Abstract
The antithrombotic surface of endothelium is regulated in a coordinated manner. Tissue factor pathway inhibitor (TFPI) localized at the endothelial cell surface regulates the production of FXa by inhibiting the TF/VIIa complex. Systemic homozygotic deletion of the first Kunitz (K1) domain of TFPI results in intrauterine lethality in mice. Here we define the cellular sources of TFPI and their role in development, hemostasis, and thrombosis using TFPI conditional knockout mice. We used a Cre-lox strategy and generated mice with a floxed exon 4 (TFPIFlox) which encodes for the TFPI-K1 domain. Mice bred into Tie2-Cre and LysM-Cre lines to delete TFPI-K1 in endothelial (TFPITie2) and myelomonocytic (TFPILysM) cells resulted in viable and fertile offspring. Plasma TFPI activity was reduced in the TFPITie2 (71% ± 0.9%, P < .001) and TFPILysM (19% ± 0.6%, P < .001) compared with TFPIFlox littermate controls. Tail and cuticle bleeding were unaffected. However, TFPITie2 mice but not TFPILysM mice had increased ferric chloride–induced arterial thrombosis. Taken together, the data reveal distinct roles for endothelial- and myelomonocytic-derived TFPI.
Introduction
The endothelium provides an antithrombotic interface with circulating blood which is generated in part by the coordinated expression of endothelial-derived anticoagulants. The regulated expression of these endothelial-derived proteins may account in part for differences in thrombotic phenotype among vessels.1 Tissue factor pathway inhibitor (TFPI) is a Kunitz-type serine protease inhibitor expressed in endothelial cells and regulates the initiation of coagulation by inhibiting tissue factor (TF)/factor VIIa activation of factor X.
TFPI, originally known as lipoprotein associated coagulation inhibitor, was isolated from a hepatoma cell line.2 TFPI circulates at low (nmol/L) levels in humans largely associated with lipoproteins.3 Infusion of heparin increases circulating levels of TFPI in humans, and this increase has been attributed to displacement of TFPI from glycosoaminoglycans on the surface of endothelial cells.4,5 As such, the endothelium has been thought to be the dominant source of circulating TFPI. However, TFPI is also expressed in platelets, vascular smooth muscle, cardiac myocytes, and monocyte/macrophages.4,6-10 The physiologic importance of TFPI is confirmed in that no known human deficiencies of TFPI have been reported. Homozygotic deletion of exon 4 in mice (which encodes the Kunitz 1 domain) resulted in embryonic lethality.11 Heterozygotic deletion results in an increased response to acute and chronic vascular injury.12-14 Conversely, vascular-directed overexpression of TFPI attenuates this response.15
To better define the cellular sources of TFPI and their role in development, hemostasis, and thrombosis, we generated mice with endothelial and monocytic-restricted deletion of exon 4, which encodes for the TFPI-K1 domain. We also used bone marrow transplantation as a means to generate mice with a TFPI-K1 deficiency in circulating cells. Based on studies in these mice, we conclude that neither endothelial nor myelomonocytic-derived TFPI is necessary for murine development or fertility. Furthermore, we define the contributions of each to mouse plasma TFPI activity. Finally, we demonstrate the regulatory role of endothelial-derived TFPI in response to acute injury.
Methods
Generation of floxed TFPI mice
Conditional deletion of TFPI in a cell type–specific manner was accomplished using the Cre/loxP recombination system. The strategy targeted exon 4, which encodes the Kunitz 1 domain (Figure 1A) and results in a K1 deleted form of TFPI containing the remainder of the TFPI protein and would affect all isoforms containing this domain.11 Three fragments of genomic DNA from mouse strain 129Sv/J were amplified from the TFPI gene locus using polymerase chain reaction (PCR). These fragments included a 2110-bp sequence upstream of exon 4, a 350-bp sequence including exon 4, and a 2300-bp sequence downstream of exon 4. A conditional targeting vector (pNTKV1901-frt/loxP) was used.16 The final targeting vector included 2 loxP sites situated in introns 3 and 4 that floxed exon 4. A neomycin resistance gene (NEO) fused to the phosphoglycerokinase (PGK) promoter was inserted upstream of exon 4 in intron 3 and was surrounded by flippase recombination target sites for removal by flippase-mediated recombination.17 The final targeting vector contained the 3 fragments totaling 4760 bp and the 1750-bp pGK-NEO resistance cassette for a complete length of 6500 bp. The construct was linearized and recovered from an agarose gel using the Qiaex ll (QIAGEN) gel extraction kit. The targeting vector was electroporated into pluripotent 129Sv/J embryonic stem cells, and clones carrying the construct were identified by neomycin selection.
To examine the efficacy of the loxP/cre system, we replicated a knockout mouse by crossing TFPIFlox/Flox mice with B6.C-Tg(CMV-cre)1Cgn/J males (CMV-Cre) that constitutively express the Cre recombinase.
Removal of the pGK-NEO cassette was accomplished by crossing TFPIFlox/Flox pGK-NEO+/+ mice with 129S4/SvJaeSor-Gt(ROSA)26Sortm1(FLP1)Dym/J mice (FLP) purchased from The Jackson Laboratory. The presence of the flippase recombinase was evaluated using the primers oIMR 1348, TCCGCCTCAGAAGCCATAGA; and oIMR 1349, GATAGCCGCGCTGCCT. Removal of the pGK-NEO resistance cassette was confirmed using a pair of primers that targeted NEO (Neo 1123, TCCGCCTCAGAAGCCATAGA; Neo 1990, GATAGCCGCGCTGCCT). After removal of the resistance cassette, progeny were successfully backcrossed to TFPIFlox/Flox homozygosity.
