Abstract
Immunophenotyping by flow cytometry or immunohistochemistry is a clinical standard procedure for diagnosis, classification, and monitoring of hematologic malignancies. Antibody-based cell surface phenotyping is commonly limited to cell surface proteins for which specific antibodies are available and the number of parallel measurements is limited. The resulting limited knowledge about cell surface protein markers hampers early clinical diagnosis and subclassification of hematologic malignancies. Here, we describe the mass spectrometry based phenotyping of 2 all-trans retinoic acid treated acute myeloid leukemia model systems at an unprecedented level to a depth of more than 500 membrane proteins, including 137 bona fide cell surface exposed CD proteins. This extensive view of the leukemia surface proteome was achieved by developing and applying new implementations of the Cell Surface Capturing (CSC) technology. Bioinformatic and hierarchical cluster analysis showed that the applied strategy reliably revealed known differentiation-induced abundance changes of cell surface proteins in HL60 and NB4 cells and it also identified cell surface proteins with very little prior information. The extensive and quantitative analysis of the cell surface protein landscape from a systems biology perspective will be most useful in the clinic for the improved subclassification of hematologic malignancies and the identification of new drug targets.
Introduction
Current diagnosis of hematologic malignancies relies on a combination of the immunophenotype, morphology, cytochemistry, and karyotype according to the World Health Organization classification1 and the previously used French-American-British (FAB) classification.2,3 The current World Health Organization classification divides neoplasms according to their lineage into myeloid neoplasms, lymphoid neoplasms, histiocytic neoplasms, and mast cell disorders. As a neoplasm of the myeloid lineage, acute myeloid leukemia (AML) is one of the most frequently diagnosed types of leukemia in adults. Acute promyelocytic leukemia (APL) is a distinct subtype of AML and it seems to be the most malignant form of AML characterized by a life threatening coagulopathy.4 APL is cytogenetically characterized by a specific reciprocal translocation t(15;17), which results in the fusion between the promyelocytic leukemia gene and the retinoic acid receptor α gene. All-trans retinoic acid (ATRA) binds to retinoic acid receptors and it induces differentiation of APL cells into granulocyte-like cells. Since the 1980s, ATRA has been successfully applied in the clinic and the combination of ATRA and anthracycline-based chemotherapy is highly effective to induce complete remission in APL patients.5 However, approximately 25% of APL patients treated with ATRA suffer from the differentiation syndrome.6 The differentiation syndrome is characterized by increased leukocyte transmigration, for which cell surface proteins play a crucial role.7
Cell surface protein markers are commonly used in clinical immunohistochemistry and flow cytometry to immunophenotype hematologic malignancies, such as AML. The process of immunophenotyping malignant cells implies the availability of affinity reagents, such as antibody-based probes. However, for the detection of cell surface exposed proteins, only a limited number of specific anti-CD antibodies are available for consistent, clinical-grade immunophenotyping. The CD nomenclature for cell surface molecules was devised in 1984 by the Human Leukocyte Differentiation Antigen (HLDA) Workshop to better characterize and study cell surface molecules. The HLDA Workshops have up to now assigned 350 CD numbers to cell surface molecules, against which 2 independent antibodies are available.8 For example, to distinguish myeloid from lymphoid neoplasms, monoclonal antibodies against the pan-myeloid cell surface markers CD13, CD33, and CD65 are used.9 Although a few hundred anti-CD antibodies are available, fewer than 50 anti-CD antibodies are used to immunophenotype hematologic neoplasms by flow cytometry.10
Antibody arrays are a promising technology toward a more complete immunophenotype of cells, since they allow the concurrent determination of 130 different surface antigens.11 However, all antibody-based technologies are limited to the number of available antibodies and therefore they are not suitable for the discovery of new cell surface markers. Mainly due to the lack of suitable affinity reagents, the cell surface proteome of most cells, including AML cells, is not well known. Several thousand proteins of the human proteome are predicted to contain a transmembrane segment12 and many of these transmembrane proteins are expected to be exposed on the cell surface of different cell types, influencing the communication potential of these cell types with their particular microenvironments.
The currently limited knowledge about the cell surface protein phenotype of malignant cells hampers the precise clinical disease subclassification on the protein level, the predictive management of the disease, and the selection of therapeutic target proteins.10 To predict and efficiently treat hematologic malignancies, it is necessary to understand cellular behavior on the protein level and an essential step is to gain a qualitative and quantitative overview of the cell surface proteome. To reach these goals, techniques supporting an extensive analysis of the cell surface proteome are required.
