Abstract
Activation of plasminogen, the zymogen of the primary thrombolytic enzyme, plasmin, is markedly promoted when plasminogen is bound to cell surfaces, arming cells with the broad spectrum proteolytic activity of plasmin. In addition to its role in thrombolysis, cell surface plasmin facilitates a wide array of physiologic and pathologic processes. Carboxypeptidase B-sensitive plasminogen binding sites promote plasminogen activation on eukaryotic cells. However, no integral membrane plasminogen receptors exposing carboxyl terminal basic residues on cell surfaces have been identified. Here we use the exquisite sensitivity of multidimensional protein identification technology and an inducible progenitor cell line to identify a novel differentiation-induced integral membrane plasminogen receptor that exposes a C-terminal lysine on the cell surface, Plg-RKT (C9orf46 homolog). Plg-RKT was highly colocalized on the cell surface with the urokinase receptor, uPAR. Our data suggest that Plg-RKT also interacts directly with tissue plasminogen activator. Furthermore, Plg-RKT markedly promoted cell surface plasminogen activation. Database searching revealed that Plg-RKT mRNA is broadly expressed by migratory cell types, including leukocytes, and breast cancer, leukemic, and neuronal cells. This structurally unique plasminogen receptor represents a novel control point for regulating cell surface proteolysis.
Introduction
Localization of plasminogen on cell surfaces is a crucial control point for positive regulation of cell surface plasmin proteolytic activity that facilitates both physiologic and pathologic processes,1,2 including macrophage recruitment during the inflammatory response,3-6 tissue remodeling,7 wound healing,8,9 tumor cell invasion and metastasis,10-12 skeletal myogenesis,13 neuroendocrine prohormone processing,14,15 and neurite outgrowth.16,17 Cell surface plasminogen binding sites promote plasminogen activation by reducing the Km (11- to 60-fold) for plasminogen activation.18-24 Active plasmin also associates with the cell surface, where its activity is protected from inhibitors.25,26
Plasminogen binding sites are very broadly distributed on both eukaryotic and prokaryotic cells.27 Of the many eukaryotic cells examined to date, only erythrocytes do not bind plasminogen.28 The interactions of plasminogen with eukaryotic cells are mediated by lysine binding sites within the disulfide-bonded kringle domains of plasminogen.18,29 Therefore, plasminogen binding to eukaryotic cells is blocked in the presence of lysine and lysine analogs, including ϵ-aminocaproic acid (EACA).27 Because most cell types have a very high capacity for plasminogen, no single molecule can account for the entire plasminogen binding capacity of a given cell type.27 However, a subset of plasminogen binding proteins exposing C-terminal basic residues on cell surfaces are predominantly responsible for the ability of eukaryotic cells to enhance plasminogen activation because carboxypeptidase B (CpB) treatment abrogates cell surface–dependent plasminogen activation.24 Correspondingly, plasminogen-dependent macrophage recruitment in vivo is mediated by CpB-sensitive plasminogen receptors, and plasminogen binding to recruited macrophages is increased, compared with peripheral blood monocytes.6,30 Therefore, we probed the monocyte proteome as a source of an inducible integral membrane plasminogen receptor(s) exposing a C-terminal basic residue on the cell surface.
Several plasminogen binding proteins with established intracellular functions that are synthesized with C-terminal lysines associate with the monocytoid cell surface (eg, α-enolase,29,31 TIP49a,32 histone H2B, and p1133 ). Other functional plasminogen binding proteins that are not synthesized with C-terminal basic residues are present on monocytoid cells, including annexin II,34 amphoterin,35 tissue factor,36 and αMβ2.37 However, no integral membrane plasminogen binding proteins that are synthesized with C-terminal basic residues have been identified to date. The existence of a receptor with such a structure would constitute a novel mechanism for stimulating plasminogen activation because its induction would endow cells with the ability to bind plasminogen and promote plasminogen activation, without requiring release and rebinding of intracellular proteins or proteolytic cleavage of a membrane protein to reveal C-terminal basic residues. Therefore, we used the exquisite sensitivity of multidimensional protein identification technology (MudPIT) to search for an integral membrane plasminogen receptor(s), exposing a C-terminal basic residue on the cell surface and up-regulated during differentiation.
Methods
Proteins
Glu-plasminogen was purified from fresh human blood as described.38,39 Lys-plasminogen was from Enzyme Research Laboratories. Single-chain tissue plasminogen activator (t-PA) was from Calbiochem. Polyclonal antibodies were raised in rabbits and monoclonal antibodies were raised in rats and mice against the synthetic peptide, CEQSKLFSDK, thiol coupled to keyhole limpet hemocyanin. Antibodies were selected for direct binding to immobilized CEQSKLFSDK coupled to bovine serum albumin (BSA) and for the ability to inhibit specific plasminogen binding to CEQSKLFSDK. Anti–α-enolase monoclonal antibody (mAb) 9-C1231 and polyclonal antiplasminogen40 were prepared in our laboratory. Anti–urokinase plasminogen activator receptor (uPAR; product no. 3936) was from American Diagnostica. Fluorescein isothiocyanate (FITC)–conjugated CD marker antibodies and the relevant FITC-conjugated isotype control antibodies were from Pharmingen. The mouse macrophage differentiation antibody rat anti–mouse F4/80 FITC conjugate was from Serotec. Goat anti–mouse FITC-conjugated polyclonal antibody and goat anti–rabbit FITC-conjugated polyclonal antibody were from Calbiochem. Alexa 488-F(ab′)2 of goat anti–rabbit immunoglobulin G (IgG) and Alexa 568-F(ab′)2 fragment of goat anti–mouse IgG were from Invitrogen.
Cells
For differentiation experiments, monocytes were further isolated from the mononuclear cell population by plating onto tissue culture dishes (Corning), before the addition of human recombinant macrophage colony stimulating factor (M-CSF; Calbiochem).
Hoxa9-ER4 cells were cultured as described41 and were differentiated either with murine M-CSF (Calbiochem) or with conditioned media produced by LADMAC cells (ATCC), as a source of M-CSF.
Quantitative flow cytometry
Quantitative flow cytometric equilibrium binding of fluorescein isothiocyanate (FITC)–plasminogen to monocytoid cells was analyzed using beads impregnated with FITC as described.42 Briefly, the output from the flow cytometer was standardized into mean equivalent standard fluorescence units using beads impregnated with different mean equivalent standard fluorescence units of FITC. The fluorescence intensity change of the EACA-inhibitable FITC-plasminogen conformational change (Q = ΔIf/ΔIi; where Ii and If are the initial and final fluorescence intensities of FITC-plasminogen, respectively) was approximated using a F500 fluorometric plate reader (Bio-Rad Model 680; Bio-Rad) equipped with a FITC filter set (excitation, 490 nm; emission, 520 nm). Binding parameters were determined using the program, NLREG, Version 4.1. (Phillip H. Sherrod; www.nlreg.com)
Laser scanning confocal microscopy
Confocal images were captured using a Zeiss 710 laser confocal scanning microscope (LCSM), with a 63 × objective (1.4 na), running the latest Zen 2009 Zeiss software suite (Carl Zeiss Inc). All images were then imported and further analyzed for quantitative colocalization using 2 independent software packages: LSM examiner (Zeiss Inc) and ImageJ (National Institutes of Health [NIH] imaging; http://rsb.info.nih.gov/ij). Colocalization between fluorescently labeled aggregates of Plg-RKT with either uPAR or plasminogen was quantified by obtaining the threshold range of real over background signal and then using the average real threshold range to calculate the correlation coefficients (M values) of at least 40 cells in 2 separate experiments. To define the number and size of each labeled aggregate, images were imported into Image Pro Plus (Media Cybernetics Inc) where each cell was outlined and a similar threshold range was used to define a real signal within each cell. Once this range was defined the software then automatically extracts parameters such as area, perimeter total number, and average fluorescence intensity of the fluorescently labeled aggregates per cell.