Generation of mice with TFPI Kunitz 1 deletion
Throughout the development of mice with K1 TFPI deletions, the Cre recombinase transgene was introduced exclusively through the male germ line.18 To generate mice with an endothelial specific deletion of TFPI, homozygously TFPI floxed mice were crossed with B6.Cg-Tg(Tek-cre)12Flv/J males (Tie2-Cre) obtained from The Jackson Laboratory. The resulting offspring were backcrossed to floxed homozygosity while maintaining the Tie2-Cre transgene (TFPITie2). This same process was used to develop mice with a monocyte and macrophage specific deletion of TFPI. TFPIFlox/Flox females were crossed with B6.129P2-Lyz2tm1(cre)Ifo/J males (LysM-Cre) from The Jackson Laboratory, and offspring were backcrossed to TFPIFlox/Flox homozygosity. Mice used for experiments were littermates on a mixed C57/129Sv background. Additional backcrosses were performed to determine whether the effects on TFPI activity were maintained in the C57BL/6 background.
PCR genotyping of targeted mice
Previous studies have demonstrated that the actions of the Tie2-Cre and LysM-Cre transgene can be monitored by the detection of deleted products in the blood stream.19,20 We therefore monitored the status of TFPI floxed DNA and the Cre transgene through PCR analysis on whole blood. To detect the TFPI wild-type allele, the following primers were used: TFPI Short, AAGAGTTGGTGTG; and TFPI Rev, TTCAATGCCAAAGCTAGA. These primers were intentionally designed to be interrupted by loxP sites surrounding exon 4 and, therefore, developed no product in the presence of the construct. To assess the flox status of the construct, we used the primers TFPI 2793 F, CCGGGATAACTTCGTATAGC; and TFPI 3894 R, GATACATCTACCCCCTCAAA. This set of primers encompassed exon 4 and through a size shift enabled us to determine whether exon 4 had been deleted or remained floxed. To detect the Tie2-Cre transgene, we used the primers Cre F, GCGGTCTGGCAGTAAAAACTATC; and Cre R, GTGAAACAGCATTGCTGTCACTT. To detect the LysM-Cre transgene, the following primers were used: Cre 8, CCCAGAAATGCCAGATTACG; and Mlys1, CTTGGGCTGCCAGAATTTCTC.
RT-PCR to determine transcriptional efficiency of the Tie2 and LysM transgenes
RNA was isolated from whole blood using a Mouse RiboPure-Blood RNA Isolation Kit (Applied Biosystems/Ambion) per the manufacturer protocol. Peritoneal lavage cells were isolated from TFPILysM and TFPIFlox littermates 72 hours after an intraperitoneal injection of 1 mL of a 3% thyoglycolate solution prepared in sterile saline. Peritoneal cells were isolated by flushing the peritoneal cavity with 3 × 1 mL of sterile PBS. Cells were pelleted by centrifugation for 10 minutes at 300g. RNA was isolated from aortic endothelial cells, bone marrow, and peritoneal lavage cells using an RNeasy Plus MiniKit (QIAGEN) per the manufacturer protocol. First-strand cDNA synthesis was carried out using SuperScript First-Strand Synthesis System for reverse transcriptase (RT)-PCR (Invitrogen) using 1 μg of total RNA for each reaction per the manufacturer protocol. PCR was then performed using Hot-Start Taq supermix (Denville Scientific) with 2 μL of cDNA template for 40 cycles and an annealing temperature of 56°C. The products were then separated on 2% agarose gels containing ethidium bromide, and products were visualized by UV transillumination. Primers used were: TFPI Forward, TGTATCACTTTCGGGACCTGTCTC; TFPI Reverse, GAGTGGACTGG-ATTCTCACAGATC; HPRT Forward, GGAGCGGTAGCACCTCCT; and HPRT Reverse, CTGGTTCATCATCGCTAATCAC.
Measurement of plasma TFPI
Blood was collected at baseline or 60 minutes after intravenous bolus of heparin (100 U) via cheek pouch puncture into a tube containing 0.1M sodium citrate (one-tenth the volume of blood collected), centrifuged at 10 000g for 10 minutes at 4°C, and the resulting plasma was transferred to new tubes and frozen at −80°C until analysis. As murine TFPI is known to inhibit human clotting factors,21 TFPI activity was measured using an ACTICHROME TFPI Activity Assay kit (American Diagnostica Inc) per the manufacturer's protocol. An ELISA was performed to detect TFPI immunoreactivity as previously described.22
Blood analysis
Blood samples (100 μL) were collected via cheek pouch puncture into tubes containing 10 μL of 3.2% sodium citrate solution. Blood samples were analyzed within 2 hours of collection for complete blood count with differential and prothrombin (PT) and activated partial thromboplastin times. For analysis, the VetScan HMT Hematology System and an SCA2000 Veterinary Coagulation Analyzer were used according to the manufacturer instructions. The calibration was verified before each analysis for quality control. As the effects of TFPI on PT times can depend on the concentration of TF used in the assay,23 we further evaluated PT times using serial dilutions of TF (Innovin, Dade Behring) using a modified turbidimetric assay for the measurement of clotting times.24 Plasma samples were diluted 1:4 in PBS containing 0.1% bovine serum albumin, and 50 μL of the mixture was dispensed to a 96-well plate. Fifty microliters of serially diluted TF (1:200, 1:400, 1:800, 1:1600, and 1:3200) was added to the plasma samples, and all samples were incubated at 37°C before the addition of 50 μL of 25mM CaCl3 to initiate clotting. Optical density at 340 nm was monitored approximately every 10 seconds, and clotting times were determined from plots of optical density versus time.
Analysis of hemostasis
Induction of arterial thrombosis
Mice used in these experiments were 8 to 12 weeks old. The FeCl3 induced model of vascular thrombosis has been previously described.15 Mice were anesthetized by an intramuscular injection of approximately 0.02 mL of a solution containing 83.3 mg/mL ketamine and 16.7 mg/mL xylazine. After the left common carotid artery was exposed, a flow probe (model 0.5 VB, Transonic Systems) was placed on the artery. The probe was connected to a flowmeter (Transonic model T106), and data were collected via connection to a PC-driven PowerLab with ChartPro software (ADInstruments). A strip of filter paper saturated with 10% FeCl3 solution was applied to the adventitial surface of the surgically exposed carotid artery for 3 minutes. Carotid artery blood flow was then followed by the T106 Small Animal Blood Flow Meter for 20 minutes or until complete occlusion (0 flow for at least 10 seconds) occurred.
