Abstract
Treatment of acute promyelocytic leukemia (APL) with all-trans retinoic acid and/or arsenic trioxide represents a paradigm in targeted cancer therapy because these drugs cause clinical remission by affecting the stability of the fusion oncoprotein promyelocytic leukemia (PML)/retinoic acid receptor alpha (RARA). The authors of previous studies have implicated the ubiquitin-proteasome pathway as the main mechanism involved in therapy-induced PML/RARA degradation. Here we have investigated a role of autophagy, a protein degradation pathway that involves proteolysis of intracellular material within lysosomes. We found that both all-trans retinoic acid and arsenic trioxide induce autophagy via the mammalian target of rapamycin pathway in APL cells and that autophagic degradation contributes significantly both to the basal turnover as well as the therapy-induced proteolysis of PML/RARA. In addition, we observed a correlation between autophagy and therapy-induced differentiation of APL cells. Given the central role of the PML/RARA oncoprotein in APL pathogenesis, this study highlights an important role of autophagy in the development and treatment of this disease.
Introduction
The promyelocytic leukemia/retinoic acid receptor alpha (PML/RARA) oncoprotein, which is expressed by the acute promyelocytic leukemia (APL)–specific t(15;17) chromosomal translocation, is known to inhibit granulocyte development at the promyelocytic stage of differentiation and to promote malignant transformation of hematopoietic progenitor cells mainly by acting as a repressor of gene expression.1-3 All-trans retinoic acid (ATRA), the first of 2 drugs that was found to cause disease regression specifically in APL patients, interacts with the ligand binding domain present on the RARA moiety of the chimeric oncoprotein and causes both its transcriptional activation, as well as its proteolytic degradation, an effect that phenotypically leads to granulocyte differentiation of APL cells.3,4
In later years, arsenic trioxide (ATO) also has been shown to be remarkably effective as an agent that cures APL.4 Compared with ATRA, this drug appears to have a more limited capacity to induce differentiation and is thought to induce disease regression mainly by causing proteolytic degradation of PML/RARA.4,5 The mechanism by which ATO causes PML/RARA proteolysis was recently shown to involve PML small ubiquitin-like modifier (SUMO)–ylation and subsequent poly-ubiquitination and proteolytic degradation of PML and PML/RARA by a protein called RNF4.6,7 The ability of ATRA and ATO to activate disease regression through distinct molecular mechanisms that lead to PML/RARA degradation is further underscored by recent studies in which the authors showed a synergy effect of these 2 drugs both in their ability to induce clinical remission of APL patients,8,9 as well as their capacity to cause PML/RARA degradation.5 Thus, the ability of ATRA and ATO to activate PML/RARA degradation appears to represent a critical parameter for successful APL treatment.
Two major routes for degradation of intracellular proteins exist: the ubiquitin-proteasome system (UPS) and the autophagy-lysosome pathway. The UPS generally is used for degradation of short-lived soluble proteins and misfolded proteins after their labeling with poly-ubiquitin chains. However, misfolded proteins that accumulate to form larger aggregates generally are poor substrates for the UPS.10 Macro-autophagy, hereafter referred to as autophagy, involves the turnover of cytoplasmic macromolecules, protein aggregates, and defective organelles by their sequestration by double-layered membranes that form autophagosomes, which eventually fuse with lysosomes in which the sequestered contents are degraded.11 Mice and flies lacking essential autophagy genes are characterized by increased tumorigenesis12-14 and encompass neurodegenerative phenotypes,15-17 indicating that autophagy is an important tumor-suppressive and neuroprotective mechanism.
The authors of previous studies have implicated an important role of the UPS in therapy-induced degradation of PML/RARA. This role is evident from experiments showing that ATRA and ATO–induced PML/RARA degradation is blocked by proteasome inhibitors.5,18,19 Furthermore, SUMO-conjugated PML, RNF4, and components of the UPS are corecruited to nuclear compartments called PML nuclear bodies (PML NBs) in the presence of ATO.6 Finally, the proteasome inhibitor bortezomib has been found to cause reduced ATO and ATRA–induced disease regression in an APL mouse model, indicating that a functional UPS is important for disease remission.5
Although a role of the UPS in therapy-induced clearance of PML/RARA has been established, a contributing role of autophagy in the degradation of this oncoprotein has not previously been determined. In the present study, we demonstrate that the catabolism of PML/RARA is largely affected also by this protein degradation pathway and that the clearance of this oncoprotein in both ATO and ATRA–treated cells depends upon and correlate with increased autophagic activity. Finally, we observed a stimulatory role of autophagy on the differentiation of the APL cell line NB4, suggesting that this degradation pathway potentiate therapy-induced differentiation of APL cells.
Methods
Cell culture
HeLa cells were obtained from ATCC. The APL NB4 cell line was a kind gift from Dr Michel Lanotte at Hôpital Saint-Louis. HeLa cells were grown in Dulbecco modified Eagle medium (Invitrogen) containing 10% fetal bovine serum (Sigma-Aldrich). NB4 cells were cultured in Iscove modified Dulbecco medium (Lonza) containing 10% fetal bovine serum. For starvation experiments, cells were incubated in Earle balanced salt solution (EBSS; Invitrogen) for 4 hours.