Plasminogen receptor isolation
Progenitor and M-CSF–differentiated Hoxa9-ER4 cells (5 × 108) were separately biotinylated, using EZ-Link Amine-PEO3-Biotin (Pierce). The cells were then subjected to dead cell removal on annexin V–coated magnetic microspheres (Miltenyi) that resulted in a 99% enrichment of viable cells (as determined in fluorescence-activated cell sorting [FACS] analysis with propidium iodide and annexin V). Membrane fractions were prepared from the viable cells by Dounce homogenization in the presence of Complete Protease Inhibitor Cocktail (Roche) in Invitrosol (Invitrogen), followed by centrifugation steps as used in our laboratory32,43 and 3 mg was applied to a 1-mL plasminogen-Sepharose affinity column as described.29 The column was washed in phosphate-buffered saline (PBS) containing 1 × Invitrosol until no protein was detected at 280 nm followed by elution with the washing buffer containing 0.2M of EACA. The eluant from the plasminogen-Sepharose column was incubated with 50 μL of immobilized avidin for 30 minutes at 4°C. Proteins bound to the immobilized avidin were resuspended in 5 μL of Invitrosol and heated at 60°C for 5 minutes. Then, 45 μL of 80% acetonitrile was added and the samples were digested with trypsin at 37°C for 18 hours. After 24 hours, the solvent was evaporated in a speedvac (ThermoFisher Scientific Model), and peptides were dissolved in 50 μL of buffer A (95% H2O, 5% acetonitrile, and 0.1% formic acid).
Multidimensional chromatography and tandem mass spectrometry
The protein digest was subjected to MudPIT (reviewed in Eng et al44 ). Peptide mixtures were resolved by strong cation exchange liquid chromatography followed by reversed-phase liquid chromatography.45-48 Eluting peptides were electrosprayed into an LTQ ion trap mass spectrometer equipped with a nano–liquid chromatography electrospray ionization source (ThermoFinnigan). Full mass spectrometry (MS) spectra were recorded over a 400- to 1600-m/z range, followed by 3 tandem mass spectrometry (MS/MS) events sequentially generated in a data-dependent manner on the first, second, and third most intense ions selected from the full MS spectrum (at 35% collision energy). Mass spectrometer scan functions and high-performance liquid chromatography solvent gradients were controlled by the Xcalibur data system (ThermoFinnigan).
Database search and interpretation of MS/MS datasets
Tandem mass spectra were extracted from raw files, and a binary classifier,49 previously trained on a manually validated dataset, was used to remove low-quality MS/MS spectra. Remaining spectra were searched against a Mus musculus protein database containing 50 370 protein sequences downloaded as FASTA-formatted sequences from European Bioinformatics Institute's International Protein Index (EBI-IPI; database Version 3.23, released on November 2, 2006), and 124 common contaminant proteins, for a total of 66 743 target database sequences.50,51 To calculate confidence levels and false-positive rates, we used a decoy database containing the reverse sequences of the 66 743 proteins appended to the target database, and the SEQUEST algorithm52 to find the best matching sequences from the combined database.
SEQUEST searches were done on an Intel Xeon 80-processor cluster running under the Linux operating system. The peptide mass search tolerance was set to 3 Da. No differential modifications were considered. No enzymatic cleavage conditions were imposed on the database search, so the search space included all candidate peptides whose theoretic mass fell within the 3-Da mass tolerance window, despite their tryptic status.
The validity of peptide/spectrum matches was assessed in DTASelect253 (Scripps Research Institute; http:/fields.scripps.edu/?q=content/software) using SEQUEST-defined parameters, the cross-correlation score (XCorr), and normalized difference in cross-correlation scores (DeltaCN). The search results were grouped by charge state (+1, +2, and +3) and tryptic status (fully tryptic, half-tryptic, and nontryptic), resulting in 9 distinct subgroups. In each one of the subgroups, the distribution of XCorr and DeltaCN values for direct and decoy database hits was obtained, and the 2 subsets were separated by quadratic discriminant analysis. Outlier points in the 2 distributions (for example, matches with very low Xcorr but very high DeltaCN) were discarded. Full separation of the direct and decoy subsets is not generally possible; therefore, the discriminant score was set such that a false-positive rate of 5% was determined based on the number of accepted decoy database peptides. This procedure was independently performed on each data subset, resulting in a false-positive rate independent of tryptic status or charge state.
Plasminogen and t-PA binding assays
Either the C-terminal peptide of Plg-RKT (CEQSKLFSDK coupled to BSA), the reverse peptide (KDSFLKSQEC), or BSA alone was coated onto wells of microtiter plates at 10 μg/mL. The wells were blocked with 5% BSA. Either Glu-plasminogen or Lys-plasminogen was then incubated with the wells, followed by antiplasminogen mAb51 raised in our laboratory,54 and detection with horseradish peroxidase (HRP)–conjugated goat anti–mouse IgG (Biosource). Binding of single-chain t-PA was determined using anti–t-PA mAb (Calbiochem) followed by HRP-conjugated goat anti–mouse IgG (Biosource).
Plasminogen activation assay
Cells (8 × 104/mL) were preincubated with rat anti–Plg-RKT mAb 35B10 or rat IgG2a for 30 minutes at 37°C. Glu-plasminogen (2.7 μM) was then added and incubated for an additional 30 minutes at 37°C. Then 20 nM of single-chain recombinant t-PA was added. Plasmin activity was measured after 3 minutes by diluting the reaction mixture 1:10 into S-2251 (DiaPharma Group) to a final concentration of 1mM and monitoring absorbance at 405 nm as described previously.24,42
Reagents
Immuno-Fluore Mounting Medium was from ICN Biomedicals Inc.
Results
Plasminogen binding capacity is enhanced when monocytes differentiate
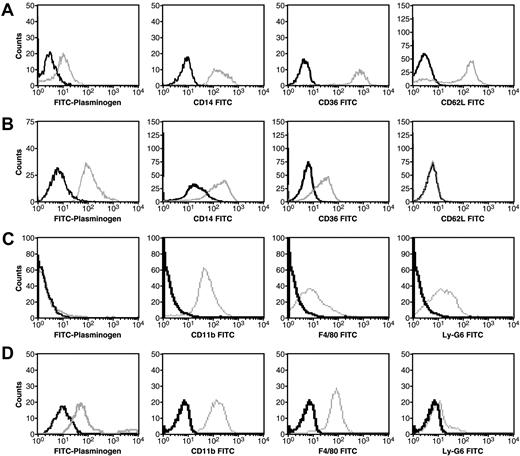
To establish a system whereby plasminogen receptors could be induced, we examined the effect of differentiation on plasminogen binding to monocytes. Peripheral blood monocytes were obtained from mononuclear cells isolated from freshly donated human blood as described.55 In FACS analysis, as described,10 the cells were identified by their distinct forward scatter/side scatter properties compared with the other cells within the mononuclear cell preparations. Plasminogen binding was analyzed by dual-color FACS analysis using FITC-plasminogen and propidium iodide to gate for only viable cells, as described.10 Specific binding was defined as binding that was inhibited by EACA. The viable peripheral blood monocytes bound plasminogen specifically, as defined by the shift in fluorescence in the presence of EACA (Figure 1A left-most column, [FITC-plasminogen]). Monocytes were further purified from the mononuclear cell preparation by adherence as described under “Cells” and then treated with the differentiation-inducing cytokine, M-CSF. The fluorescence signal indicating specific FITC-plasminogen binding to the viable M-CSF–differentiated cells was 11-fold greater than that observed with the unstimulated monocytes (compare Figure 1A and B left-most columns [FITC-plasminogen]). The CD antigen profiles were consistent with differentiation into macrophages (compare Figure 1A and B), and the morphology of the cells changed from a rounded shape to a spindle-type morphology (data not shown). Thus, differentiation of the monocytes toward the macrophage phenotype markedly up-regulated plasminogen binding.