Mass spectrometry (MS) based proteomics enables the multiplexed identification and quantification of peptides in a discovery-driven analysis of the proteome.13 The analysis of the cell surface proteome by MS is hampered by factors, such as the hydrophobicity of transmembrane proteins and the low relative abundance of cell surface proteins compared with intracellular proteins.14 We recently circumvented most of these issues by developing the MS-based CSC technology,15 which enables the reproducible identification and quantification of cell surface proteins.16 The CSC technology is based on hydrazide chemistry and it enables the specific enrichment of glycopeptides through the biotinylation of glycans of cell surface proteins.
Here, we applied a range of complementary CSC technology variants selecting overlapping segments of the cell surface proteome for the multiplexed identification and relative quantification of AML cell surface proteins upon ATRA stimulation. Using HL60 and NB4 cells as differentiation model systems, we identified more than 500 membrane proteins, including 137 CD annotated proteins and detected distinct molecular phenotypes of drug-induced differentiation processes independent of antibodies.
Methods
Cell culture
HL60 and NB4 cells were kindly provided by Dr Juerg Gertsch (Institute of Pharmaceutical Sciences, ETH Zurich, Switzerland) and Dr Udo Doebbeling (Department of Dermatology, University Hospital Zurich, Switzerland), respectively. HL60 and NB4 cells were cultured in RPMI 1640 (10% fetal calf serum, 100 units/mL penicillin, and 100 μg/mL streptomycin; Sigma-Aldrich). For ATRA stimulation experiments, cells were cultured for at least 6 cell doublings in RPMI 1640 containing 10% dialyzed fetal calf serum (BioConcept) and either L-arginine and L-lysine (light medium; Sigma-Aldrich) or L-[13C6,15N4]arginine and L-[13C6,15N2]lysine (heavy medium; Sigma-Aldrich). HL60 and NB4 cells were stimulated by adding 1μM ATRA (Sigma-Aldrich) for 4 days. Three independent ATRA stimulation experiments were carried out for each AML cell line and the Cys-Glyco-CSC, Glyco-CSC, and Lys-CSC technology variant were applied in each experiment. In 2 experiments of each cell line, cells cultured in heavy medium were stimulated with ATRA and cells cultured in light medium were unstimulated. In 1 experiment of each cell line, cells cultured in light medium were stimulated with ATRA and cells cultured in heavy medium were unstimulated.
CSC technology and sample preparation
Equal cell numbers (∼ 108 cells) of ATRA stimulated and unstimulated cells that were cultured in light and heavy medium, respectively, were used for 1 CSC experiment. Using variants of the original CSC technology, 3 partially overlapping cell surface subproteomes consisting of glycoproteins, cysteine containing glycoproteins, and lysine containing proteins, respectively, were selectively isolated and analyzed by MS.
Cys-Glyco-CSC/Glyco-CSC
The protocol for the isolation of cysteine containing peptides and N-linked glycopeptides was adapted from the initial CSC protocol.15 Briefly, cells were treated for 15 minutes at 4°C in the dark with 2mM sodium meta-periodate (Pierce) in phosphate-buffered saline (PBS) pH 6.5. Afterward, cells were washed and incubated with 6.5mM biocytin hydrazide (Biotium) in PBS pH 6.5 for 60 minutes to biotinylate oxidized carbohydrates of cell surface glycoproteins. Then, cells were washed again and incubated on ice in hypotonic lysis buffer (10mM Tris pH 7.5, 0.5mM MgCl2, and 10mM iodoacetamide) for 10 minutes. Cells were homogenized with forty strokes using a Dounce homogenizer. Afterward, cell debris and nuclei were removed by centrifugation at 1700g for 7 minutes and the supernatant was centrifuged again in an ultracentrifuge at 35 000 rpm (∼ 150 000g) for 1 hour to generate a membrane pellet. The membrane pellet was solubilized in 400 μL of digestion buffer (100mM ammonium bicarbonate, 1mM iodoacetamide, 1mM 2,2′-thiodiethanol, and 0.1% RapiGest; Waters) by indirect sonication with a VialTweeter (Hielscher). Proteins were digested overnight with trypsin in a protease:protein ratio of 1:100. Iodoacetamide was added to the digestion buffer to preserve protein disulfide bridges and 2,2′-thiodiethanol was added to prevent overalkylation of peptides.17 After protein digestion, the peptide mixture was heated at 95°C for 10 minutes to inactivate trypsin and then biotinylated glycopeptides were bound to Streptavidin Plus UltraLink Resin (SA beads; Pierce). After extensive washing, cysteine containing peptides, which were bound via a disulfide bridge to the biotinylated glycopeptides, were eluted from the SA beads by incubation with elution buffer (100mM ammonium bicarbonate, 10mM TCEP, and 1mM DTT) for 1h at room temperature (Cys-Glyco-CSC). Afterward, SA beads were washed again and in a second elution step N-linked glycopeptides were enzymatically released from the SA beads overnight by PNGase F (Glyco-CSC). Produced free thiols of cysteine containing peptides were alkylated with iodoacetamide. Peptides were desalted on Ultra MicroTIP Columns (The Nest Group) and dried in a SpeedVac concentrator. Finally, peptides were solubilized in LC-MS grade water containing 0.1% formic acid and 5% acetonitrile.