Transfection of small interfering RNA oligonucleotides and plasmids
For combined transfection of HeLa cells with plasmid and siRNAs, cells were first transfected with 100nM ON-TARGETplus SMARTpool control or siRNAs against human ULK1 (NM_003565; Dharmacon) by use of the oligofectamine transfection reagent (Invitrogen). Three days later, the cells were transfected with a plasmid encoding PML/RARA (a kind gift from Dr Pier Giuseppe Pelicci at the Istituto Europeo di Oncologia) by use of the FuGENE transfection reagent (Roche) and left for another 24 or 48 hours for immunofluorescence microscopy or Western blot analysis, respectively.
Antibodies
The following antibodies were used: rabbit anti-RARA (Santa Cruz Biotechnology; 1:1000), rabbit anti-ULK1 (Santa Cruz Biotechnology; 1:500), mouse anti-tubulin (Sigma-Aldrich; 1:1000), mouse anti–FK1 ubiquitin (Biomol; 1:400), rabbit anti–P-p70S6K (Cell Signaling; 1:1000), guinea pig anti-p62 (Progen; 1:1000), rabbit anti-LC3 (Cell Signaling; 1:1000), mouse anti-LC3 (Nanotools; 1:100), mouse anti-PML (Santa Cruz Biotechnology; 1:200), rabbit anti-PML (Santa Cruz Biotechnology; 1:200), and mouse anti–CD11b fluorescein isothiocyanate (FITC; Abcam; 1:50).
Reagents
All drugs were purchased from Sigma-Aldrich and were used at the following final concentrations: 1μM ATRA (stock 10mM in 100% dimethyl sulfoxide), 1μM ATO (0.1M stock in 0.1M NaOH), 100nM bafilomycin A (Baf A; stock 200μM in EtOH), 5 to 10mM 3-MA (stock 100mM in EBSS), and 100nM rapamycin (stock 200μM in EtOH). Protease and phosphatase inhibitors were obtained from Roche.
Confocal microscopy analyses
Cells were fixed in 3% paraformaldehyde, permeabilized with 0.05% saponin, and stained for fluorescence microscopy as described previously.20 Before fixation, HeLa cells were grown on coverslips and NB4 cells were subjected to cytospin centrifugation and subsequently air dried for 1 hour. Fluorescently labeled cells were examined by the use of a Zeiss LSM 510 META microscope equipped with a 63× oil-immersion objective. Image processing and analysis were done with Zeiss LSM 510 software version 3.2, ImageJ Version 1.42 (National Institutes of Health), and Adobe Photoshop Version 7.0 (Adobe Systems).
Differential detergent extraction and Western blot analysis
To analyze the cellular levels of different proteins and their solubility, cells were first extracted in ice-cold Triton X-100–containing lysis buffer (50mM NaCl, 10mM Tris, 5mM ethylenediaminetetraacetic acid, 1% Triton X-100 + protease and phosphatase inhibitor cocktails). Cell lysates were then centrifuged (18 000g) for 10 minutes and the supernatants (soluble proteins) collected. The remaining protein pellets were washed with phosphate-buffered saline before extraction with an SDS-containing buffer (2% SDS, 1mM dithiothreitol, 50mM Tris + protease and phosphatase inhibitor cocktails) or an urea-containing lysis buffer (8M urea, 0.5% Triton, 10mM dithiothreitol + protease and phosphatase inhibitor cocktails) to obtain SDS- or urea-extracted proteins, respectively. Unless otherwise stated, the cells were lysed in the urea-containing lysis buffer for Western blot analysis. Protein extracts were loaded and resolved on 12% gels (Pierce) followed by electroblotting to Immobilon-P membranes (Millipore). The blots were probed with specific antibodies, which were detected by the use of standard ECL reagents. The intensities of the different bands obtained were quantified by the use of the software Image quant (GE Healthcare) and relative amounts quantified by the use of antitubulin as a loading control.
Flow cytometry
NB4 cells were incubated with or without ATRA for 24 hours, then incubated for another 24 hours with 3MA, BafA1, or rapamycin in the presence or absence of ATRA. The cells were then washed twice in phosphate-buffered saline with 0.01% bovine serum albumin before incubation with FITC-conjugated anti-CD11b antibody for 30 minutes, washed 3 times, and analyzed by the use of a FACSCalibur flow cytometer (BD Biosciences).
Statistical analysis
Data are expressed as mean plus or minus SEM or mean plus or minus SD quantified from at least 3 independent experiments. Statistical significance was determined by use of the Student t test. Differences were considered significant when P was less than .05.