Bone marrow transplantation and analysis
C57BL/6 mice were purchased from The Jackson Laboratory. Mice with heterozygous TFPI deficiency (ie, TFPI+/−) were a gift from Dr George Broze (Washington University, St Louis, MO).11 These mice, in which the first Kunitz domain of TFPI was deleted resulting in a nonfunctional TFPI protein, were backcrossed for at least 8 generations into the C57BL/6 genetic background. All animal care and experimental procedures complied with the Principles of Laboratory Animal Care established by the National Society for Medical Research and were approved by the University of Michigan and the Mayo Clinic Committees on Use and Care of Animals.
Male and female TFPI+/− mice were mated. Sixteen days postcoitus, the pregnant females were killed and individual fetuses were recovered. Genotyping of embryos and isolation of fetal liver cells for transplant was performed according to a previously published method, with minor modification, as described below.27 All steps were performed at 4°C, unless otherwise stated. Fetal livers were placed in 2 mL of RPMI media (GIBCO/BRL) supplemented with 2% fetal calf serum (FCS). Cell suspensions were prepared by cutting livers into small fragments, then aspirating and ejecting fragments several times through an 18-gauge needle. Cell suspensions were washed twice with RPMI/2% FCS and resuspended in RPMI. Cell counts of suspensions were determined by using a hemacytometer. Recipient, wild-type (ie, TFPI+/+) C57BL/6 mice (specific pathogen–free, 8 weeks old, weight approximately 25 g, n = 22) were subjected to marrow-ablative irradiation (1300 rads, delivered in equal divided doses 3 hours apart) after having received acidified water (pH 2.0) for 1 week to reduce intestinal bacterial flora. After irradiation, mice were sedated by inhalation of methoxyflurane. Fetal liver cells (4 × 106 trypan blue–positive cells/0.3 mL RPMI) were injected into the retro-orbital sinus with a 25-gauge needle. One-half of the irradiated TFPI+/+ mice were transplanted with TFPI−/− fetal liver cells while the other half received TFPI+/+ fetal liver cells (n = 11/group). Animals received acidified water for 2 weeks after transplantation. They were fed normal mouse chow. TFPI+/+ mice receiving transplants from TFPI−/− liver cells were termed BMTFPI−/− mice, whereas the control group was termed BMTFPI+/+ mice. All experiments were performed 24 weeks after transplantation.
To confirm reconstitution of bone marrow, 20 weeks after transplantation, PCR was performed on DNA extracted from peripheral blood obtained from BMTFPI−/− (n = 11) and BMTFPI+/+ (n = 11) mice. The mutant allele size was 330 bp and the wild-type allele was 170 bp.28
To evaluate differences in the cellular regeneration after bone marrow transplantation between the 2 groups, hemoglobin, WBC, RBC, hematocrit, and platelets were analyzed in blood drawn from BMTFPI−/− (n = 11) and BMTFPI+/+ (n = 11) mice using an automated cell counter.
Results
Generation of mice with cell-specific deletion of the TFPI-K1 domain
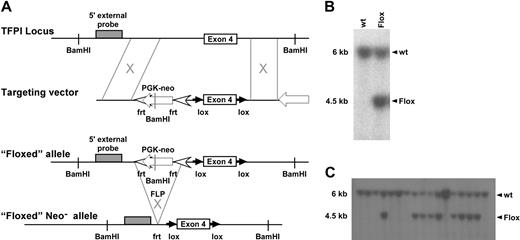
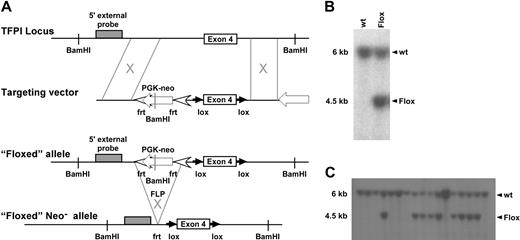
To elucidate the biologic significance of endothelial or monocytic cell derived TFPI, we used a Cre-loxP mediated gene targeting approach.29 A targeting vector was constructed in which exon 4 of the murine TFPI gene is flanked by loxP sites, TFPIFlox (Figure 1A). This strategy results in expression of a K1-deleted form of TFPI. Cell screening was determined by Southern blot using a 5′ external probe on BamHI-cut genomic DNA (Figure 1B), which generated a 6-kb restriction fragment from the wild-type allele and a 4.5-kb restriction fragment from the mutated allele. Three TFPIFlox/wt clones were identified and microinjected into mouse blastocysts. Chimeric mice derived from these embryos successfully transmitted the mutation to the germ line as demonstrated by Southern blot analysis (Figure 1C). These mice provided founders for subsequent inbreeding.
TFPI targeting vector with 5′ DNA probe Southern blot screening. (A) Schematic representation of the introduction of the targeting vector and subsequent removal of the PGK-NEO cassette using flippase-mediated flippase recombination target site. (B) 129Sv/J embryonic stem cells were screened for successful targeting of TFPI exon 4 using Southern blot analysis that used a 5′ DNA probe differentiating wild-type and floxed alleles. (C) Germline transmission of the targeting vector was also assessed using Southern blot screening.
TFPI targeting vector with 5′ DNA probe Southern blot screening. (A) Schematic representation of the introduction of the targeting vector and subsequent removal of the PGK-NEO cassette using flippase-mediated flippase recombination target site. (B) 129Sv/J embryonic stem cells were screened for successful targeting of TFPI exon 4 using Southern blot analysis that used a 5′ DNA probe differentiating wild-type and floxed alleles. (C) Germline transmission of the targeting vector was also assessed using Southern blot screening.