Lys-CSC
The Lys-CSC protocol differed from the above described Cys-Glyco-CSC/Glyco-CSC protocol only in few steps: biotinylation of cell surface proteins was carried out in HL60 cells by incubation with 2mM sulfosuccinimidyl-2-(biotinamido)-ethyl-1, 3′-dithiopropionate (sulfo-NHS-SS-biotin; Pierce) and in NB4 cells by incubation with 2mM sulfosuccinimidyl-6-(biotinamido) hexanoate (sulfo-NHS-LC-biotin; Pierce) for 30 minutes at 4°C on a MACSmix tube rotator (Miltenyi Biotec), a sucrose gradient centrifugation step was used for the enrichment of membranes as described elsewhere,18 and N-glycosylated proteins were deglycosylated by PNGase F for 2 hours at room temperature before protein digestion. The samples generated by CSC technology variants were stored frozen and analyzed individually.
MS analysis
Each peptide sample generated from HL60 cells was analyzed twice on an Eksigent Nano LC system (Eksigent Technologies) connected to a hybrid linear ion trap LTQ Orbitrap (Thermo Scientific), which was equipped with a nanoelectrospray ion source (Thermo Scientific). Peptide separation was carried out on a RP-HPLC column (75 μm inner diameter and 10 cm length) packed in-house with C18 resin (Magic C18 AQ 3 μm; Michrom Bioresources) using a linear gradient from 90% solvent A (water, 0.2% formic acid, and 1% acetonitrile) and 10% solvent B (water, 0.2% formic acid, and 80% acetonitrile) to 65% solvent A and 35% solvent B over 39 minutes at a flow rate of 0.2 μL/minute. The data acquisition mode was set to acquire 1 high resolution MS scan in the ICR cell followed by 3 collision induced dissociation MS/MS scans in the linear ion trap. For a high resolution MS scan, 2 × 106 ions were accumulated over a maximum time of 400 ms and the FWHM resolution was set to 60 000 (at m/z 300). Only MS signals exceeding 500 ion counts triggered a MS/MS attempt and 104 ions were acquired for a MS/MS scan over a maximum time of 200 ms. The normalized collision energy was set to 35%. Singly charged ions were excluded from triggering MS/MS scans. Each peptide sample generated from NB4 cells was analyzed similarly (for details see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the article).
MS data analysis
Raw data files from the MS instruments were converted with ReAdW into mzXML files19 and mzXML files were searched with Sorcerer-SEQUEST20 (for details see supplemental Methods) against a concatenated protein database combining the human protein database of the UniProtKB/Swiss-Prot Protein Knowledgebase (Version 56.9), the reversed sequences of all proteins, and common contaminants (41 104 protein entries). Statistical analysis of each search result for each LC-MS analysis was performed using the Trans-Proteomic Pipeline TPP21 v4.0 JETSTREAM rev 2 including PeptideProphet22 and ProteinProphet.23 The ProteinProphet probability score was set to 0.9, which resulted in an average protein false discovery rate of less than 1% for all search results estimated by ProteinProphet. Proteins, which were identified with only single peptide identifications in the whole dataset, were excluded from subsequent analyses. If peptide identifications led to an indistinguishable group of proteins, the alphabetically first UniProtKB/Swiss-Prot AC number was used for subsequent analyses and the remaining protein identifiers of the protein group were listed in supplemental tables. For the final dataset, a protein false discovery rate of less than 1% and a peptide false discovery rate of less than 1% were estimated by the MAYU software.24 CD numbers were mapped to UniProtKB/Swiss-Prot IDs according to the UniProtKB/Swiss-Prot Protein Knowledgebase cross-reference table (Version 57.0). Transmembrane helices of proteins were predicted with 2 algorithms, the TMHMM algorithm25 (Version 2.0) and the Phobius algorithm.26 Protein identifications were denoted as membrane proteins, if at least 1 transmembrane helix algorithm predicted 1 or more transmembrane helices for the protein sequences. Furthermore, proteins, which contained a lipid moiety-binding region according to the UniProtKB/Swiss-Prot Protein Knowledgebase, were also denoted as membrane proteins.