Results
The PML/RARA fusion protein is highly insoluble
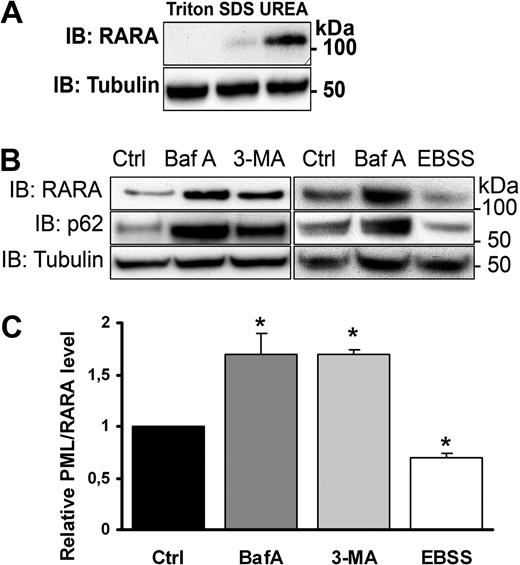
To investigate a role of autophagy in PML/RARA catabolism, we used the APL cell line NB4.21 This cell line contains the t(15;17) chromosomal translocation and mimics several features of clinically isolated APL blast cells, including the ability to undergo ATRA-stimulated differentiation along the granulocyte linage and the susceptibility of the expressed PML/RARA oncoprotein to ATRA- and/or ATO-stimulated degradation. Because both the PML protein as well as the PML/RARA fusion is known to be aggregation prone, we first assessed the ability of different solubilization agents, including Triton X-100, SDS, and urea for their abilities to extract PML/RARA. We found that extraction buffers containing 8M urea were markedly more effective in extracting PML/RARA from NB4 cells than buffers containing 0.5% Triton X-100 or 2% SDS (Figure 1A). Interestingly, both PML and RARA were found to be considerably more amenable to extraction by Triton compared with PML/RARA (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), suggesting that the chimeric protein is more susceptible to aggregate formation than its respective fusion partners expressed from nonrearranged alleles. The protein species detected at 110 kDa, which in the present study is used as diagnostic for the PML/RARA oncoprotein, was not observed in non-APL cells, including HL60 and T2 (data not shown). Because aggregated proteins are poor substrates for the UPS10 and as such proteins are known targets of autophagy,22 this result is consistent with the notion that the oncogenic fusion protein may be subjected to autophagic degradation.
PML/RARA is degraded by autophagy. (A) Immunoblot (IB) showing optimal extraction efficiency of PML/RARA by a buffer containing UREA. NB4 cells were first solubilized in a lysis buffer containing Triton X-100. The pelleted material was further dissolved in either SDS containing or UREA containing buffers. (B) NB4 cells grown in nutrient-rich medium were treated or not with Baf A (100nM) for 12 hours or with 3-MA (10mM) for 4 hours, or cells were incubated in starvation media (EBSS) for 4 hours, followed by IB with anti-RARA, anti-p62, or anti-tubulin antibodies. The band corresponding to PML/RARA (110 kDa) is shown. The whole blot, including RARA (50 kDa), is shown in supplemental Figure 2. (C) Quantification of relative protein level from 3 independent experiments. Data are mean ± SEM. *P < .05, Student t test.
PML/RARA is degraded by autophagy. (A) Immunoblot (IB) showing optimal extraction efficiency of PML/RARA by a buffer containing UREA. NB4 cells were first solubilized in a lysis buffer containing Triton X-100. The pelleted material was further dissolved in either SDS containing or UREA containing buffers. (B) NB4 cells grown in nutrient-rich medium were treated or not with Baf A (100nM) for 12 hours or with 3-MA (10mM) for 4 hours, or cells were incubated in starvation media (EBSS) for 4 hours, followed by IB with anti-RARA, anti-p62, or anti-tubulin antibodies. The band corresponding to PML/RARA (110 kDa) is shown. The whole blot, including RARA (50 kDa), is shown in supplemental Figure 2. (C) Quantification of relative protein level from 3 independent experiments. Data are mean ± SEM. *P < .05, Student t test.
PML/RARA is degraded by autophagy
We next determined the expression level of PML/RARA under conditions that stimulate or inhibit autophagic degradation. To stimulate autophagy, NB4 cells were exposed to nutrient deprivation (starvation media, EBSS), and, to inhibit autophagic degradation, cells were incubated in the presence of the phosphatidylinositol 3-kinase inhibitor 3-methyladenine (3-MA), which inhibits sequestration of cargo, or the lysosomal proton pump inhibitor Baf A. Whereas induction of autophagy by starvation caused increased PML/RARA degradation (Figure 1B-C), inhibition of autophagic degradation by Baf A or 3-MA caused a significant accumulation of PML/RARA in NB4 cells (Figure 1B-C). Importantly, p62, which represents a known target for autophagic degradation,23 displayed a similar expression pattern as PML/RARA in response to autophagic stimulation or inhibition (Figure 1B), demonstrating that the cells are sensitive for the inhibitors used and that the starvation treatment caused increased autophagy.