To demonstrate the efficiency of the Cre-loxP gene targeting system for deletion of TFPI-K1, we crossed TFPIFlox mice with mice that constitutively express Cre recombinase via the CMV promoter (CMV-Cre). The resulting offspring containing the CMV-Cre transgene and a single floxed TFPI-K1 allele were then backcrossed. Among more than 200 live births, no mice were born containing the CMV-Cre transgene and 2 deleted TFPI-K1 alleles. This confirms that ubiquitous deletion of the TFPI-K1 domain using a loxP-Cre gene targeting approach is inconsistent with viability as seen with systemic deletion.11
Endothelial and hematopoietic deletion of TFPI-K1 was achieved by crossing the TFPIFlox mice with transgenic mice expressing Cre recombinase under the control of the Tie2 promoter/enhancer (Tie2-Cre). The resulting offspring were backcrossed to TFPIFlox homozygosity while maintaining the Tie2-Cre transgene (TFPITie2). Numerous studies have demonstrated the effectiveness of using the Tie2-Cre mice to selectively delete floxed genes in cells of endothelial lineage.30-32
Myelomonocytic-specific deletion of TFPI-K1 was achieved by crossing the TFPIFlox mice with transgenic mice expressing Cre recombinase under control of the LysM promoter (LysM-Cre). The resulting offspring were backcrossed to TFPIFlox homozygosity while maintaining the LysM-Cre transgene (TFPILysM). This gene targeting approach has previously been used for the conditional deletion of floxed genes in monocytes, macrophages, and granulocytes.20
Efficiency and specificity of the Tie2-Cre– and LysM-Cre–directed deletion of TFPI-K1
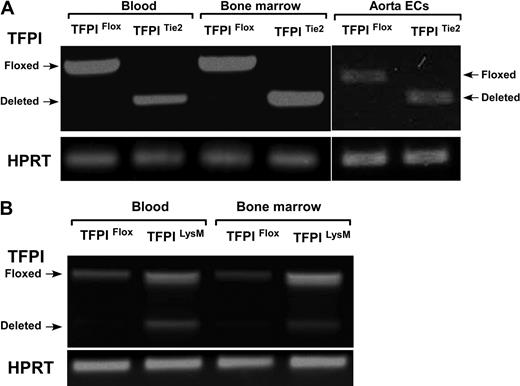
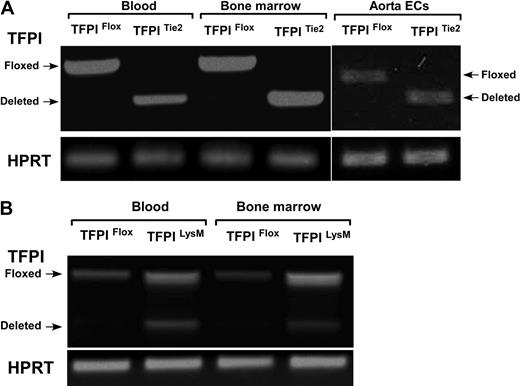
We examined the efficiency and endothelial specificity of the Tie2-Cre TFPI-K1 targeting approach. As there is not a combination of reagents and methods available to distinguish K1 deleted and wild-type TFPI, we used RT-PCR to detect deletion. RNA isolated from primary isolated aortic endothelial cells, bone marrow cells, and circulating blood cells from either TFPIFlox or TFPITie2 mice were subjected to RT-PCR. Using primers that span exon 4 of the mRNA encoding mouse TFPI, we were able to detect the PCR products for either the floxed transcript or the deleted transcript. In TFPIFlox mice, a single PCR product (630 bp) corresponding to the floxed transcript is observed in aortic endothelial cells, bone marrow cells, and circulating blood cells (Figure 2A). In TFPITie2 mice, a single PCR product (430 bp) corresponding to the deleted transcript is observed in aortic endothelial cells, bone marrow, and blood (Figure 2A). This demonstrates that Cre recombinase driven by the Tie2 promoter is expressed in endothelial, bone marrow, and blood cells and that Tie2-Cre directed deletion of TFPI-K1 is detected without detection of the floxed band after 40 cycles.
Transcriptional efficiency of Cre-mediated deletion. (A) Tie2-directed deletion of TFPI-K1 had high-efficiency deletion of TFPI-K1 in vascular endothelial cells, bone marrow cells, and circulating blood cells. (B) LysM-directed deletion resulted in partial deletion of TFPI-K1 in cells from whole blood and bone marrow.
Transcriptional efficiency of Cre-mediated deletion. (A) Tie2-directed deletion of TFPI-K1 had high-efficiency deletion of TFPI-K1 in vascular endothelial cells, bone marrow cells, and circulating blood cells. (B) LysM-directed deletion resulted in partial deletion of TFPI-K1 in cells from whole blood and bone marrow.
Next, we examined the efficiency and specificity of the LysM-Cre TFPI-K1 targeting approach. RNA isolated from blood or bone marrow cells from TFPIFlox or TFPILysM mice were used for RT-PCR analysis using the same primers described above. As expected, a single band corresponding to the floxed transcript is observed in blood and bone marrow cells from TFPIFlox mice (Figure 2B). In blood and bone marrow from TFPILysM mice, 2 products are observed, one corresponding to the floxed transcript and the other to the deleted transcript (Figure 2B). This demonstrates that expression of LysM-Cre and the subsequent Cre mediated TFPI-K1 deletion in blood cells is not complete (40.7% ± 2.6% deleted) or in bone marrow (41.7% ± 0.8% deleted).