To quantify protein abundance changes between ATRA stimulated and unstimulated AML cells, identical Sorcerer-SEQUEST search criteria and TPP settings were applied as above, with the exception of 2 required tryptic termini per peptide. Proteins, which were not identified in the database search with at least 1 tryptic terminus per peptide and which were only identified with single peptide identifications in the database search with 2 tryptic termini per peptide, were excluded from subsequent analyses. Protein ratios of ATRA stimulated to unstimulated cells were determined for each LC-MS analysis by applying the XPRESS software27 and protein ratios were normalized by the mean protein ratio of each experiment. The mean protein ratio of an experiment was determined by a Gaussian curve fit to the histogram of all log10 protein ratios of the experiment. For the comparison of ATRA stimulated cells to unstimulated cells, the average log2 XPRESS protein ratios were calculated over the 2 LC-MS analyses of each CSC technology variant of each experiment, then the average values were calculated over all CSC technology variants of each experiment, and then the average values were calculated over all 3 experiments for each AML cell line. The sequence coverage of identified proteins was determined with the Protein Coverage Summarizer (Version 1.2.3365), which was downloaded from the website of the Biologic MS Data and Software Distribution Center of the Pacific Northwest National Laboratory.
Molecular functions of identified proteins were assigned by the PANTHER database (Version 6.1.1) based on the alphabetically first UniProtKB/Swiss-Prot AC number. Hierarchical cluster analysis was carried out with the Spotfire DecisionSite 9.1.1 software (TIBCO Software Inc) based on average log2 XPRESS protein ratios (log2 protein ratios) of quantified membrane proteins. For the hierarchical cluster analysis, log2 protein ratios above 3 or below −3 were trimmed to 3 and −3, respectively. Membrane proteins were excluded from the hierarchical cluster analysis, if the trimmed log2 protein ratios differed by more than 2 between HL60 and NB4 cells. Unweighted pair-group method with arithmetic mean was chosen for clustering method and Euclidean distance for similarity measure.
Flow cytometry
CD proteins in HL60 and NB4 cells were analyzed by direct immunofluorescence. First, cells were washed with PBS and then incubated for 30 minutes at 4°C in PBS containing 10% human serum to block Fc-receptors. After subsequent washing, 400 000 cells were incubated for 20 minutes at 4°C in 25 μL of a 1:12.5 dilution of anti-CD11b-FITC (Sigma-Aldrich), anti-CD54–PE (BD Biosciences), anti-CD55-FITC (Abcam) or anti-CD71-FITC (BD Biosciences) antibody. For each antibody staining, 10 000 cells were analyzed on a FACSCalibur System (BD Biosciences) and FlowJo7 (TreeStar) was used for data analysis. ATRA stimulated AML cells showed consistently in both fluorescence channels (FL-1 and FL-2) a slightly increased fluorescence intensity (FI) compared with unstimulated cells. Therefore, the geometric mean FI values of the antibody stainings of ATRA stimulated cells were corrected by subtracting the difference between the geometric mean FI values of unstained, ATRA stimulated control cells and unstained, unstimulated control cells. For each antibody staining in each experiment, the log2 ratio of the geometric mean FI values of ATRA stimulated to unstimulated cells was calculated and then the average value over all experiments was calculated for each antibody staining for each cell line.
Results
Cellular differentiation of AML cells
HL60 and NB4 cells are well characterized in vitro model systems for AML and myeloid differentiation.28,29 Only NB4 cells carry the specific reciprocal translocation t(15;17), the hallmark of APL. Although HL60 and NB4 cells carry different genetic aberrations, ATRA induces differentiation of both AML cell lines into morphologically mature granulocytes.29,30 To relatively quantify protein abundance changes of ATRA stimulated compared with unstimulated AML cells by MS, both cell lines were cultured in SILAC medium containing isotope labeled amino acids.31 AML cells were stimulated with 1μM ATRA for 4 days and cellular differentiation was controlled by flow cytometric analysis of CD11b.29,32
Complementary CSC technology variants
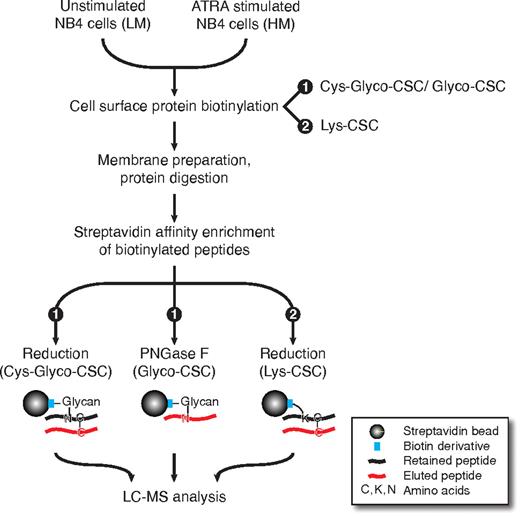
We adapted and expanded the original CSC glycopeptide selection technology15 to increase the sensitivity and the range of captured peptides. Here, we used complementary CSC technology variants (Figure 1) for the first time to identify and quantify the cell surface proteome of AML cells after ATRA stimulation. Equal cell numbers (∼ 108 cells) of ATRA stimulated and unstimulated AML cells were combined and cell surface exposed proteins were chemoselectively biotin-tagged either by the Cys-Glyco-CSC/Glyco-CSC or the Lys-CSC approach. With the Cys-Glyco-CSC and Glyco-CSC technology variant, N-linked glycopeptides (Glyco-CSC) and cysteine containing peptides (Cys-Glyco-CSC) bound via disulfide bridges to cysteine containing glycopeptides were concurrently identified. Furthermore, we used amine-reactive biotin derivatives to tag cell surface proteins (Lys-CSC).34 After tagging of cell surface proteins, cell membranes were isolated and proteins were digested with trypsin. Biotinylated peptides were then isolated by affinity chromatography with streptavidin beads. Cysteine containing peptides bound via disulfide bridges to biotinylated peptides were eluted from the streptavidin beads by chemical reduction. These cysteine containing peptides were coined “piggyback” peptides, since their enrichment was mediated by the biotinylated peptides. Biotinylated N-glycosylated peptides were enzymatically released from streptavidin beads by PNGase F. Finally, peptides were analyzed by LC-MS.