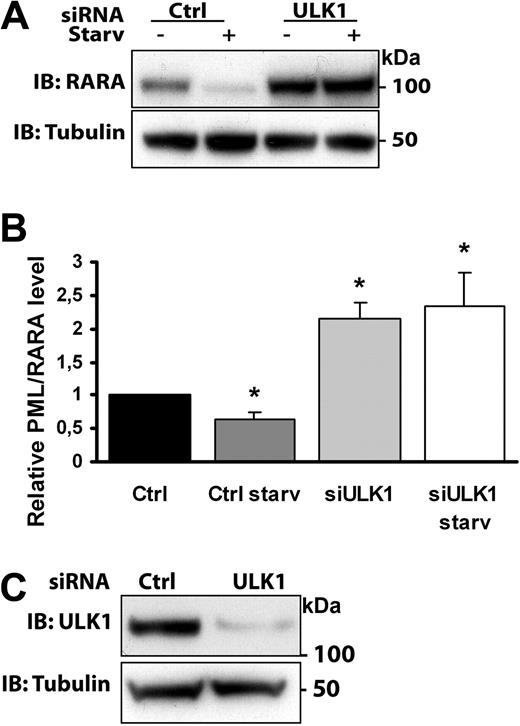
To further assess whether autophagy contributes to degradation of PML/RARA, HeLa cells transiently expressing this chimeric protein were treated with control siRNA or siRNA against ULK1, a protein kinase required for induction of autophagy downstream of mTOR (mammalian target of rapamycin).24 Cell lysates from control and ULK1-depleted PML/RARA-expressing HeLa cells (Figure 2C) were analyzed for the levels of PML/RARA after starvation (Figure 2A-B). Whereas PML/RARA levels decreased significantly in cells transfected with a control siRNA, ULK1-depleted cells were irresponsive to amino acid deprivation, which is consistent with a defect in the catabolic degradation of the oncoprotein expressed in these cells. Furthermore, the levels of PML/RARA were significantly greater in nonstarved ULK1-depleted cells compared with control cells, suggesting a reduced capacity of these cells to perform PML/RARA proteolysis in the absence of ULK1 also under basal conditions (Figure 2A-B).
ULK1 depletion inhibits PML/RARA degradation. (A) HeLa cells transfected with control RNA or ULK1-targeted siRNA were retransfected with PML/RARA cDNA and treated or not with starvation media for 4 hours. Lysates were analyzed by Western blotting by use of the indicated antibodies. (B) Quantification of data from 3 independent experiments. Error bars represent SEM. *P < .05, Student t test. (C) Confirming depletion of ULK1: Lysates from control or ULK1-depleted HeLa cells were examined by Western blotting by the use of anti-ULK1 and anti-tubulin antibodies.
ULK1 depletion inhibits PML/RARA degradation. (A) HeLa cells transfected with control RNA or ULK1-targeted siRNA were retransfected with PML/RARA cDNA and treated or not with starvation media for 4 hours. Lysates were analyzed by Western blotting by use of the indicated antibodies. (B) Quantification of data from 3 independent experiments. Error bars represent SEM. *P < .05, Student t test. (C) Confirming depletion of ULK1: Lysates from control or ULK1-depleted HeLa cells were examined by Western blotting by the use of anti-ULK1 and anti-tubulin antibodies.
Consistent with these data, an increase in PML/RARA levels in ULK1-depleted HeLa cells also was observed when we performed immunofluorescence analysis (supplemental Figure 3). In addition, confocal immunofluorescence analysis of the transiently transfected cells with the use of the anti-RARA antibody revealed the presence of PML/RARA-positive cytoplasmic foci that colocalized with endogenous p62, ubiquitin, and the autophagic membrane marker LC3 (supplemental Figure 4). No signal was detected in mock-transfected cells when we used the anti-RARA antibody (supplemental Figure 4B). p62, which possesses a ubiquitin-binding domain, as well as an LC3-interacting region, has been found to target ubiquitinated aggregate-prone proteins for autophagic degradation,25 and its colocalization with PML/RARA might suggest that p62 is involved in autophagy-mediated PML/RARA degradation. Taken together, our data indicate that PML/RARA is an autophagy substrate and that both its basal turnover and starvation-induced degradation is inhibited when autophagy is compromised.
ATRA and ATO induce autophagy in NB4 cells
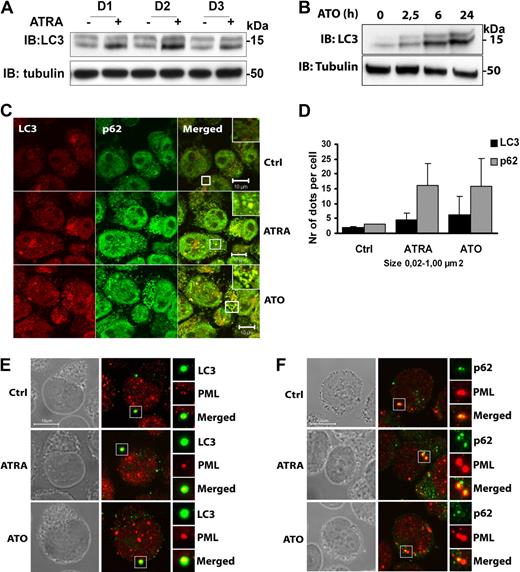
Because ATRA and ATO–induced clinical remission is associated with degradation of PML/RARA,18 we next examined the ability of these drugs to promote autophagy in NB4 cells. For these experiments we probed for the autophagy marker protein LC3 by Western blotting. Upon induction of autophagy, cytosolic LC3 (LC3-I, top band) is cleaved and subsequently conjugated to phosphatidylethanolamine within autophagic membranes (LC3-II, bottom band).26 Because LC3-II remains bound to autophagic membranes throughout the pathway, it is a useful marker to study autophagy, and the LC3-II level is a good measure of autophagic activity. As can be seen in Figure 3, LC3-II levels were induced in NB4 cells treated with ATRA (Figure 3A) or ATO (Figure 3B), suggesting that these drugs cause increased autophagic activity. By using quantitative real-time polymerase chain reaction, we found that NB4 cells treated with ATRA or ATO had increased LC3 mRNA levels (supplemental Figure 5A), indicating that these drugs also induce LC3 transcription. This newly synthesized LC3 is rapidly conjugated to LC3-II because there is no increase in the LC3-I levels.