Specific plasminogen binding is enhanced by M-CSF treatment of human peripheral blood monocytes and Hoxa9-ER4 cells. Human peripheral blood monocytes were either untreated (A) or treated with 0.44 nM of M-CSF for 8 days (B) and Hoxa9-ER4 cells were either untreated (C) or treated with 20% LADMAC conditioned media (a source of M-CSF) for 2 days (D). The cells were analyzed by dual-color fluorescence-activated cell sorting (FACS) analysis for specific plasminogen binding and CD antigen expression as described.10 Viable (propidium iodide negative and annexin V negative) cells were gated from the nonviable cells. Histogram plots of FITC-plasminogen (left columns) or specific anti-CD antibody binding to viable cells are shown. Gray tracings indicate either FITC-plasminogen or specific anti-CD antibody; black tracings, either FITC-plasminogen + 0.2 M EACA or isotype controls.
Specific plasminogen binding is enhanced by M-CSF treatment of human peripheral blood monocytes and Hoxa9-ER4 cells. Human peripheral blood monocytes were either untreated (A) or treated with 0.44 nM of M-CSF for 8 days (B) and Hoxa9-ER4 cells were either untreated (C) or treated with 20% LADMAC conditioned media (a source of M-CSF) for 2 days (D). The cells were analyzed by dual-color fluorescence-activated cell sorting (FACS) analysis for specific plasminogen binding and CD antigen expression as described.10 Viable (propidium iodide negative and annexin V negative) cells were gated from the nonviable cells. Histogram plots of FITC-plasminogen (left columns) or specific anti-CD antibody binding to viable cells are shown. Gray tracings indicate either FITC-plasminogen or specific anti-CD antibody; black tracings, either FITC-plasminogen + 0.2 M EACA or isotype controls.
Our search for an integral membrane plasminogen receptor(s), exposing a C-terminal basic residue on the cell surface and up-regulated during monocyte differentiation, required large quantities of cells. Therefore, we tested whether the Hoxa9-ER4 cell line would behave as peripheral blood monocytes with respect to plasminogen binding capacity. The Hoxa9-ER4 cell line is derived from primary murine bone marrow myeloid precursors immortalized with an estrogen-regulated conditional oncoprotein, Hoxa9-ER4.56 The Hoxa9-ER4 line is factor dependent (granulocyte-macrophage colony-stimulating factor), and differentiates to monocytes when estrogen is removed from the medium, thereby inactivating the Hoxa9-ER protein. The mature monocytes respond to M-CSF.41 We compared plasminogen binding to undifferentiated and M-CSF–differentiated Hoxa9-ER4 progenitor cells. No specific binding of plasminogen to the Hoxa9-ER4 progenitor cells was detected (Figure 1C left-most column [FITC = Plasminogen]). However, as the cells differentiated along the monocytic pathway, specific plasminogen binding to the cells was observed (Figure 1D left-most column [FITC = Plasminogen]). The CD antigen profiles were consistent with differentiation into the macrophage phenotype (compare Figure 1 C and D).
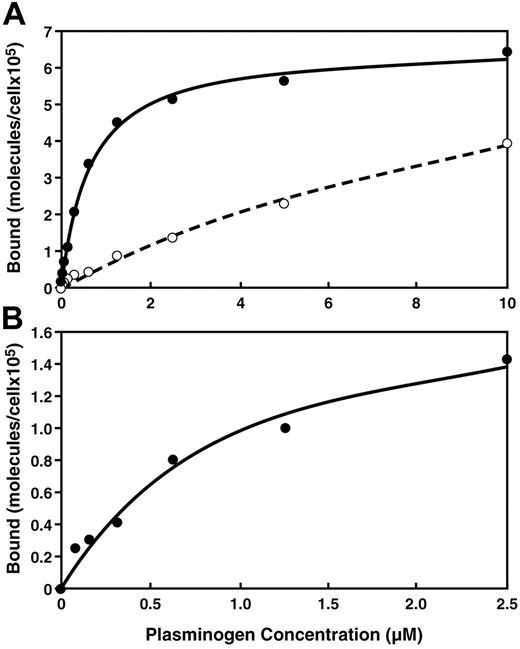
Binding isotherms were constructed for the specific interaction of plasminogen with the monocytoid cells using dual-color quantitative flow cytometry (Figure 2). FACS data from the gated viable cell populations that were treated with different concentrations of FITC-plasminogen, in the presence or absence of EACA, were used to derive the apparent Kd and Bmax of the interactions by fitting these data to the single site binding equation (LR) = ([L]Bmax)/([L]+Kd). (The FACS signal that was inhibitable by EACA was calculated as specific binding.) The plasminogen binding isotherms with M-CSF–treated monocytes and M-CSF–treated Hoxa9-ER4 cells were saturable (Figure 2A-B). The binding parameters determined for the M-CSF–differentiated Hoxa9-ER4 cells were apparent Kd = 0.95 μM and Bmax = 1.9 × 105 sites/cell. An apparent Kd of 0.57 μM and a Bmax of 9.1 × 105 molecules/cell were calculated for M-CSF–treated monocytes. In contrast, with peripheral blood monocytes, the specific plasminogen binding isotherms did not reach saturation at a plasminogen concentration of 10μM (Figure 2A), suggesting that the Kd is more than 5μM and the Bmax is more than 4 × 105 molecules/cell. (Based on the data in Figure 2A, at blood and extracellular concentrations of plasminogen [2 μM57 ], ∼ 105 molecules of plasminogen are predicted to be bound to each circulating peripheral blood monocyte, whereas ∼ 5 × 105 molecules of plasminogen are predicted to be bound per macrophage.) Thus, differentiation increased both the affinity and capacity of monocytes and progenitor cells for plasminogen.
Equilibrium binding of plasminogen is observed to viable human peripheral blood monocytes and viable Hoxa9-ER4 cells. Human peripheral blood monocytes (A) were either untreated (○) or treated with M-CSF (●) as in Figure 1 and Hoxa9-ER4 cells (B) were treated with M-CSF as in Figure 1 and subjected to quantitative flow cytometry with increasing concentrations of FITC-plasminogen as described in under “Quantitative flow cytometry.”
Equilibrium binding of plasminogen is observed to viable human peripheral blood monocytes and viable Hoxa9-ER4 cells. Human peripheral blood monocytes (A) were either untreated (○) or treated with M-CSF (●) as in Figure 1 and Hoxa9-ER4 cells (B) were treated with M-CSF as in Figure 1 and subjected to quantitative flow cytometry with increasing concentrations of FITC-plasminogen as described in under “Quantitative flow cytometry.”
We examined whether the M-CSF up-regulated plasminogen binding sites were accessible to CpB. The M-CSF–differentiated Hoxa9-ER4 cells were incubated with 100 U/mL CpB and washed, before determining FITC-plasminogen binding to the viable cells in FACS analysis. CpB treatment decreased specific FITC-plasminogen binding to the viable cells by 78%, consistent with our results with other monocytoid cells.24,29,58
Isolation of a regulated integral membrane plasminogen receptor exposing a C-terminal basic residue on the cell surface
We used MudPIT to probe the membrane proteome of M-CSF–differentiated Hoxa9-ER4 cells for the presence of a regulated integral membrane plasminogen receptor(s), exposing a C-terminal basic residue on the cell surface. Intact M-CSF–differentiated Hoxa9-ER4 cells were biotinylated as described in “Plasminogen receptor isolation.” Initially, using FACS analysis we verified that (1) viable cells were effectively biotinylated using antibiotin FITC-labeled monoclonal antibodies, (2) specific plasminogen binding by the cells was not decreased when cells were treated with the biotinylating reagent, and (3) biotinylated cells exhibited the same reduction in plasminogen binding capacity in response to CpB as nonbiotinylated cells, as criteria that the biotinylated plasminogen receptors behaved as native plasminogen receptors. The cell membranes were then isolated, subjected to affinity chromatography on plasminogen-Sepharose, bound to immobilized avidin, digested with trypsin, and subjected to MudPIT analysis, as described in “Multidimensional chromatography and tandem mass spectrometry.” Using this method, only 1 protein with a predicted transmembrane sequence and a C-terminal basic residue was identified: the hypothetical protein, C9orf46 homolog (IPI00136293), homologous to the protein predicted to be encoded by human chromosome 9, open reading frame 46. Table 1 lists the peptides corresponding to C9orf46 homolog that were obtained when the membrane fraction from M-CSF–treated Hoxa9-ER4 cells was subjected to MudPIT. Our isolation of peptides corresponding to C9orf46 homolog is, to our knowledge, the first demonstration of the existence of this protein. We have designated the protein, Plg-RKT, to indicate a plasminogen receptor with a C-terminal lysine and having a transmembrane domain.
