSF.
G-CSF promotes growth and differentiation in responsive cells, but the signaling pathways that mediate these distinct cytokine functions remain incompletely defined and are probably complex.36,37 In Gfi1-null myeloid cells, where little or no Erk1/2 is activated by G-CSF, we found that G-CSF promotes exaggerated cell proliferation (shown here) and colony formation5,11 but minimal neutrophil differentiation. At least partial neutrophil maturation by G-CSF occurred with reconstitution of Ras/MEK/Erk signaling, supporting the notion that the Ras/MEK/Erk pathway contributes to G-CSF–induced neutrophil differentiation. A previous study with 32D cells bearing a mutant G-CSFR suggested a potential role for Ras activation in maintaining G-CSF–induced proliferation rather than differentiation.38 It is possible that different Ras activators may lead to distinct downstream pathways and that RasGRP1 activates Ras in unique ways, as it was proposed in the case of T cells.39 The Raf-1/MAPK pathway is one of several pathways regulated by active Ras, which can bind to several effector proteins, including phosphatidylinositol 3 kinase and the small GTPase Ral.40 It is also possible that proliferative and differentiation responses in myeloid cells share the same signaling pathway, but the duration of Erk activation differs. In PC12 cells, short-lived Ras-Erk activation leads to proliferation, whereas persistent activation of Ras-Erk leads to differentiation.41
Previously, the monopoietic cytokine CSF-1 (colony stimulating factor-1/M-CSF),11 the transcription factor PU.1,42 and the microRNAs miR21 and miR-196b14 were reported to contribute to defective myelopoiesis in Gfi1-null mice. It is not surprising that defective granulopoiesis in the context of Gfi1 deficiency is multifactorial. Consistent with this possibility, granulopoiesis from Gfi1-null precursors was only partially reconstituted by neutralization of CSF-1,11 heterozygosity at the PU.1 locus,42 silencing miR21 and miR-196b,14 and transduction of RasGRP1, shown here.
Thus far, transcriptional repression of target genes has been identified as a principal mechanism of Gfi1 function as a regulator of granulopoiesis. This has contributed to models in which commitment to granulopoiesis is a default pathway.1 Here, we find evidence that Gfi1 can stimulate the expression of RasGRP1 and induce granulopoiesis G-CSF/G-CSFR differentiation, but the underlying mechanisms need investigation. It is possible that Gfi1 could function as a transcriptional activator of RasGRP1 expression. However, because we did not find conserved Gfi1 binding sequences in the putative promoter region of RasGRP1, Gfi1 may indirectly modulate RasGRP1 expression by repressing negative regulators. Other transcription factors can both activate and repress transcription of target genes. Myc, for example, activates transcription directly when bound to its DNA consensus sequence, but it represses transcription when tethered to target sequences by MiZ1 or other cofactors.43 Besides expanding the spectrum of Gfi function as a regulator of hematopoiesis, the current observations will probably lead to important insights in myeloid cell development, maturation, and transformation that will inform therapies aimed at manipulating myelopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Cynthia A. Pise-Masison, LCO, CCR, NCI, for performing microarray analysis; the Clinical Pathology Laboratory of the Clinical Center, National Institutes of Health and the Lab Animal Science Program, NCI; and Drs Linda Wolff and Juraj Bies for advice.
This work was supported by the Intramural Research Program at NIH, NCI, Center for Cancer Research and the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. S.S. is the recipient of a JSPS fellowship; M.S. is the recipient of a Ministerio de Educacion y Ciencia (MEC)–Fulbright fellowship.
National Institutes of Health
Authorship
Contribution: G.T. and M.D.L.L.S. designed research, performed the experiments, analyzed data, and wrote the paper; S.S., P.G., O.S., M.S., K.J., and D.M. performed some experiments; P.J.M., J.S., and D.R.L. provided critical intellectual input to experiments and manuscript; and J.Z. and X.Q. provided critical reagents and experimental suggestions.
Conflict-of-interest disclosure: The authors declare no competing financial interests and no involvement of medical writers or researchers in the writing of the article.