Autophagy is activated by ATRA and ATO in NB4 cells. (A) NB4 cells were treated with or without ATRA (1μM) for 1, 2, or 3 days (D1, D2, and D3, respectively), followed by IB analysis by the use of anti-LC3 and antitubulin antibodies. (B) Cell lysates from NB4 cells treated with or without ATO (1μM) for the indicated time were subjected to immunoblotting by the use of anti-LC3 and anti-tubulin antibodies. (C) NB4 cells with or without ATRA (1μM) or ATO (1μM) for 24 hours were stained with anti-LC3 (red) and anti-p62 (green) antibodies. Cells were examined by confocal microscopy. Scale bars, 10 μm. (D) The number and size (0.02-1.00 μm2) of p62- and LC3-positive structures were estimated by the use of the ImageJ quantification program from 2 separate experiments (total 150 cells). The graphs show average number of particles per cell ± SEM. (E-F) NB4 cells treated or not with ATRA or ATO were fluorescently labeled with antibodies against (E) PML (red) and LC3 (green) or (F) PML (red) and p62 (green). Cells were examined by confocal microscopy. Phase contrast images in grayscale are shown to visualize the nuclear boundary. Scale bars, 10 μm.
Autophagy is activated by ATRA and ATO in NB4 cells. (A) NB4 cells were treated with or without ATRA (1μM) for 1, 2, or 3 days (D1, D2, and D3, respectively), followed by IB analysis by the use of anti-LC3 and antitubulin antibodies. (B) Cell lysates from NB4 cells treated with or without ATO (1μM) for the indicated time were subjected to immunoblotting by the use of anti-LC3 and anti-tubulin antibodies. (C) NB4 cells with or without ATRA (1μM) or ATO (1μM) for 24 hours were stained with anti-LC3 (red) and anti-p62 (green) antibodies. Cells were examined by confocal microscopy. Scale bars, 10 μm. (D) The number and size (0.02-1.00 μm2) of p62- and LC3-positive structures were estimated by the use of the ImageJ quantification program from 2 separate experiments (total 150 cells). The graphs show average number of particles per cell ± SEM. (E-F) NB4 cells treated or not with ATRA or ATO were fluorescently labeled with antibodies against (E) PML (red) and LC3 (green) or (F) PML (red) and p62 (green). Cells were examined by confocal microscopy. Phase contrast images in grayscale are shown to visualize the nuclear boundary. Scale bars, 10 μm.
Because elevated levels of autophagy have been shown to correlate with increased numbers of p62 and LC3-containing cytoplasmic structures,27 we examined the effect of ATRA and ATO on the subcellular distribution of these proteins in NB4 cells. Immunofluorescence labeling revealed an increased number of p62 and LC3-positive cytoplasmic dots in ATRA and ATO–treated cells compared with untreated cells (Figure 3C-D). We also performed immunofluorescence analysis of untreated and drug-treated cells by using antibodies against the PML protein, which is expected to react with PML/RARA as well as PML expressed from the nonrearranged PML allele. Although the PML-reactive antibodies primarily distributed to the nucleus, a significant portion of PML also was detected within the cytoplasm. Interestingly, a subset of the PML-positive cytoplasmic structures was observed to colocalize with LC3 and p62 both in untreated and ATRA- or ATO-treated cells (Figure 3E-F). Together, these data suggest that ATRA and ATO stimulate autophagy and that PML/RARA expressed in NB4 cells has the potential to contact the autophagy machinery within the cytoplasm.