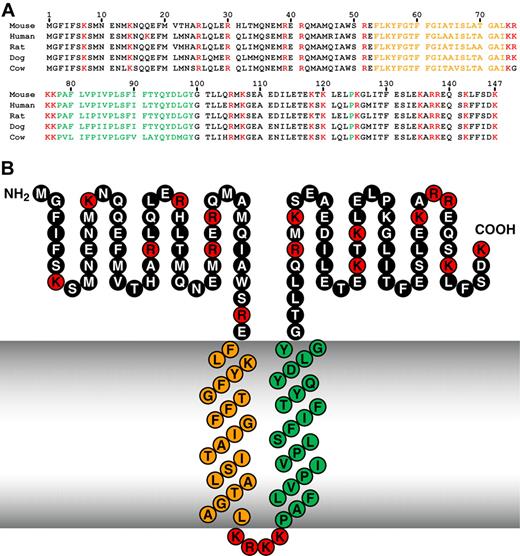
The C9orf46 homolog/Plg-RKT murine DNA sequence encodes a protein of 147 amino acids with a calculated molecular mass of 17 261 Da (Figure 3A top line). Notably, a C-terminal lysine is encoded. We blasted the C9orf46 homolog/Plg-RKT sequence against all species using NCBI Blast59 and obtained unique human, rat, dog, and cow predicted orthologs, with high homology (eg, human vs mouse = 94% similarity), high identity, and no gaps in the sequence (Figure 3A and Table 2). Of key importance, a C-terminal lysine was predicted for all of the mammalian orthologs obtained in the blast search (Figure 1A). In a query of the Ensembl Gene Report,60 DNA sequences of all 9 other fully sequenced mammalian orthologs (Table 3) and 13 other partially sequenced mammalian orthologs encoded C-terminal lysines.
High interspecies homology of Plg-RKT. Alignment of predicted amino acid sequences of mouse, human, rat, dog, and cow orthologs of Plg-RKT (A) and the structural model of Plg-RKT (B). Green indicates amino acids within the predicted primary transmembrane helix; orange, amino acids within the predicted secondary transmembrane helix; and red, basic amino acids.
High interspecies homology of Plg-RKT. Alignment of predicted amino acid sequences of mouse, human, rat, dog, and cow orthologs of Plg-RKT (A) and the structural model of Plg-RKT (B). Green indicates amino acids within the predicted primary transmembrane helix; orange, amino acids within the predicted secondary transmembrane helix; and red, basic amino acids.
In addition, the primary sequence of C9orf46/Plg-RKT is apparently tightly conserved in humans, with no polymorphisms (cSNPs) within the 6 exons encoded by the gene (on chromosome 9p24.1) yet identified in the NCBI human genome sequence variation database (dbSNP, http://www.ncbi.nlm.nih.gov/SNP).61
The C9orf46 homolog/Plg-RKT sequence was analyzed in the TMpred site (Swiss Institute of Bioinformatics; http://www.ch.embnet.org/software/TMPRED_form.html). The strongly preferred model predicted 2 transmembrane helices extending from F53-L73 (secondary helix, oriented from outside the cell to inside) and P78-Y99 (primary helix, oriented from inside the cell to outside; Figure 3B). The 2 transmembrane helices with the same orientation were also predicted using the HMMTOP62 site (Institute of Enzymology, Biological Research Center, Hungarian Institute of Science; http://www.enzim.hu/hmmtop. This model takes into account the “positive inside” rule in which positively charged residues in membrane proteins (eg, residues K74R75K76K77) are found predominantly on the cytoplasmic side of the membrane.63 Notably, a highly positively charged loop (residues 74-77) between the 2 predicted transmembrane domains is conserved in the predicted protein sequences of all mammalian orthologs for which either full or partial sequences are available (eg, Table 3). When analyzed in the Split 4.0 Server site (Membrane Protein Secondary Structure Prediction Server; http://split.pmfst.hr/split/4),64 only the region extending from F80-Q94 was identified as a predicted transmembrane helix. Thus, the possibility that the N-terminus may be embedded in the cytoplasm cannot be excluded. Nonetheless, the conservation of the highly charged positive loop (residues 74-77) strongly favors the model in which F53-L73 also is a transmembrane domain (with the N-terminus outside) because the presence of positively charged residues at the C-terminus of apolar domains in proteins favors the insertion of the upstream apolar domains into the cell membrane.65
Several key aspects of our experimental paradigm support the topologic model shown in Figure 3B. First, recovery of peptides corresponding to C9orf46 homolog relied on accessibility to the biotinylation reagent in the context of intact cells, supporting the exposure of Plg-RKT domains on the cell surface. The biotinylation reagent we used reacts with the carboxyl moieties of the R-groups of either Asp or Glu. Thus, biotinylation of Plg-RKT on intact cells would not occur if the highly basic loop between the transmembrane helices (K74-K77) was exposed on the cell surface. Hence, a model in which both the amino and C-termini are on the cytoplasmic face of the membrane can be excluded.
Second, a key prediction from the model in Figure 3B is the presence of a 48 amino acid C-terminal tail with a C-terminal lysine exposed on the cell surface. We addressed this prediction experimentally. When intact M-CSF–treated Hoxa9-ER4 cells were incubated with CpB before subcellular fractionation, purification, and MudPIT analysis, no peptides corresponding to C9orf46 homolog were detected. These data are consistent with exposure of the C-terminal lysine of Plg-RKT on the cell surface and, hence, accessibility to CpB,32 so that CpB treatment of intact cells resulted in loss of the ability of Plg-RKT to bind to the plasminogen-Sepharose column.
Up-regulation of Plg-RKT in membranes of M-CSF–treated cells
To assess whether expression of C9orf46 homolog/Plg-RKT was increased during cellular differentiation, biotinylated membrane fractions from undifferentiated progenitor Hoxa9-ER4 cells were subjected to the purification method described in “Plasminogen receptor isolation.” In the absence of M-CSF treatment, no peptides corresponding to C9orf46 homolog/Plg-RKT were detected, suggesting that membrane expression of Plg-RKT was markedly up-regulated by M-CSF treatment.
To further assess the M-CSF–responsiveness and membrane localization of Plg-RKT we raised a monoclonal antibody in rats against the synthetic peptide, CEQSKLFSDK (corresponding to the 9 C-terminal amino acids of murine Plg-RKT with an amino terminal cysteine added for coupling). Membrane and cytoplasmic fractions from progenitor and M-CSF–differentiated Hoxa9-ER4 cells were electrophoresed and Western blotted with either anti–Plg-RKT mAb or isotype control. A specific immunoreactive band migrating with an Mrapp of approximately 17 000, consistent with the predicted molecular mass of C9orf46 homolog/Plg-RKT, was detected in membranes from M-CSF–differentiated cells (Figure 4A), clearly demonstrating the existence of this novel protein. The protein was not detected in membrane fractions from progenitor Hoxa9-ER4 cells or in cytoplasmic fractions from either progenitor or differentiated cells (Figure 4A). These data further demonstrate the responsiveness of the C9orf46 homolog gene to a maturation-inducing signal, and verify the presence of C9orf46 homolog/Plg-RKT in the cell membrane.