Correspondence: Giovanna Tosato, Laboratory of Cellular Oncology, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bldg 37, Rm 4124, Bethesda, MD 20892; e-mail: tosatog@mail.nih.gov.





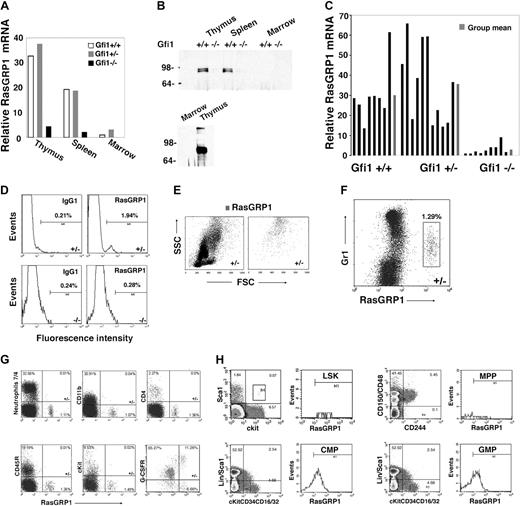
 . (D) Flow cytometric detection of RasGRP1 in a population of bone marrow MNCs from a Gfi1+/− but not Gfi1−/− mouse. After fixation and permeabilization, the cells were immunostained with a mouse monoclonal antibody to RasGRP1 or isotype control antibody (mouse IgG1) followed by a goat anti–mouse IgG Alexa Fluor 488–conjugated antibody. The results reflect the percentage of RasGRP1-positive cells and are representative of 5 independent experiments. (E) Forward scattering counter (FSC) and SSC of bone marrow MNCs from a Gfi1+/− mouse after RasGRP1 immunostaining (as described in panel D). Left panel shows total; right panel shows RasGRP1 only. The results are representative of Gfi1+/+ (n = 4) and Gfi1+/− (n = 3) mice. (F) Flow cytometric analysis of Gr1 and RasGRP1 expression within bone marrow MNCs from a Gfi1+/− mouse, showing that RasGRP1-positive cells display low-level surface Gr1. The MNCs were first immunostained for surface Gr1 (PE-labeled) and after fixation/permeabilization immunostained for RasGRP1 (as described in panel D). Representative of 5 independent experiments is shown. The percentage of cells in the box is shown. (G) Flow cytometric detection of the surface markers neutrophils 7/4, CD11b, CD4, CD45R/B220, cKit, G-CSFR, and intracellular RasGRP1in bone marrow MNCs from a Gfi1+/− mouse. G-CSFR was detected by PE-labeled G-CSF cross-linked to the cell surface with Bis(sulfosuccinimidyl)suberate
. (D) Flow cytometric detection of RasGRP1 in a population of bone marrow MNCs from a Gfi1+/− but not Gfi1−/− mouse. After fixation and permeabilization, the cells were immunostained with a mouse monoclonal antibody to RasGRP1 or isotype control antibody (mouse IgG1) followed by a goat anti–mouse IgG Alexa Fluor 488–conjugated antibody. The results reflect the percentage of RasGRP1-positive cells and are representative of 5 independent experiments. (E) Forward scattering counter (FSC) and SSC of bone marrow MNCs from a Gfi1+/− mouse after RasGRP1 immunostaining (as described in panel D). Left panel shows total; right panel shows RasGRP1 only. The results are representative of Gfi1+/+ (n = 4) and Gfi1+/− (n = 3) mice. (F) Flow cytometric analysis of Gr1 and RasGRP1 expression within bone marrow MNCs from a Gfi1+/− mouse, showing that RasGRP1-positive cells display low-level surface Gr1. The MNCs were first immunostained for surface Gr1 (PE-labeled) and after fixation/permeabilization immunostained for RasGRP1 (as described in panel D). Representative of 5 independent experiments is shown. The percentage of cells in the box is shown. (G) Flow cytometric detection of the surface markers neutrophils 7/4, CD11b, CD4, CD45R/B220, cKit, G-CSFR, and intracellular RasGRP1in bone marrow MNCs from a Gfi1+/− mouse. G-CSFR was detected by PE-labeled G-CSF cross-linked to the cell surface with Bis(sulfosuccinimidyl)suberate