ATRA and ATO–induced PML/RARA degradation is autophagy-dependent
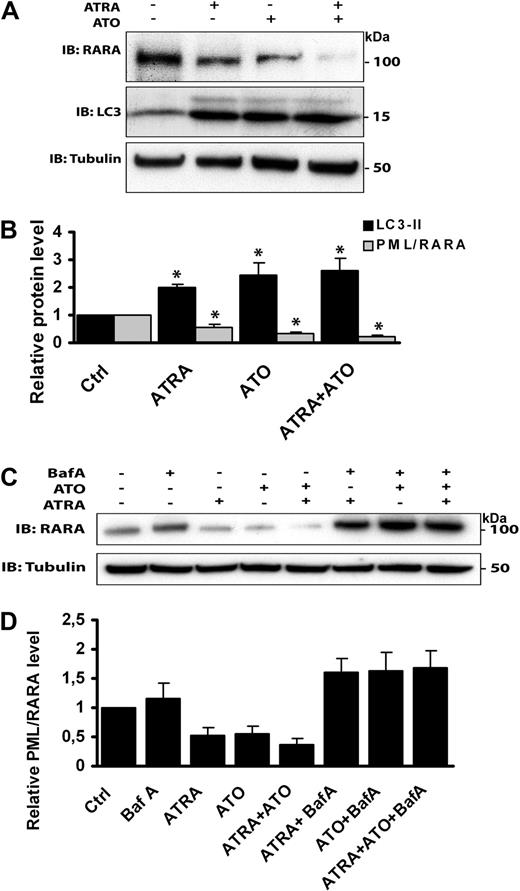
To explore whether ATRA and/or ATO induce clearance of PML/RARA through autophagy, the expression level of this chimeric protein was monitored in NB4 cells incubated in drug-free medium, medium containing 1 of the 2 drugs alone, or medium containing both ATRA and ATO combined. In line with previous studies,5 we found that ATRA and ATO induce degradation of PML/RARA and that the 2 drugs together synergize to stimulate further degradation (Figure 4A-B). The level of autophagy induced by the different drug combinations appeared to be comparable, as determined by LC3-II expression levels (Figure 4A-B). To assess whether autophagy contributes to ATRA and ATO–mediated PML/RARA degradation, control and drug-treated cells were incubated in the presence or absence of Baf A. We found that the ability of ATRA and ATO to induce PML/RARA degradation, both alone and in combination, was considerably reduced in the presence of this autophagy inhibitor (Figure 4C-D), further indicating that these drugs induce clearance of the fusion protein through autophagy.
ATRA and ATO induce autophagy-dependent degradation of PML/RARA. (A) NB4 cells treated with ATRA (1μM) or ATO (1μM) alone or in combination for 24 hours were subjected to SDS-PAGE followed by immunoblotting with anti-RARA, anti-LC3, and antitubulin antibodies. (B) Quantification of the data shown in panel A. Relative protein levels are presented as mean ± SEM from 3 independent experiments. *P < .05, Student t test. (C) Western blot analysis of PML/RARA in NB4 cells treated with the drugs indicated. Incubation of cells in the presence of ATRA (1μM) or ATO (1μM) was performed for 24 hours. Baf A (100nM) was present during the last 12 hours of the incubation. (D) Quantification of the data shown in panel C. Relative PML/RARA protein levels are presented as mean ± SEM from 3 independent experiments.
ATRA and ATO induce autophagy-dependent degradation of PML/RARA. (A) NB4 cells treated with ATRA (1μM) or ATO (1μM) alone or in combination for 24 hours were subjected to SDS-PAGE followed by immunoblotting with anti-RARA, anti-LC3, and antitubulin antibodies. (B) Quantification of the data shown in panel A. Relative protein levels are presented as mean ± SEM from 3 independent experiments. *P < .05, Student t test. (C) Western blot analysis of PML/RARA in NB4 cells treated with the drugs indicated. Incubation of cells in the presence of ATRA (1μM) or ATO (1μM) was performed for 24 hours. Baf A (100nM) was present during the last 12 hours of the incubation. (D) Quantification of the data shown in panel C. Relative PML/RARA protein levels are presented as mean ± SEM from 3 independent experiments.
ATRA and ATO induce autophagy through the mTOR pathway
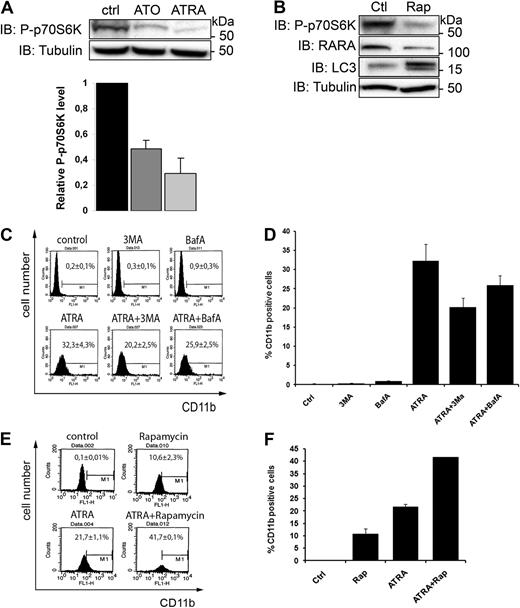
Because we found that degradation of ectopically expressed PML/RARA is sensitive to depletion of ULK1 (Figure 2), a downstream target of mTOR, we determined whether ATRA and ATO–induced autophagy is mTOR-dependent. Under nutrient-rich conditions, mTOR inhibits autophagy by inhibitory phosphorylation of ULK1 and its associated partner Atg13. Upon induction of autophagy by nutrient deprivation, mTOR dissociates from the ULK1 complex, which becomes activated to initiate autophagy.28-30 Furthermore, mTOR-mediated phosphorylation of p70 ribosomal S6 kinase (p70S6K) is prevented, and a reduced level of phosphorylated p70S6K (P-p70S6K) is therefore used as an indicator of mTOR-dependent autophagy.31 Cell extracts from ATRA- or ATO-treated NB4 cells were analyzed by immunoblotting with an antibody against P-p70S6K. Interestingly, the level of phosphorylated p70S6K decreased significantly in both ATRA-treated and ATO-treated cells (Figure 5A), suggesting that ATRA and ATO activate autophagy via an mTOR-dependent pathway. In line with these results we found that treatment of NB4 cells with rapamycin, a drug known to inhibit mTOR and induce autophagy,31 led to increased PML/RARA degradation, induction of autophagy (increased LC3-II levels), and inhibition of p70S6K phosporylation (Figure 5B) but had no effect on cell survival (supplemental Figure 5B). Taken together, our results show that PML/RARA clearance is stimulated upon inhibition of mTOR and that ATO and ATRA–induced PML/RARA degradation correlate with inhibited mTOR activity.