Plg-RKT behaves as a regulated integral membrane protein. (A) Membrane fractions or cytoplasmic fractions from either undifferentiated or M-CSF–treated Hoxa9-ER4 cells (30 μg/lane) were electrophoresed on 12% sodium dodecyl sulfate polyacrylamide gels under reducing conditions and Western blotted with either anti–Plg-RKT mAb, anti–α-enolase mAb as a loading control, or isotype control IgG. (B) M-CSF–treated Hoxa9-ER4 cell membranes were solubilized in 3% Triton X-114. After heating at 37°C and separation of the phases by centrifugation, an aliquot of both phases was electrophoresed and Western blotted with anti–Plg-RKT mAb 35B10. In controls for the method, when the cell lysates were spiked with BSA and subjected to phase partitioning, BSA was detected in the aqueous, but not the detergent phase (data not shown).
Plg-RKT behaves as a regulated integral membrane protein. (A) Membrane fractions or cytoplasmic fractions from either undifferentiated or M-CSF–treated Hoxa9-ER4 cells (30 μg/lane) were electrophoresed on 12% sodium dodecyl sulfate polyacrylamide gels under reducing conditions and Western blotted with either anti–Plg-RKT mAb, anti–α-enolase mAb as a loading control, or isotype control IgG. (B) M-CSF–treated Hoxa9-ER4 cell membranes were solubilized in 3% Triton X-114. After heating at 37°C and separation of the phases by centrifugation, an aliquot of both phases was electrophoresed and Western blotted with anti–Plg-RKT mAb 35B10. In controls for the method, when the cell lysates were spiked with BSA and subjected to phase partitioning, BSA was detected in the aqueous, but not the detergent phase (data not shown).
Plg-RKT partitions to the detergent phase
To test the prediction that Plg-RKT is an integral membrane protein, M-CSF–differentiated Hoxa9-ER4 cell membranes were subjected to phase separation in Triton X-114 as described.66,67 In this technique, integral membrane proteins form mixed micelles with the nonionic detergent and are recovered in the Triton X-114 detergent phase, whereas hydrophilic proteins remain in the aqueous phase. An immunoreactive band migrating with a Mrapp of approximately 17 000 was detected in the detergent phase in Western blotting with anti–Plg-RKT mAb, but was not detected in the aqueous phase (Figure 4B). These data further support the prediction that Plg-RKT is an integral membrane protein.
Plg-RKT is dispersed over the cell surface and colocalizes with uPAR
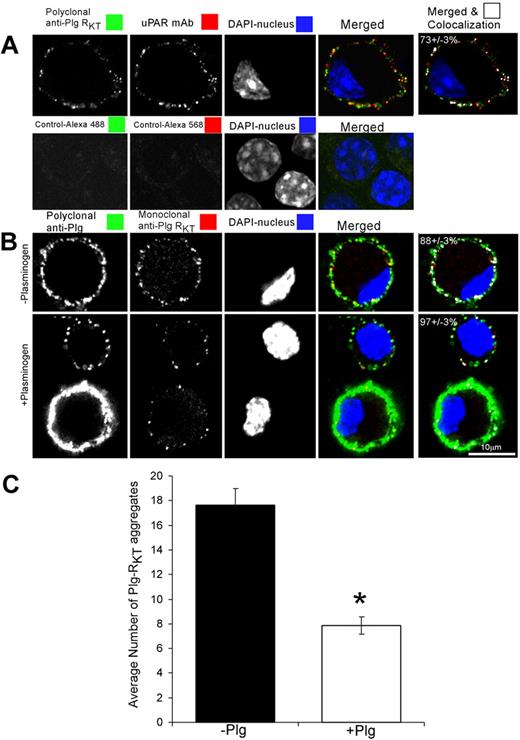
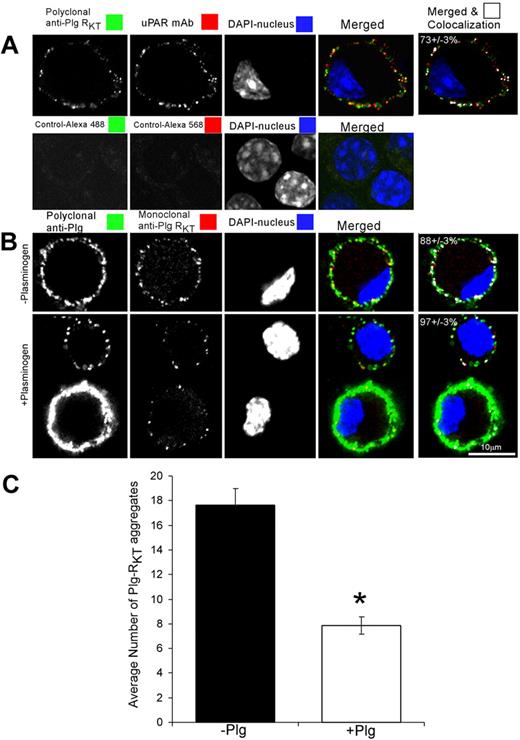
Confocal microscopy was performed to examine the subcellular expression of Plg-RKT and to examine whether Plg-RKT colocalized with uPAR, an additional key component of the cell surface plasminogen activation system. Plg-RKT was immunodetected in small aggregates (∼ 0.2μm2) dispersed over the cell surface (Figure 5A green). uPAR was also immunodetected in small aggregates (∼ 0.2 μm2) dispersed over the cell surface (Figure 5A red), consistent with published data.68 Plg-RKT colocalized with uPAR as revealed by merged images (Figure 5A). The extent of colocalization of Plg-RKT with uPAR was 73% (± 3%). These results suggest that Plg-RKT and uPAR are present in very close proximity on the cell surface in an orientation to promote plasminogen activation.
Plg-RKT is dispersed over the cell surface and colocalizes with uPAR. M-CSF–differentiated Hoxa9-ER4 cells were grown on coverslips and incubated with a combination of polyclonal rabbit anti–Plg-RKT IgG (20 μg/mL) and mouse monoclonal anti-uPAR (20 μg/mL; A). Cells were washed, fixed in 1% formaldehyde, and then stained with a combination of Alexa 488-F(ab′)2 fragment of goat anti–rabbit IgG and Alexa 568-F(ab′)2 fragment of goat anti–mouse IgG for 60 minutes at 20°C in PBS containing 0.001% Triton X-100. Controls are samples incubated without first antibody. In panel B, cells were preincubated with either PBS (− plasminogen) or 2μM plasminogen (+ plasminogen) for 10 minutes at 4°C. Then, the cells were fixed in 1% formaldehyde, washed, and then stained with polyclonal anti-Plg IgG or mouse anti–Plg-RKT mAb and stained with a combination of Alexa 488-F(ab′)2 fragment of goat anti–rabbit IgG and Alexa 568-F(ab′)2 fragment of goat anti–mouse IgG. Cells were washed and mounted in Immuno-Fluore Mounting Medium. Images were captured using a Zeiss laser confocal scanning microscope, then imported into LSM Examiner and ImageJ for further processing as described in “Scanning confocal microscopy.” The data in panel C were quantified and the number and size of each labeled aggregate were determined as described in “Scanning confocal microscopy.” The results reflect counts (C) and colocalization correlation coefficient (M1) values (last column in panels A-B) from more than 40 cells in 2 independent experiments. Data represent mean ± SEM. *P < .001.
Plg-RKT is dispersed over the cell surface and colocalizes with uPAR. M-CSF–differentiated Hoxa9-ER4 cells were grown on coverslips and incubated with a combination of polyclonal rabbit anti–Plg-RKT IgG (20 μg/mL) and mouse monoclonal anti-uPAR (20 μg/mL; A). Cells were washed, fixed in 1% formaldehyde, and then stained with a combination of Alexa 488-F(ab′)2 fragment of goat anti–rabbit IgG and Alexa 568-F(ab′)2 fragment of goat anti–mouse IgG for 60 minutes at 20°C in PBS containing 0.001% Triton X-100. Controls are samples incubated without first antibody. In panel B, cells were preincubated with either PBS (− plasminogen) or 2μM plasminogen (+ plasminogen) for 10 minutes at 4°C. Then, the cells were fixed in 1% formaldehyde, washed, and then stained with polyclonal anti-Plg IgG or mouse anti–Plg-RKT mAb and stained with a combination of Alexa 488-F(ab′)2 fragment of goat anti–rabbit IgG and Alexa 568-F(ab′)2 fragment of goat anti–mouse IgG. Cells were washed and mounted in Immuno-Fluore Mounting Medium. Images were captured using a Zeiss laser confocal scanning microscope, then imported into LSM Examiner and ImageJ for further processing as described in “Scanning confocal microscopy.” The data in panel C were quantified and the number and size of each labeled aggregate were determined as described in “Scanning confocal microscopy.” The results reflect counts (C) and colocalization correlation coefficient (M1) values (last column in panels A-B) from more than 40 cells in 2 independent experiments. Data represent mean ± SEM. *P < .001.