Plasma TFPI activity in TFPIFlox, TFPITie2, and TFPILysM mice
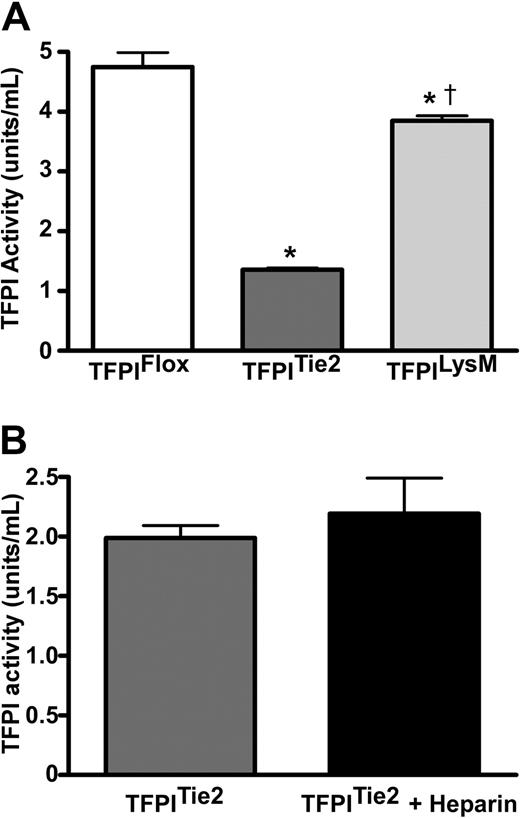
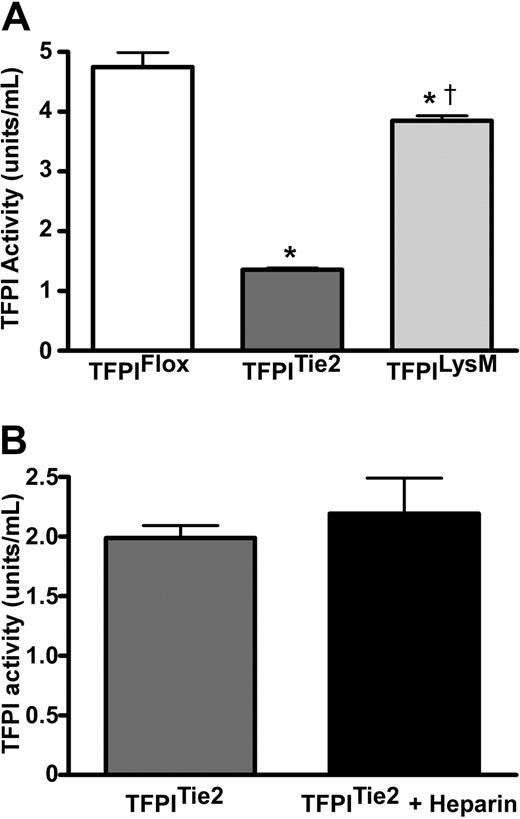
We examined the effects of endothelial or myeloid cell deletion of the TFPI-K1 domain on plasma TFPI activity to reflect the deletion of the K1 domain. Plasma TFPI activity of TFPITie2 mice was 71% lower (P < .001) than those of TFPIFlox mice (Figure 3A). Plasma TFPI activity of TFPILysM mice was 19% lower (P < .001) compared with their TFPIFlox littermates (Figure 3A). TFPILysM mice also had significantly higher plasma TFPI activity than the TFPITie2 mice. An ELISA using anti-murine TFPI antiserum detected differences in TFPI immunoreactivity between TFPITie2 and TFPIFlox mice (supplemental Figure 1; available on the Blood Web site; see the Supplemental Materials link at the top of the online article). This difference likely reflects the difference in affinity of our antisera between wild-type TFPI and the K1 deleted form.
Plasma TFPI activities. (A) TFPI activities were measured in plasma from mice that contain a floxed TFPI-K1 domain and either do not express Cre recombinase (TFPIFlox) or express the Cre recombinase via the endothelial specific promoter for Tie2 (TFPITie2) or the myelomonocytic specific promoter for LysM (TFPILysM). Animals that express Cre recombinase have a TFPI K1 domain deletion, which is the domain necessary for TFPI inhibition of the TF coagulation pathway. TFPITie2 mice have significantly reduced circulating TFPI activity levels compared with TFPIFlox (*P < .001) and TFPILysM mice (†P < .001). The TFPILysM mice have significantly lower circulating TFPI activities than TFPIFlox littermate controls (*P < .001; panel A). (B) Heparin infusion fails to increase plasma TFPI activity in TFPITie2 mice.
Plasma TFPI activities. (A) TFPI activities were measured in plasma from mice that contain a floxed TFPI-K1 domain and either do not express Cre recombinase (TFPIFlox) or express the Cre recombinase via the endothelial specific promoter for Tie2 (TFPITie2) or the myelomonocytic specific promoter for LysM (TFPILysM). Animals that express Cre recombinase have a TFPI K1 domain deletion, which is the domain necessary for TFPI inhibition of the TF coagulation pathway. TFPITie2 mice have significantly reduced circulating TFPI activity levels compared with TFPIFlox (*P < .001) and TFPILysM mice (†P < .001). The TFPILysM mice have significantly lower circulating TFPI activities than TFPIFlox littermate controls (*P < .001; panel A). (B) Heparin infusion fails to increase plasma TFPI activity in TFPITie2 mice.
Additional activity assays in TFPITie2 mice following 7 backcrosses into the C57Bl/6 background suggest that the differences are maintained in the C57 background (TFPIFlox; 5.0 ± 1.2 vs TFPITie2 1.4 ± 0.1 U/mL, P < .0001; supplemental Figure 2). Furthermore, Tie2 mediated deletion removes the previously demonstrated heparin-mediated increase in plasma TFPI in mice22 (Figure 3B).
To further determine the role of bone marrow-derived cells in determining circulating levels of TFPI, we performed hematopoietic stem cell transplantation with fetal liver cells obtained from E16.5 wild-type (TFPI+/+) or TFPI−/− embryos. Wild-type male mice (C57BL/6) were used as recipients and received lethal doses of radiation before rescue by fetal liver cell transplantation. Reconstitution of transplant-recipient bone marrow with donor-derived cells was confirmed by PCR analysis of peripheral blood performed 20 weeks after transplant. 2 groups of transplanted mice were analyzed: those that received TFPI−/− hematopoietic stem cells (BMTFPI−/−) and those that received wild-type cells (BMTFPI+/+). In BMTFPI−/−mice, plasma TFPI activity was decreased by 20% compared with BMTFPI+/+ mice (P < .01). These data, taken together, suggest that endothelial cells account for approximately 50% of circulating TFPI while bone marrow–derived cells account for an additional 20%.