ATRA and ATO induce autophagy through the mTOR pathway. (A) Lysates from NB4 cells treated with or without ATO (1μM) or ATRA (1μM) were analyzed by IB by the use of antibodies against P-p70S6K. Tubulin was used as a loading control. Data from 3 independent experiments were quantified. Data are expressed as the mean ± SEM. (B) Rapamycin inhibits mTOR and induces autophagy and PML/RARA degradation. Lysates from control or rapamycin-treated (200nM for 4 hours) NB4 cells were examined by Western blotting by the use of anti-P-p70S6K, anti-RARA, anti-LC3, or anti-tubulin antibodies. (C-F) Measurement of CD11b by was performed with fluorescence-activated cell sorting. NB4 cells were incubated or not with ATRA (1μM) for 24 hours, then incubated for another 24 hours with 3MA (5mM) or BafA1 (50nM) (C-D) or rapamycin (100nM; E-F) in the presence or absence of ATRA. The cells were then incubated with FITC-conjugated anti-CD11b antibody and analyzed by flow cytometry. (C-D) Representative flow diagrams are shown. The graph represents mean of 4 independent experiments done in duplicate ± SEM. (E-F) Representative flow diagrams are shown. The graph represents mean of 2 independent experiments done in duplicates ± SEM.
ATRA and ATO induce autophagy through the mTOR pathway. (A) Lysates from NB4 cells treated with or without ATO (1μM) or ATRA (1μM) were analyzed by IB by the use of antibodies against P-p70S6K. Tubulin was used as a loading control. Data from 3 independent experiments were quantified. Data are expressed as the mean ± SEM. (B) Rapamycin inhibits mTOR and induces autophagy and PML/RARA degradation. Lysates from control or rapamycin-treated (200nM for 4 hours) NB4 cells were examined by Western blotting by the use of anti-P-p70S6K, anti-RARA, anti-LC3, or anti-tubulin antibodies. (C-F) Measurement of CD11b by was performed with fluorescence-activated cell sorting. NB4 cells were incubated or not with ATRA (1μM) for 24 hours, then incubated for another 24 hours with 3MA (5mM) or BafA1 (50nM) (C-D) or rapamycin (100nM; E-F) in the presence or absence of ATRA. The cells were then incubated with FITC-conjugated anti-CD11b antibody and analyzed by flow cytometry. (C-D) Representative flow diagrams are shown. The graph represents mean of 4 independent experiments done in duplicate ± SEM. (E-F) Representative flow diagrams are shown. The graph represents mean of 2 independent experiments done in duplicates ± SEM.
Autophagy contributes to differentiation of APL cells
Because therapy-induced clearance of the PML/RARA oncoprotein is associated with increased differentiation of APL cells, we wanted to assess whether autophagy may affect maturation of NB4 cells along the granulocyte linage. For this experiment we used a flow cytometry assay that monitors the expression of the granulocyte surface marker protein CD11b. As expected, we detected a considerable increase in CD11b surface expression after incubation of NB4 cells with ATRA for 48 hours (Figure 5C-D). Interestingly, the ATRA-induced CD11b expression was partially inhibited upon incubation of NB4 cells with the autophagy inhibitors 3-MA or Baf A during the last 24 hours of ATRA stimulation (Figure 5C-D). Because the aforementioned experiments demonstrated a role of mTOR in ATRA and ATO–induced autophagy, we also examined whether down-regulation of mTOR activity by rapamycin would affect differentiation of NB4 cells. We found that rapamycin alone was capable of inducing a modest increase of CD11b expression and that this drug potentiated ATRA-mediated CD11b expression (Figure 5E-F). Together, our results indicate that autophagy contributes to the ATRA-induced differentiation of APL cells and may suggest that basal autophagy aids in the process of APL cell differentiation.
Discussion
In the present work we have demonstrated a role of autophagy in degradation of the APL-associated oncoprotein PML/RARA and shown that ATRA and ATO, both drugs that induce clinical remission in APL patients, stimulate mTOR-dependent autophagy and concomitant autophagic degradation of PML/RARA. Further, our study also implicates a function of autophagy in differentiation of APL cells along the granulocyte linage because autophagy inhibitors were found to partially prevent ATRA-stimulated differentiation of NB4 cells and because the mTOR inhibitor rapamycin was found to stimulate differentiation both in the absence and in the presence of ATRA.