To determine whether the epitope recognized by anti–Plg-RKT mAb functioned as a plasminogen binding site, the cells were either preincubated with buffer or plasminogen, fixed, and then stained with polyclonal antiplasminogen or anti–Plg-RKT mAb. As with the polyclonal anti–Plg-RKT, Plg-RKT was immunodetected in small aggregates (0.16 ± 0.03 to 0.26 ± 0.03 μm2) dispersed over the cell surface (Figure 5B red). However, after preincubation with plasminogen, the ability to immunodetect Plg-RKT was reduced by half (Figure 5B-C). These results suggest that plasminogen and the anti–Plg-RKT mAb (directed against the C-terminal peptide) share binding sites on the cell and that plasminogen binds to the C-terminal domain of Plg-RKT on the cell surface.
Plasminogen binds specifically to the carboxyl terminal peptide of Plg-RKT
To further explore the functional importance of the C-terminal lysine, we tested whether the synthetic peptide, corresponding to the C-terminus of Plg-RKT could bind plasminogen. The peptide, CEQSKLFSDK, was coupled to BSA and then coated onto wells of microtiter plates. Glu-plasminogen was incubated with the wells, followed by our antiplasminogen mAb54 and detection with HRP-conjugated goat anti–mouse IgG. Glu-plasminogen bound to the peptide in a concentration-dependent manner, achieving half saturation at a concentration of 7.6nM (Figure 6A). The binding was specific because it was blocked in the presence of EACA, consistent with the ability of EACA to inhibit plasminogen binding to differentiated Hoxa9-ER4 cells (Figure 1D). Lys-plasminogen also bound specifically to the peptide (Figure 6B) and the concentration for half saturation was less than 2.7nM, consistent with the higher affinity of Lys-plasminogen compared with Glu-plasminogen for cell surfaces.69,70 We investigated the interaction of t-PA with the C-terminal peptide because t-PA and plasminogen share binding sites on monocytoid cells and t-PA binding to monocytoid cells is sensitive to CpB.58 Concentration-dependent binding of t-PA to the peptide was observed (Figure 6C) and the concentration for 50% saturation was 3.2nM, consistent with the relative affinities obtained when comparing Glu-plasminogen and t-PA binding to cell surfaces.71 We noted that the concentration for 50% saturation of plasminogen binding to immobilized CEQSKLFSDK coupled to BSA was much less than the Kd value we had determined for plasminogen binding to cells. These differences in apparent affinity when plasminogen binds to the immobilized peptide compared with the cell surface may be due to our use of BSA-conjugated peptide to coat the plate. If multiple peptides were conjugated to the BSA, that may provide a higher affinity surface than the cell surface receptor, because plasminogen has multiple kringle domains that may interact in a cooperative lysine-dependent manner with multiple Plg-RKT peptides on a single BSA molecule. To resolve this issue, we tested the ability of the soluble C-terminal peptide to inhibit Glu-plasminogen binding under solution phase equilibrium conditions. The soluble peptide competed for Glu-plasminogen binding in a dose-dependent manner with an IC50 of 2μM (Figure 6D), similar to the Kd values we had determined for Glu-plasminogen binding to both human M-CSF–treated monocytes and M-CSF–treated Hoxa9-ER4 cells (see “Plasminogen binding is enhanced when monocytes differentiate”). In addition, a mutated peptide with the C-terminal lysine substituted with alanine did not compete for plasminogen binding at concentrations up to 1mM (Figure 6D), further supporting the role of the C-terminal lysine in the interaction of Plg-RKT with plasminogen.
Plasminogen binds to the C-terminal peptide of Plg-RKT. The peptide, CEQSKLFSDK, corresponding to the amino terminus of Plg-RKT was coupled to BSA and coated onto wells of microtiter plates. Either Glu-plasminogen (A) or Lys-plasminogen (B) or t-PA (C) was then incubated with the wells, followed by antiplasminogen mAb54 (A-B) or anti–t-PA mAb (C) and detection with HRP-conjugated goat anti–mouse IgG (●) as described in “Plasminogen and t-PA binding assay.” The binding was specific because it was blocked in the presence of 0.2M EACA (△), consistent with the ability of EACA to inhibit plasminogen binding to differentiated Hoxa9-ER4 cells. In additional controls, nonspecific binding to either BSA (▲), or to the reverse peptide (○) was < 10% of binding to CEQSKLFSDK. (At high input concentrations of t-PA, nonspecific binding increased, but was < 10% of binding to CEQSKLFSDK at the concentration required for 50% saturation [3.2nM]). In controls for the detection method, optical density at 490 nm (OD490) values obtained using an isotype control antibody or in the absence of added plasminogen or t-PA were < 5% of the values for plasminogen or t-PA binding to immobilized CEQSKLFSDK. (D-E) Biotinylated Glu-plasminogen (25nM) was incubated with immobilized CEQSKLFSDK in the presence of increasing concentrations of (D) the C-terminal peptide, CEQSKLFSDK (●) or a mutated C-terminal peptide with K147 substituted with alanine, CEQSKLFSDA (○) or (E) anti–Plg-RKT mAb 35B10 (●) or isotype control (○). Biotinylated Glu-plasminogen binding was detected with HRP-streptavidin and was 96% inhibited in the presence of 0.2M EACA (not shown). Data are mean ± SEM; n = 3, for each determination.
Plasminogen binds to the C-terminal peptide of Plg-RKT. The peptide, CEQSKLFSDK, corresponding to the amino terminus of Plg-RKT was coupled to BSA and coated onto wells of microtiter plates. Either Glu-plasminogen (A) or Lys-plasminogen (B) or t-PA (C) was then incubated with the wells, followed by antiplasminogen mAb54 (A-B) or anti–t-PA mAb (C) and detection with HRP-conjugated goat anti–mouse IgG (●) as described in “Plasminogen and t-PA binding assay.” The binding was specific because it was blocked in the presence of 0.2M EACA (△), consistent with the ability of EACA to inhibit plasminogen binding to differentiated Hoxa9-ER4 cells. In additional controls, nonspecific binding to either BSA (▲), or to the reverse peptide (○) was < 10% of binding to CEQSKLFSDK. (At high input concentrations of t-PA, nonspecific binding increased, but was < 10% of binding to CEQSKLFSDK at the concentration required for 50% saturation [3.2nM]). In controls for the detection method, optical density at 490 nm (OD490) values obtained using an isotype control antibody or in the absence of added plasminogen or t-PA were < 5% of the values for plasminogen or t-PA binding to immobilized CEQSKLFSDK. (D-E) Biotinylated Glu-plasminogen (25nM) was incubated with immobilized CEQSKLFSDK in the presence of increasing concentrations of (D) the C-terminal peptide, CEQSKLFSDK (●) or a mutated C-terminal peptide with K147 substituted with alanine, CEQSKLFSDA (○) or (E) anti–Plg-RKT mAb 35B10 (●) or isotype control (○). Biotinylated Glu-plasminogen binding was detected with HRP-streptavidin and was 96% inhibited in the presence of 0.2M EACA (not shown). Data are mean ± SEM; n = 3, for each determination.