Hemostasis and thrombosis in TFPIFlox, TFPITie2, and TFPILysM mice
We used a tail bleeding model to assess hemostasis in TFPIFlox, TFPITie2, and TFPILysM mice. TFPITie2 or TFPILysM mice had no significant differences in tail bleeding times or cuticle bleeding times compared with TFPIFlox mice (data not shown). Standard prothrombin times and activated partial thromboplastin times did not differ compared with TFPIFlox littermate controls. Serial dilutions of TF also failed to demonstrate differences in plasma clotting times (supplemental Figure 3).
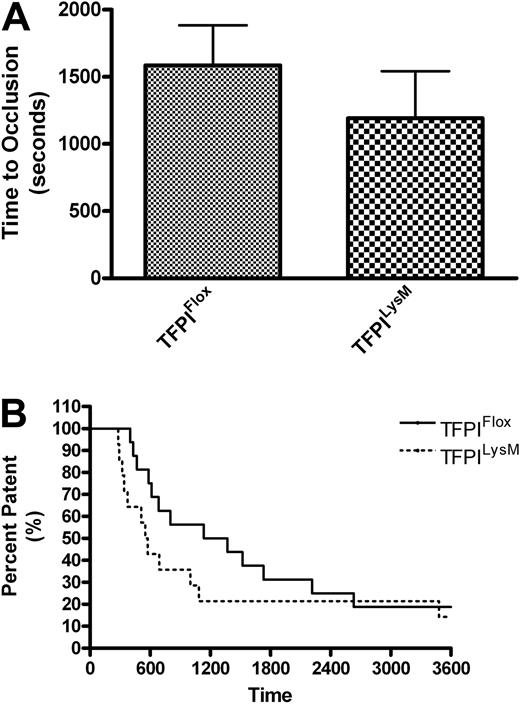
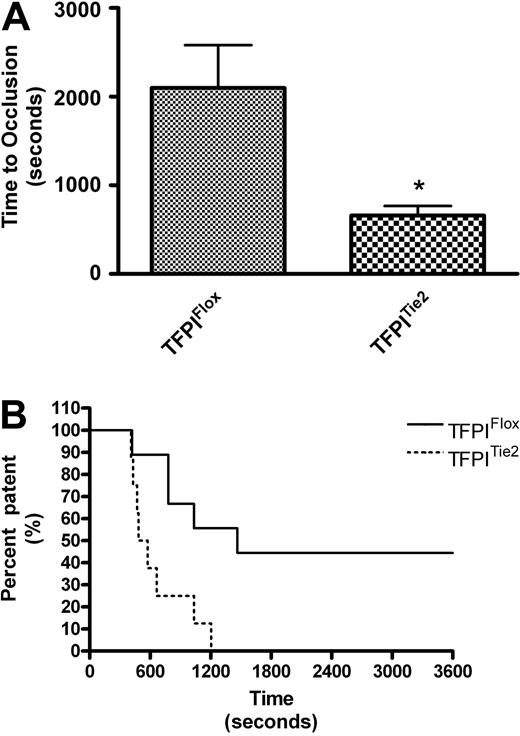
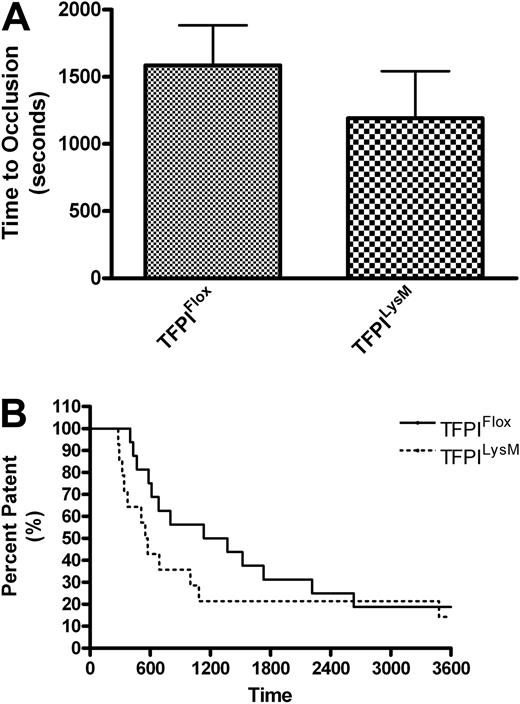
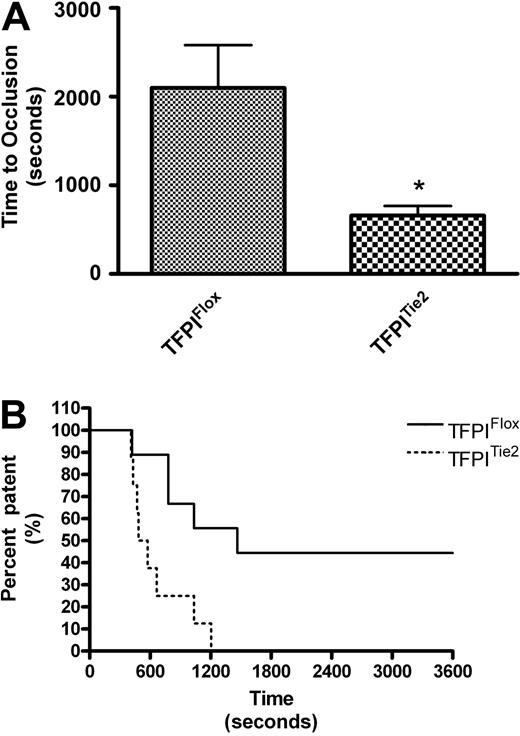
To assess arterial thrombosis in TFPIFlox, TFPITie2, and TFPILysM mice, we used a FeCl3-induced model of thrombosis. Times to occlusion were not significantly different between TFPILysM mice and TFPIFlox mice (1193 ± 349 vs 1586 ± 298 seconds, Figure 4A). Chi-square analysis of Kaplan-Meier survival plots of percent patent versus time also revealed no significant differences between these 2 groups of mice (Figure 4B). In contrast, times to occlusion in TFPITie2 mice were 69% shorter than their TFPIFlox littermate controls (659 ± 106 vs 2098 ± 484 seconds, Figure 5A). A χ2 analysis of Kaplan-Meier survival plots of percent patent versus time revealed a significant difference between the plots for TFPITie2 and TFPIFlox mice (Figure 5B, P = .007).