The PML/RARA oncoprotein is known to be prone to aggregation, a feature that makes it a good substrate for autophagic degradation. The aggregation-prone characteristics of PML/RARA are further substantiated by our data showing that this protein chimera is considerably more effectively extracted by buffers containing the protein denaturation reagent UREA compared with buffers containing the detergents SDS or Triton X-100. Its ability to form protein aggregates is possibly attributable to the PML moiety of the fusion protein because PML contains a tripartite motif domain that together with SUMO-conjugated residues and a SUMO-interacting domain facilitates the formation of protein-protein interaction networks.32,33 Further, the PML/RARA chimera may adapt a different (and possibly misfolded) conformation that renders it more aggregation prone than PML or RARA expressed from nonrearranged genes. Because of its highly insoluble conformation, the PML/RARA oncoprotein may be subjected to clearance by the same or similar mechanisms that degrades other types of misfolded or aggregation-prone proteins in the cell, including those responsible for causing neurodegenerative diseases such as Alzheimer and Huntington disease.34 Consistent with this notion is our observation that cytoplasmic PML/RARA-containing particles formed by ectopically expressed PML/RARA, as well as cytoplasmic PML-positive structures in NB4 cells, were found to colocalize with ubiquitin, p62 and LC3, proteins known to be involved in autophagy-dependent degradation of protein aggregates. p62 contains an ubiquitin-binding UBA domain and an LC3-interacting region and is therefore able to link ubiquitinated cargo to the core autophagic machinery.25,35 Thus, it will be important to elucidate whether ubiquitin is a signal for autophagic degradation of PML/RARA.
Because both PML and RARA represent proteins that are targeted primarily to the nucleus, and because the PML/RARA chimera is detected mainly within the nucleus of interphase cells, therapy-induced degradation of PML/RARA is largely thought to occur within the nuclear compartment. This notion is also supported by studies in which the authors demonstrated that PML/RARA, together with ubiqutin and components of the UPS, become recruited to PML NBs during the first few hours of ATO treatment.6,36 However, because autophagy represents a cytoplasmic degradation mechanism, our results indicate that proteolytic clearance of this oncoprotein also can take place in the cytoplasm. Consistent with this notion is the localization of PML/RARA to cytoplasmic structures, seen in this as well as in previous studies, both in NB4 cells and in primary APL blasts.37-41 Interestingly, a cytoplasmic compartment referred to as cytoplasmic assemblies of PML and nucleoporins, which appear after mitotic cell division and contain PML or PML/RARA, was found to be cleared from NB4 cells in the presence of ATRA, suggesting a possible link between the stability of these cytoplasmic structures and autophagic activity.40
Our results showing the involvement of the mTOR pathway in the autophagy-mediated clearance of PML/RARA is supported by a recent study in which the authors demonstrated that ATRA-induced growth arrest and differentiation of acute myelogenous leukemia cells is mTOR dependent and that mTOR inhibitors further potentiates the effects of ATRA.42 It has further been shown that this effect of ATRA is mediated through the stress response gene RTP801, an early RA target gene in myeloid cells, which acts as a negative regulator of the mTOR pathway.42,43 Further, autophagic death of acute lymphoblastic lymphoma cells treated with dexamethasone was recently shown to be dependent on the PML protein, which is known to represent an inhibitor of mTOR activity.44,45 Thus, mTOR-regulated autophagy may play a general role in development and treatment of leukemia.
Because the authors of previous studies have implicated a major role of the UPS in ATRA and ATO–mediated clearance of PML/RARA6,19,46 autophagy and proteasome dependent degradation may cooperate in therapy induced clearance of the APL-associated oncoprotein. Consistent with this finding, some proteolytic cross-talk clearly exists between autophagy and the UPS.47 Furthermore, proteolytic degradation of PML/RARA by caspases,48 neutrophil elastase,49 and lysosomal proteases50 also has been reported, suggesting the existence of multiple proteolytic pathways with a potential to target PML/RARA for degradation. However, because autophagy appears to represent a pathway that becomes markedly induced in the presence of ATRA and ATO, we propose that this degradation pathway may have a determinant role in therapy-induced PML/RARA clearance. Most importantly, because PML/RARA proteolysis appears to be critical for APL remission, our data may provide new options for therapy-resistant patients because drugs known to induce autophagy already are used in the clinic.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Soheil Naderi and Martine Kloster for help with the flow cytometry experiments and Heidi Kiil Blomhoff for critical reading of the manuscript.
This work was supported by grants from the Research Council of Norway (FUGE and Nevronor) and the Norwegian Cancer Society.
Authorship
Contribution: P.I. was responsible for most of the experimental work and created the figures; M.B. was involved in data analysis; and S.O.B. and A.S., with the help of the other authors, were involved in project planning, data analysis, and writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Anne Simonsen, Department of Biochemistry, Institute of Basic medical Sciences, University of Oslo, PB 1112 Blindern, 0317 Oslo, Norway; e-mail: anne.simonsen@medisin.uio.no.
References
Author notes
S.O.B. and A.S. contributed equally to this work.