CSC technology. Unstimulated and ATRA stimulated NB4 cells were cultured in light medium (LM) and heavy medium (HM), respectively. Equal cell numbers of unstimulated and ATRA stimulated NB4 cells were combined and cell surface proteins were biotinylated on living cells either by the Cys-Glyco-CSC/Glyco-CSC protocol or the Lys-CSC protocol. After membrane preparation and protein digestion, biotinylated peptides were isolated by streptavidin affinity chromatography. “Piggyback” peptides were bound via disulfide bridges to biotinylated peptides on streptavidin beads, because protein disulfide bridges had been protected during cell lysis and protein digestion. In the Cys-Glyco-CSC and Lys-CSC protocol, piggyback peptides were eluted from streptavidin beads by chemical reduction. In the Glyco-CSC protocol, peptides with N-glycosylation were enzymatically released from streptavidin beads by PNGase F. HL60 cells were processed analogously except for the Lys-CSC protein biotinylation (for details see “Lys-CSC”).
CSC technology. Unstimulated and ATRA stimulated NB4 cells were cultured in light medium (LM) and heavy medium (HM), respectively. Equal cell numbers of unstimulated and ATRA stimulated NB4 cells were combined and cell surface proteins were biotinylated on living cells either by the Cys-Glyco-CSC/Glyco-CSC protocol or the Lys-CSC protocol. After membrane preparation and protein digestion, biotinylated peptides were isolated by streptavidin affinity chromatography. “Piggyback” peptides were bound via disulfide bridges to biotinylated peptides on streptavidin beads, because protein disulfide bridges had been protected during cell lysis and protein digestion. In the Cys-Glyco-CSC and Lys-CSC protocol, piggyback peptides were eluted from streptavidin beads by chemical reduction. In the Glyco-CSC protocol, peptides with N-glycosylation were enzymatically released from streptavidin beads by PNGase F. HL60 cells were processed analogously except for the Lys-CSC protein biotinylation (for details see “Lys-CSC”).
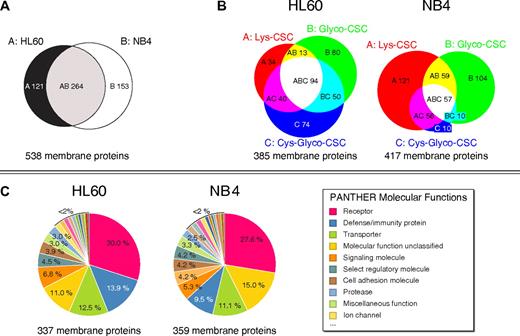
Overall, these LC-MS measurements led to the identification of more than 500 membrane proteins, including 137 CD annotated cell surface proteins (Table 1 and supplemental Tables 1-3). Nearly half of the identified membrane proteins were shared between both AML cell lines (Figure 2A). Furthermore, 188 and 235 membrane proteins were identified in HL60 and NB4 cells, respectively, with only 1 CSC technology variant showing that the 3 enrichment strategies were complementary to extensively analyze the cell surface proteome (Figure 2B). In addition, if distinct peptides of a protein were identified with different CSC technology variants, confidence of correct protein identification was further increased. Because different amine-reactive biotin derivatives were used (for details see “Ly-CSC”), it was not possible to directly compare the distribution of identified membrane proteins in the 2 AML cell lines. However, similar functional protein classes were found in the PANTHER database35 for the identified membrane proteins in both AML cell lines (Figure 2C; supplemental Table 4), including receptor proteins, cell adhesion proteins, signaling proteins, and transporters. Furthermore, identified peptides showed a high specificity of up to more than 90% in LC-MS analyses (supplemental Table 5). Specific peptides for the Cys-Glyco-CSC and Lys-CSC technology variant contained cysteine residues. Specific peptides for the Glyco-CSC technology variant contained deamidated glycosylation motives (NXS/T → DXS/T; N is asparagine, X is any amino acid except proline, S/T is either serine or threonine, and D is aspartic acid) due to the PNGase F treatment.