Plg-RKT regulates cell surface plasminogen activation
We found that plasminogen activation was stimulated 12.7-fold in the presence of M-CSF–treated Hox9-ER4 cells, compared with the reaction in the absence of cells and that cell-dependent plasminogen activation was stimulated 4.4-fold on differentiated cells, compared with undifferentiated cells (Figure 7). To verify the role of Plg-RKT in plasminogen activation, we tested the effect of a monoclonal antibody (anti–Plg-RKT mAb 35B10) raised in rats against the synthetic peptide, CEQSKLFSDK. The IgG fraction reacted with the C-terminal peptide of murine Plg-RKT, blocked plasminogen binding to CEQSKLFSDK (Figure 6E), and specifically recognized a protein with a Mrapp of approximately 17 000 in Western blotting of M-CSF–treated Hoxa9-ER4 cells (Figure 4). In FACS analysis of M-CSF–treated Hox9-ER4 cells, anti–Plg-RKT mAb 35B10 reacted specifically with the cell surface giving a mean fluorescence intensity value of 93.2 compared with the mean fluorescence intensity value for the rat IgG2a isotype control of 32.8. Anti–Plg-RKT mAb 35B10 substantially suppressed cell-dependent plasminogen activation by 46% and suppressed cell differentiation–dependent plasminogen activation by 58% (Figure 7). In controls, plasminogen activation in the absence of cells or on undifferentiated cells was not affected by anti–Plg-RKT mAb.
Plg-RKT regulates cell surface plasminogen activation. Plasminogen activation was determined after 12 minutes as described in “Plasminogen activation assay” in either the presence or absence of either undifferentiated Hoxa9-ER4 progenitor cells or M-CSF–differentiated Hoxa9-ER4 cells and in the presence of either rat anti–Plg-RKT mAb 35B10 (■) or isotype control rat IgG2a (□). ***P < .001, compared with the corresponding isotype control.
Plg-RKT regulates cell surface plasminogen activation. Plasminogen activation was determined after 12 minutes as described in “Plasminogen activation assay” in either the presence or absence of either undifferentiated Hoxa9-ER4 progenitor cells or M-CSF–differentiated Hoxa9-ER4 cells and in the presence of either rat anti–Plg-RKT mAb 35B10 (■) or isotype control rat IgG2a (□). ***P < .001, compared with the corresponding isotype control.
Discussion
Localization of plasminogen on cell surfaces is a crucial control point for positive regulation of cell surface plasmin proteolytic activity that facilitates both physiologic and pathologic processes.1,2 Plasminogen binding sites, with C-terminal basic residues, are predominantly responsible for the ability of eukaryotic cells to bind plasminogen and promote plasminogen activation.24 In the current study, we identified a novel protein, Plg-RKT, the first integral membrane plasminogen receptor, synthesized with a C-terminal lysine.
Our results support a mechanism in which Plg-RKT binds plasminogen via an interaction requiring the C-terminal lysine of Plg-RKT that is exposed on the cell surface. When intact, differentiated cells were pretreated with CpB before membrane fractionation, Plg-RKT was not detectable in the MudPIT analysis, indicating that the C-terminal lysine of Plg-RKT was accessible to CpB on the cell surface and, thus, lost the ability to bind plasminogen-Sepharose after CpB treatment of intact cells. Second, Glu-plasminogen bound specifically to a synthetic peptide corresponding to the C-terminus of Plg-RKT and the binding interaction was inhibited by the lysine analog, EACA, that inhibits the interaction of plasminogen with the cell surface. Lys-plasminogen also bound specifically to the peptide consistent with the ability of Lys-plasminogen to also bind to the cell surface.69,70 Third, plasminogen binding to a mutated peptide with the C-terminal lysine substituted with alanine was not detected, further supporting the key role of the C-terminal lysine in the interaction with plasminogen. Of key importance, anti–Plg-RKT antibodies directed against the C-terminal peptide of Plg-RKT detected Plg-RKT dispersed over the cell surface in small aggregates. A similar immunolocalization pattern of small aggregates dispersed over the cell surface has been published for other cell surface plasminogen binding proteins, α-enolase, histone 2B, p11, and annexin 2.6 Of key importance, after preincubation of the cells with plasminogen, the ability to immunodetect Plg-RKT was reduced by half (Figure 5B-C). These results suggest that plasminogen and the anti–Plg-RKT mAb share binding sites on the cell and that plasminogen binds to the C-terminal domain of Plg-RKT on the cell surface. Notably, a C-terminal lysine is encoded for all 26 mammalian orthologs of Plg-RKT for which the C-terminal sequence is available, consistent with a fundamental role of Plg-RKT to bind plasminogen in mammals.
t-PA also bound specifically to the C-terminal peptide of Plg-RKT, consistent with sharing of binding sites with plasminogen and with the CpB sensitivity of the interaction of t-PA with cells.58 Despite potentially sharing a binding site on Plg-RKT, the relative concentrations of t-PA and plasminogen in the circulation should permit simultaneous binding of both ligands to the cell surface, and each t-PA molecule should be bound proximally to several plasminogen molecules.58 We also found that Plg-RKT was spatially highly colocalized with the uPAR. These results suggest a mechanism for colocalization of plasminogen with plasminogen activators to promote cell surface plasminogen activation. Of key importance, treatment of the cells with anti–Plg-RKT mAb markedly reduced the ability of M-CSF–treated Hoxa9-ER4 cells to stimulate plasminogen activation.
Our experimental data support the prediction that Plg-RKT is a multipass transmembrane protein, exposing a C-terminal lysine on the cell surface. Our results demonstrating CpB-sensitivity of Plg-RKT on the cell surface supported the cell surface exposure of the C-terminus. Regarding transmembrane topology, Plg-RKT was immunochemically detected in the membrane fraction, but not the cytoplasmic fraction of differentiated cells. And, in phase partitioning experiments, Plg-RKT partitioned to the detergent phase.
Our data also indicate that Plg-RKT is up-regulated during differentiation. In the MudPIT analysis, Plg-RKT peptides were detected in M-CSF–differentiated cells, but not in progenitor cells. Similarly, in Western blotting, Plg-RKT was detected in membranes of differentiated cells, but not in the progenitor cells.
The C9orf46 homolog/Plg-RKT transcript is broadly expressed in normal human and mouse tissues (as determined in high-throughput gene expression profiling in which RNA samples from human and murine tissues were hybridized to high-density gene expression arrays72,73 ), including spleen; thymus; lymph node; lung; intestine; bone marrow; adrenal, pituitary, and other endocrine tissues; vascular tissue; kidney; liver; stomach; bladder; and neuronal tissue (hippocampus, hypothalamus, cerebellum, cerebral cortex, olfactory bulb, and dorsal root ganglion). We examined Plg-RKT protein expression in murine tissue and detected Plg-RKT in lymph node, bone marrow, and intestine, tissues rich in leukocytes but not in adipose tissue, as a negative control (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Differences in mRNA expression data and Western blotting results may be explained by high expression by specific cells within tissues. In Western blotting, we also have detected Plg-RKT in rat hippocampal neurons, bovine adrenal tissue, and in rat pheochromocytoma PC12 cells (R.J.P. et al, unpublished observations, June 2007).
We searched for C9orf46 homolog/Plg-RKT mRNA microarray expression data at http://www.ebi.ac.uk/gxa74 (Table 4). C9orf46 homolog/Plg-RKT mRNA is present in monocytes; leukocytes; natural killer (NK) cells; T cells; myeloid, dendritic, and plasmacytoid cells; breast cancer cells; acute lymphoblastic leukemia cells; and Molt-4 acute lymphoblastic leukemia cells. These data are consistent with previous reports documenting expression of plasminogen binding sites on peripheral blood leukocytes,55 breast cancer cells,10,75 and other tissues (reviewed in Miles et al27 ). In addition, results obtained by searching the ArrayExpress Warehouse (http://www.ebi.ac.uk/gxa) indicated that the C9orf46 homolog gene is also regulated in other tissues by lipopolysaccharide, canrenoate, H2O2, and dexamethasone (Table 5). The broad distribution and regulation in tissues that express plasminogen binding sites suggest that C9orf46 homolog/Plg-RKT provides plasminogen receptor function that may serve to modulate plasmin proteolytic functions in these tissues, as well. In genome-scale quantitative image analysis, overexpression of more than 86 cDNAs, including C9orf46 homolog, conferred dramatic increases in cell proliferation, whereas knockdown of C9orf46 homolog mRNA resulted in apoptosis.76 In microarray studies, C9orf46 homolog mRNA expression has a high power to predict cervical lymph node metastasis in oral squamous cell carcinoma.77 We have previously shown that cell-bound plasminogen protects monocytoid cells from tumor necrosis factor-α–induced apoptosis.78 Future studies should elucidate the relationship of these roles of C9orf46 homolog/ Plg-RKT to its plasminogen receptor function.