Murine model of thrombosis in TFPILysM mice. Thrombosis was examined using a FeCl3 arterial injury model in TFPIFlox and TFPILysM mice. A flow probe was used to determine when the carotid arteries of TFPIFlox and TFPILysM mice became occluded. (A) No significant differences were observed in times to occlusion between TFPIFlox and TFPILysM mice. (B) Percent patency was plotted against time using a Kaplan-Meier survival plot and χ2 analysis revealed no significant differences of plots for TFPIFlox and TFPILysM mice (P = .25).
Murine model of thrombosis in TFPILysM mice. Thrombosis was examined using a FeCl3 arterial injury model in TFPIFlox and TFPILysM mice. A flow probe was used to determine when the carotid arteries of TFPIFlox and TFPILysM mice became occluded. (A) No significant differences were observed in times to occlusion between TFPIFlox and TFPILysM mice. (B) Percent patency was plotted against time using a Kaplan-Meier survival plot and χ2 analysis revealed no significant differences of plots for TFPIFlox and TFPILysM mice (P = .25).
Murine model of thrombosis in TFPITie2 mice. Thrombosis was examined using a FeCl3 arterial injury model in TFPIFlox and TFPITie2 mice. A flow probe was used to determine when the carotid arteries of TFPIFlox and TFPITie2 mice became occluded. (A) Times to occlusion were significantly reduced in TFPITie2 mice compared with TFPIFlox mice (*P < .001). (B) Percentage of patency was plotted against time using a Kaplan-Meier survival plot, and χ2 analysis revealed a significant difference between plots for TFPIFlox and TFPILysM mice (P = .007).
Murine model of thrombosis in TFPITie2 mice. Thrombosis was examined using a FeCl3 arterial injury model in TFPIFlox and TFPITie2 mice. A flow probe was used to determine when the carotid arteries of TFPIFlox and TFPITie2 mice became occluded. (A) Times to occlusion were significantly reduced in TFPITie2 mice compared with TFPIFlox mice (*P < .001). (B) Percentage of patency was plotted against time using a Kaplan-Meier survival plot, and χ2 analysis revealed a significant difference between plots for TFPIFlox and TFPILysM mice (P = .007).
Discussion
TFPI plays an important role in the regulation of coagulation via its inhibition of the TF-pathway.33 TFPI is a Kunitz-type protease inhibitor consisting of 3 Kunitz-type inhibitory domains (K1, K2, and K3) and a basic carboxyl terminus.34 The K1 domain is responsible for the binding and inhibition of the TF-FVIIa complex, the K2 domain is responsible for the binding and inhibition of FXa, and although the exact physiologic role of the K3 domain is unknown, it plays a role in the cellular binding of TFPI.33,35 The carboxyl terminus of TFPI is required for its optimal inhibition of FXa and is involved in its interaction with lipoproteins, glycosaminoglycans, and fibrin clots.36-38 Systemic homozygotic deletion of TFPI-K1 domain results in intrauterine lethality.11 As TFPI is thought to be predominantly expressed from endothelium, it might be anticipated that the loss of endothelial TFPI would be inconsistent with viability. In the present study, we used the Cre-lox strategy to examine the effects of cell-directed deletion of the TFPI-K1 domain on development, hemostasis, and thrombosis.
To clarify the role of endothelial-derived TFPI, complementary models were used due to the deletion in nonendothelial cells using the Tie2 promoter.39,40 As deletion is seen in nucleated cells of hematopoietic origin with Tie2 promoter and TFPI is known to be expressed from myelomonocytic cells, we compared Tie2-directed deletion with that from Cre expressing mice using the LysM promoter as well as radiated mice rescued with bone marrow transplantation from fetal livers of mice with a systemic deletion in the TFPI-K1 domain. In addition, using a CMV-Cre mouse, which results in complete systemic deletion of the floxed TFPI-K1 domain, we were able to confirm the findings of Huang et al11 that homozygotic systemic gene disruption of TFPI-K1 domain leads to intrauterine lethality as no live mice born from crossings with these mice had homozygous TFPI-K1 deletion.
We demonstrate that endothelial and myelomonocytic deletion of TFPI-K1 is consistent with viability and fertility. TFPI null mice succumb to intrauterine lethality and those surviving beyond E12.5 have signs of coagulopathy which include rare intravascular thrombi in the brain and liver.11 Our data suggest that this lethality does not appear due to expression of TFPI from cells which express Tie2. Intrauterine lethality observed in TFPI null embryos is likely due to unregulated TF/FVIIa activity resulting in impaired embryogenesis and placental dysfunction.41 Rescue of embryonic lethality of TFPI null mice can be achieved by breeding into mice which express low levels of TF, resulting in TFPI−/−/low-TF mice, further confirming the importance of maintaining the TF-TFPI balance in embryonic development. The origin of TFPI in placenta is likely endothelial cells and trophoblast cells.42 In mice, the terminal maternal vascular space is formed from zygote-derived trophoblast cells.43 Findings that trophoblast cells express endothelial regulators of hemostasis suggest that these cells may possess an endothelial-like ability to regulate hemostasis at the fetomaternal interface.44 TFPI from trophoblasts as well as from the maternal circulation in the TFPITie2 mice may be sufficient to maintain the TF-TFPI balance required for placental hemostasis and embryogenesis. When TFPI heterozygosity (TFPI+/−) is bred into mice with other prothrombotic genetic traits such as factor V Leiden13 or thrombomodulin deficiency,14 thrombotic disease occurs with multiple manifestations including intrauterine lethality, perinatal lethality, and hypercoagulability in surviving adult mice.