Mapping the cell surface proteome through complementary CSC technology variants. (A) Overlap of identified membrane proteins in AML cells. (B) Area-proportional Venn diagrams33 showing overlapping membrane protein identifications with complementary CSC technology variants in HL60 cells and NB4 cells, respectively. (C) PANTHER molecular functions of identified membrane proteins in AML cells.
Mapping the cell surface proteome through complementary CSC technology variants. (A) Overlap of identified membrane proteins in AML cells. (B) Area-proportional Venn diagrams33 showing overlapping membrane protein identifications with complementary CSC technology variants in HL60 cells and NB4 cells, respectively. (C) PANTHER molecular functions of identified membrane proteins in AML cells.
Multiplexed proteomic CD phenotyping
Immunophenotyping of hematologic malignancies has traditionally depended on well characterized monoclonal antibodies and it is commonly restricted to few antibodies. In contrast, the data generated with MS-based CSC technology variants contained 137 CD proteins and therefore phenotyped ATRA stimulated AML cells at an unprecedented level (Figure 3). Identified CD proteins included the hematopoietic cell surface marker CD45 as well as pan-myeloid markers such as CD13 and CD33. Well known granulocyte differentiation markers such as CD11b, CD11c, CD35, and CD66a were also identified.36 More than 75% of the CD proteins were identified with more than 10 assigned spectra per CD protein in at least 1 AML cell line, demonstrating the robustness of cell surface phenotyping with the CSC technology.
Extensive CD phenotyping of AML cells. Overview on 137 CD proteins identified in AML cells by the MS-based CSC technology. The inner color code of the squares visualizes the number of assigned spectra per CD protein for HL60 cells (top squares) and NB4 cells (bottom squares), respectively.
Extensive CD phenotyping of AML cells. Overview on 137 CD proteins identified in AML cells by the MS-based CSC technology. The inner color code of the squares visualizes the number of assigned spectra per CD protein for HL60 cells (top squares) and NB4 cells (bottom squares), respectively.
CSC technology versus flow cytometry
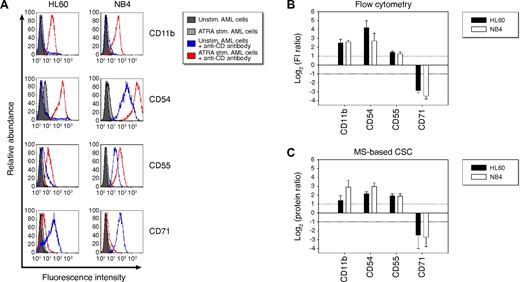
To compare the accuracy of MS-based quantification of protein abundance changes by the CSC technology with that obtained by flow cytometry, we analyzed CD11b, CD54, CD55, and CD71 by flow cytometry in ATRA stimulated and unstimulated AML cells (Figure 4). In the flow cytometric analysis, the average log2 FI ratios of CD proteins were calculated with the geometric mean FI values of ATRA stimulated and unstimulated cells (Figure 4B). The flow cytometric analysis indicated a significant increase in CD11b, CD54, and CD55 protein abundances and a sharp decrease of CD71 protein abundances in both AML cell lines after ATRA stimulation. In the MS-based CSC analysis, protein abundance changes were quantified by applying the XPRESS software27 to the generated MS datasets (supplemental Tables 6-7). Based on peptide identifications, the XPRESS software isolated chromatographic elution profiles of the unlabeled and the heavy isotope labeled peptides, respectively, and determined the area of each peptide peak. XPRESS protein ratios were then calculated by the geometric mean of the ratios of corresponding peptide peak areas. In agreement with the flow cytometric analysis, log2 protein ratios of ATRA stimulated to unstimulated AML cells indicated a significant increase in CD11b, CD54, and CD55 protein abundances and a sharp decrease of CD71 protein abundances in both AML cell lines after ATRA stimulation (Figure 4C).