In summary, regulation of the cell surface expression of plasminogen receptors is a mechanism by which cells modulate the proteolytic activity of their environment. We have identified a novel plasminogen receptor with unique characteristics: Plg-RKT is an integral membrane protein that exposes a C-terminal lysine and serves to markedly promote plasminogen activation on the cell surface.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Ray Stevens at The Scripps Research Institute and Dr Nuala Booth and Dr Ian Booth, University of Aberdeen, for helpful discussions. This is publication 19034 from The Scripps Research Institute.
This work was supported by the National Institutes of Health (grants HL 38272, HL 45934, and HL 081046 [L.A.M.], HL 50398 [R.J.P.], NIH P41 RR011823 [J.R.Y.], National Institute of Allergy and Infectious Diseases subcontract grant UCSD/MCB0237059 [E.I.C.]), and the Department of Veterans Affairs (R.J.P.).
National Institutes of Health
Authorship
Contribution: N.M.A., W.B.K., J.R.Y., R.J.P., and L.A.M. designed research; N.M.A., E.I.C., N.B., H.B., and C.M.P. performed research; M.P.K. and J.R.Y. contributed new reagents and analytic tools; N.M.A., E.I.C., N.B., H.B., C.M.P., W.B.K., R.J.P., and L.A.M. analyzed data; and N.M.A., R.J.P., and L.A.M. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
N.M.A.'s current affiliation is FD McMaster Laboratories, Armidale, NSW, Australia. E.I.C.'s current affiliation is Department of Pharmacological Sciences, Stony Brook University, Stony Brook, NY. C.M.P.'s current affiliation is Yale University, New Haven, CT.
Correspondence: Lindsey A. Miles, Department of Cell Biology, SP30-20, The Scripps Research Institute, 10550 N Torrey Pines Rd, La Jolla, CA 92037; e-mail: lmiles@scripps.edu.




![Figure 6. Plasminogen binds to the C-terminal peptide of Plg-RKT. The peptide, CEQSKLFSDK, corresponding to the amino terminus of Plg-RKT was coupled to BSA and coated onto wells of microtiter plates. Either Glu-plasminogen (A) or Lys-plasminogen (B) or t-PA (C) was then incubated with the wells, followed by antiplasminogen mAb54 (A-B) or anti–t-PA mAb (C) and detection with HRP-conjugated goat anti–mouse IgG (●) as described in “Plasminogen and t-PA binding assay.” The binding was specific because it was blocked in the presence of 0.2M EACA (△), consistent with the ability of EACA to inhibit plasminogen binding to differentiated Hoxa9-ER4 cells. In additional controls, nonspecific binding to either BSA (▲), or to the reverse peptide (○) was < 10% of binding to CEQSKLFSDK. (At high input concentrations of t-PA, nonspecific binding increased, but was < 10% of binding to CEQSKLFSDK at the concentration required for 50% saturation [3.2nM]). In controls for the detection method, optical density at 490 nm (OD490) values obtained using an isotype control antibody or in the absence of added plasminogen or t-PA were < 5% of the values for plasminogen or t-PA binding to immobilized CEQSKLFSDK. (D-E) Biotinylated Glu-plasminogen (25nM) was incubated with immobilized CEQSKLFSDK in the presence of increasing concentrations of (D) the C-terminal peptide, CEQSKLFSDK (●) or a mutated C-terminal peptide with K147 substituted with alanine, CEQSKLFSDA (○) or (E) anti–Plg-RKT mAb 35B10 (●) or isotype control (○). Biotinylated Glu-plasminogen binding was detected with HRP-streptavidin and was 96% inhibited in the presence of 0.2M EACA (not shown). Data are mean ± SEM; n = 3, for each determination.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/7/10.1182_blood-2008-11-188938/4/m_zh89990946750006.jpeg?Expires=1766382317&Signature=VGQ7BqKn~EGa1DtELW9VTRPUjRs7u0DY63I79pQrGu7Q661VtdjqlMMHuVGyJLaWroR9YJADe8cEIrex-HZU9ks3~0kODEkdcVtXMhfZHuuXFXDeebeBMOIKakgzY4lmaFrddNFIjY9B~JMrdOuNQMeTmdJP1aaujs8FPOgVvIGv8p9tBCUC2u108dQcSdCiD7HNDM0PbyjAG8~tiAx-QHljOWaebIYvW5Zp2EERj9n1N3p0K3sWn3ZowMx0~~TgsJszmBo-1oCNre575gdMxPHtqh-sCudae1N9glN~DYGXnqPqa8XxiXQtiD2Y2SeBFMemX7YjQ7qvX1Uj7Xy~yQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![Figure 6. Plasminogen binds to the C-terminal peptide of Plg-RKT. The peptide, CEQSKLFSDK, corresponding to the amino terminus of Plg-RKT was coupled to BSA and coated onto wells of microtiter plates. Either Glu-plasminogen (A) or Lys-plasminogen (B) or t-PA (C) was then incubated with the wells, followed by antiplasminogen mAb54 (A-B) or anti–t-PA mAb (C) and detection with HRP-conjugated goat anti–mouse IgG (●) as described in “Plasminogen and t-PA binding assay.” The binding was specific because it was blocked in the presence of 0.2M EACA (△), consistent with the ability of EACA to inhibit plasminogen binding to differentiated Hoxa9-ER4 cells. In additional controls, nonspecific binding to either BSA (▲), or to the reverse peptide (○) was < 10% of binding to CEQSKLFSDK. (At high input concentrations of t-PA, nonspecific binding increased, but was < 10% of binding to CEQSKLFSDK at the concentration required for 50% saturation [3.2nM]). In controls for the detection method, optical density at 490 nm (OD490) values obtained using an isotype control antibody or in the absence of added plasminogen or t-PA were < 5% of the values for plasminogen or t-PA binding to immobilized CEQSKLFSDK. (D-E) Biotinylated Glu-plasminogen (25nM) was incubated with immobilized CEQSKLFSDK in the presence of increasing concentrations of (D) the C-terminal peptide, CEQSKLFSDK (●) or a mutated C-terminal peptide with K147 substituted with alanine, CEQSKLFSDA (○) or (E) anti–Plg-RKT mAb 35B10 (●) or isotype control (○). Biotinylated Glu-plasminogen binding was detected with HRP-streptavidin and was 96% inhibited in the presence of 0.2M EACA (not shown). Data are mean ± SEM; n = 3, for each determination.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/7/10.1182_blood-2008-11-188938/4/m_zh89990946750006.jpeg?Expires=1766598592&Signature=qgqSnQg-IhsYWEtenPQFJ91dpoI~Mgd6xFVZrZFm2tzvji35PoZoZqXQUCk7fMwpbv94KPWot7Nl5KJEW7X1wy3OlMsloBq8f4imGsUC28zN0anLp0EBRQhiePvtDAHwKuxFsZxzI~QmmdQyf75LZuX4AYKiPFOlR0QUevrYavwU51C25RWC1Zf4DX-rJu-AflW9S6GR7bFcp9fACZ~8EKOV9R8PPOtRZNzHVZV-9TY8FMjgL5TIXAIQ2w32W1Y3ELjVzMXcvathyTNZ-zrj7eK-dThm6aqAaq~~whsrmWzDeaceHXOXBU6p4IsgbIhi4zHirHonWsW7I5M~IX0Fmg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