We further demonstrate that, although LysM expressing cells contribute to a portion (∼ 19%) of circulating TFPI, this contribution is not sufficient to alter development, hemostasis, or thrombosis. In bone marrow reconstitution studies in which bone marrow from TFPI+/+ or TFPI−/− was transplanted into WT mice, those transplanted with TFPI−/− cells (BMTFPI−/−) had a 20% reduction in circulating TFPI activity levels compared with those transplanted with TFPI+/+ cells (BMTFPI+/+). There was no significant decrease in vascular TFPI activities (data not shown) between these mice suggesting that the 20% reduction was due to circulating cells derived from bone marrow of the TFPI−/− mice. BMTFPI−/− mice did not have significant differences in measures of hemostasis compared with their BMTFPI+/+ counterparts. On the other hand, Tie2 expressing cells, namely endothelial cells and cells of hematopoietic origin, produce a majority (∼ 71%) of circulating TFPI; and importantly, although deletion of TFPI-K1 in this pool of TFPI does not affect development or hemostasis, it does play a role in the regulation of arterial thrombosis. These findings should be seen in the context of a growing understanding of differences in TFPI biology between mice and man. We recently demonstrated that mice have a smaller pool of heparin-releasable TFPI than humans, and our data suggest that a majority of adult murine tissue and plasma TFPI exists as the alternatively spliced TFPIβ form.22 We demonstrate here that Tie2-mediated deletion eliminates the heparin-releasable pool.
The finding that standard measures of hemostasis were not altered in the TFPITie2, TFPILysM, or BMTFPI−/− mice may reflect that the decreases observed in circulating TFPI activities are not of sufficient magnitude to cause a difference in PT and aPTT. This is similar to many coagulation disorders in which clotting abnormalities may not be detected until there is significant reduction of clotting factors. PT and aPTT measure the clotting ability of plasma and do not involve cellular elements whereas the tail vein bleeding times reflects platelet function. The lack of effects on hemostasis suggest that the absence of functional TFPI on endothelial cells or circulating blood cells and the corresponding decreases observed in plasma TFPI are not sufficient to affect hemostasis in mice.
We examined the specificity of both the Tie2 and LysM directed deletion. Tie2 directed deletion led to deletion in endothelial, whole blood, and bone marrow cells whereas LysM directed deletion led to partial deletion in both whole blood and bone marrow cells. This is consistent with the initial work using the LysM-Cre in which there was efficient deletion in monocytes and granulocytes, but no significant deletion was observed in T or B cells.20 Tie2-Cre directed deletion has previously been reported as an endothelial cell-specific gene targeting approach30,45 ; however, Tie2 is expressed not only by endothelial cells but also by hematopoietic progenitor cells as well.39,40 Given that Tie2 directed deletion of TFPI-K1 results in a 71% reduction in circulating TFPI activity, there must be a nonendothelial and nonhematopoietic cell-derived source for TFPI. Potential sources include vascular smooth muscle cells7 or cardiomyocytes.46 Examination of the LysM, Tie2, and bone marrow cell contributions to circulating TFPI activity would suggest that approximately 50% of circulating TFPI activity comes from endothelium. Mice with a heterozygotic systemic genetic disruption of TFPI-K1 have a prothrombotic phenotype in a model of arterial thrombosis47 and have enhanced neo-intimal formation in an acute model of vascular injury.12 These heterozygotic mice have a 50% reduction in circulating TFPI activities and presumably a 50% reduction in vascular TFPI activities. Therefore, it is no surprise that we observe enhanced thrombotic potential in our TFPITie2 mice with a 71% reduction in circulating TFPI activities and TFPI-K 1 deletion in endothelial cells. In a FeCl3-induced model of arterial thrombosis, TFPITie2 mice had decreased times to occlusion compared with their littermate controls.
The antithrombotic role for TFPI varies with the model used. Westrick and colleagues studied TFPI K1+/− mice using the Rose Bengal model of photochemical injury47 and showed no difference between TFPI K1+/− mice and wild-type littermates on a wild-type background. However, on an ApoE null background, the TFPI+/− mice had shorter occlusion times compared with historical controls. These data suggest that a 50% reduction in TFPI may not affect arterial thrombosis, unless TF is up-regulated. However, we have previously shown in an electrolytic model that TFPI+/− mice have enhanced venous clot volumes.14 Our data support that endothelial deletion and a 70% reduction in circulating levels does enhance arterial thrombosis on a wild-type background.
To conclude, we have examined the effects of cell directed deletion of the TFPI-K1 domain on circulating TFPI activity, hemostasis, and thrombosis. Endothelial and hematopoietic progenitor or myleomonocytic cell deletion is compatible with survival as these mice were viable and reproduced normally. In mice, the majority of circulating TFPI activity (∼ 70%) is generated from endothelial and hematopoietic progenitor cells with approximately 50% coming from endothelium. There is a source, not yet identified, for circulating TFPI in mice with Tie2-directed deletion of TFPI-K1. Neither Tie2- nor LysM-directed deletion of TFPI-K1 had significant effects on hemostasis. Endothelial-derived TFPI does play a regulatory role in arterial thrombosis. While endothelium is the primary source for circulating TFPI, endothelial deletion of TFPI-K1 results in a relatively mild phenotype albeit with primary effects on arterial thrombosis.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by National Institutes of Health grant HL65191 to R.D.S.
National Institutes of Health
Authorship
Contribution: T.A.W. performed experiments and drafted and revised the manuscript; T.J. performed experiments and drafted and revised the manuscript; N.Z. performed experiments and revised the manuscript; C.T. performed and analyzed experiments; S.D. performed and analyzed experiments and revised the manuscript; E.W.H. performed and analyzed experiments; S.A.M. performed experiments and drafted and revised the manuscript; R.S. performed experiments; S.P. performed experiments and revised the manuscript; W.P.F. performed experiments and revised the manuscript; J.v.D. designed and performed experiments; A.E.M. performed experiments and drafted and revised the manuscript; G.S.S. performed experiments and drafted and revised the manuscript; and R.D.S. designed experiments and drafted and revised the manuscript.
Conflict-of-interest disclosure: A.E.M. receives research funding from Novo Nordisk. The remaining authors declare no competing financial interests.
Correspondence: Robert D. Simari, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: simari.robert@mayo.edu.
References
Author notes
T.A.W. and T.J. contributed equally to this work.