Flow cytometric analysis and MS-based CSC analysis of CD protein abundance changes in AML cells after ATRA stimulation. (A) Representative flow cytometric analysis of CD proteins in unstimulated (blue line) and ATRA stimulated (red line) AML cells. Unstimulated and ATRA stimulated control AML cells are shown in plain dark gray and light gray, respectively. (B) Results of the flow cytometric analysis are shown as the average log2 ratios of the geometric mean FI signals of ATRA stimulated to unstimulated AML cells (n = 3). Error bars represent SDs. (C) Results of the MS-based CSC analysis are shown as the average log2 protein ratios of ATRA stimulated to unstimulated AML cells (n = 3). Error bars represent SDs.
Flow cytometric analysis and MS-based CSC analysis of CD protein abundance changes in AML cells after ATRA stimulation. (A) Representative flow cytometric analysis of CD proteins in unstimulated (blue line) and ATRA stimulated (red line) AML cells. Unstimulated and ATRA stimulated control AML cells are shown in plain dark gray and light gray, respectively. (B) Results of the flow cytometric analysis are shown as the average log2 ratios of the geometric mean FI signals of ATRA stimulated to unstimulated AML cells (n = 3). Error bars represent SDs. (C) Results of the MS-based CSC analysis are shown as the average log2 protein ratios of ATRA stimulated to unstimulated AML cells (n = 3). Error bars represent SDs.
ATRA regulated membrane proteins
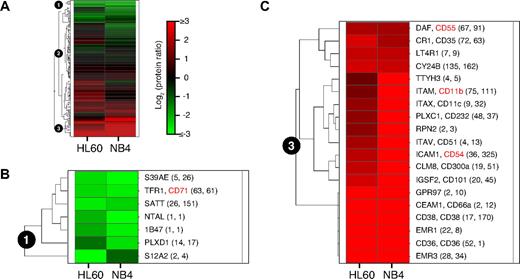
To identify commonly regulated membrane proteins after ATRA induced cellular differentiation of AML cells, hierarchical cluster analysis was applied to the log2 protein ratios of identified membrane proteins in both AML cell lines (Figure 5; supplemental Table 8). The hierarchical cluster analysis was restricted to the 165 membrane proteins that fulfilled the following criteria: firstly, the proteins had to be identified in both AML cell lines; secondly, the proteins had to be quantified with a similar log2 protein ratio of ATRA stimulated to unstimulated cells in both cell lines (for details see “MS data analysis”). 3 functional protein clusters were defined. Protein cluster number 1 (Figure 5B) contained membrane proteins, which showed decreased protein abundances in both AML cell lines after ATRA stimulation. The ATRA induced stop of cell proliferation of AML cells is known to be characterized by decreased transferrin receptor (CD71, TFR1) abundance.30 Consistent with this expectation and as described above, decreased protein abundances of CD71 were detected after ATRA stimulation by the MS-based CSC technology in both AML cell lines. In addition, several other membrane proteins were detected that showed decreased protein abundances after ATRA stimulation, such as the zinc transporter (S39AE), the amino acid transporter (SATT), and plexin D1 (PLXD1). Protein cluster number 2 contained 139 membrane proteins, which showed either no change or a differential regulation of protein abundances in the 2 AML cell lines. Finally, protein cluster number 3 contained membrane proteins, which showed increased protein abundances in both AML cell lines after ATRA stimulation (Figure 5C). Proteins in cluster number 3 included well known markers of granulocytic differentiation, such as CD66a, CD300a, CD11c, CD11b, CD35 and CD55.36,37 EMR3 and EMR1, which were described as granulocyte markers,38,39 were also identified with highly increased protein abundances after ATRA stimulation. Furthermore, cell surface proteins were identified, which are known to show increased protein abundances after ATRA stimulation, such as CD36, CD38, CD54, and CY24B.40-44 Interestingly, cluster number 3 also included proteins with very little prior information in the literature about their functional role in AML cells, such as GPR97 and TTYH3.
Membrane protein abundance changes in AML cells after ATRA stimulation. (A) Hierarchical cluster analysis of log2 protein ratios of 165 membrane proteins that were identified in both AML cell lines by the MS-based CSC technology. Each row represents the abundance changes of 1 membrane protein after ATRA stimulation and each column represents 1 AML cell line. Red indicates increased protein abundance, while green indicates decreased protein abundance after ATRA stimulation. (B) Protein cluster number 1 contained membrane proteins, which showed decreased protein abundances in both AML cell lines after ATRA stimulation. Names of proteins are shown next to the heat map and if applicable CD numbers are included. Numbers in brackets are the number of peptide identifications used for quantification of the protein in HL60 (first number) and in NB4 cells (second number), respectively. (C) Protein cluster number 3 contained membrane proteins, which showed increased protein abundances in both AML cell lines after ATRA stimulation.
Membrane protein abundance changes in AML cells after ATRA stimulation. (A) Hierarchical cluster analysis of log2 protein ratios of 165 membrane proteins that were identified in both AML cell lines by the MS-based CSC technology. Each row represents the abundance changes of 1 membrane protein after ATRA stimulation and each column represents 1 AML cell line. Red indicates increased protein abundance, while green indicates decreased protein abundance after ATRA stimulation. (B) Protein cluster number 1 contained membrane proteins, which showed decreased protein abundances in both AML cell lines after ATRA stimulation. Names of proteins are shown next to the heat map and if applicable CD numbers are included. Numbers in brackets are the number of peptide identifications used for quantification of the protein in HL60 (first number) and in NB4 cells (second number), respectively. (C) Protein cluster number 3 contained membrane proteins, which showed increased protein abundances in both AML cell lines after ATRA stimulation.
These results show that major common cell surface proteome changes in HL60 and NB4 cells upon ATRA induced differentiation were captured with the CSC technology. In addition to commonly regulated membrane proteins after ATRA stimulation, 30 quantified membrane proteins showed in the 2 AML cell lines differences between the log2 protein ratios of more than 2 (supplemental Table 7) and 274 membrane proteins were only identified in 1 cell line (Figure 2A; supplemental Table 2). Because only NB4 cells carry the specific reciprocal translocation t(15;17), HL60 and NB4 cells belong to different FAB categories, FAB-M2 and FAB-M3, respectively. Thus, the detected differences between the 2 AML cell lines in membrane protein regulation after ATRA stimulation were most likely due to different genetic backgrounds and different genetic aberrations between the cell lines.29,30
Discussion
Cell surface proteins are readily accessible and well suited as diagnostic and therapeutic targets. Extensive classification of malignancies based on cell surface proteins is often hampered by the lack of known disease specific cell surface markers and the lack of well characterized antibodies against these markers. Here, we present extensive cell surface molecular phenotypes and an extensive analysis of cell surface protein dynamics in ATRA stimulated AML cells using a MS-based strategy termed CSC technology. By enriching peptides from extracellular domains of cell surface proteins, the CSC technology allowed a multiplexed identification and quantification of hundreds of membrane proteins in AML cells. We demonstrate that complementary CSC technology variants were able to efficiently enrich for glycopeptides and cysteine containing piggyback peptides. The Glyco-CSC technology variant not only identified N-linked glycopeptides but also indicated locations of glycosylation sites of identified peptides. Through deamidated NXS/T glycosylation motives of identified peptides, we determined 775 glycosylation sites of 311 distinct membrane proteins in AML cells (supplemental Table 3). The Cys-Glyco-CSC and Lys-CSC technology variant enriched for peptides that were disulfide-linked to glycopeptides and lysine-containing peptides, respectively (Figure 1). These disulfide-linked piggyback peptides frequently increased sequence coverage of identified glycoproteins and piggyback peptides also enabled identification of membrane proteins, which were not identified by MS through glycopeptides. Thus, the combination of all 3 CSC technology variants was beneficial to increase the number of identified membrane proteins (Figure 2B) and additional valuable information about cell surface proteins was obtained, such as locations of N-glycosylation sites.
ATRA regulated membrane proteins
Changes of membrane protein abundances in AML cells after ATRA stimulation were revealed by MS-based protein quantification. The 2 most prominent biologic processes, which are mediated by ATRA stimulation in AML cells, are differentiation into granulocyte-like cells and stop of cell proliferation. Many of the identified membrane proteins, which showed changed protein abundances in the 2 AML cell lines after ATRA stimulation, were assigned to these 2 biologic processes. These proteins included CD11b, CD11c, CD35, CD36, CD38, CD54, CD55, CD66a, CD300a, CY24B, and CD71.30,36,37,40-44 Interestingly, all 4 members of the epidermal growth factor-like 7-transmembrane (EGF-TM7) domain family, which includes CD97, EMR1, EMR2, and EMR3, were identified in both AML cell lines. Only small changes in protein abundances were detected in the 2 AML cell lines after ATRA stimulation for CD97 and EMR2, which are both known to be important for granulocyte migration.45,46 In contrast, the 2 functionally less well characterized EGF-TM7 receptors, EMR1 and EMR3,38,39 were detected with highly increased protein abundances in both AML cell lines after ATRA stimulation by the CSC technology (Figure 5C). Furthermore, protein abundances of 2 members of the plexin family were detected to be differently regulated by ATRA. Decreased protein abundances of plexin D1 (PLXD1) but increased protein abundances of CD232 (PLXC1) were detected in both AML cell lines after ATRA stimulation. Plexins were originally identified in the nervous system and they are known for their crucial role in axon guidance. In recent years, it has been shown that plexins also play an important role in the immune system.47 Levels of mRNA coding for plexin C1 were shown to be decreased in blast cells of AML patients.48 This finding correlates with our results of decreased plexin C1 protein abundances in unstimulated compared with ATRA stimulated AML cells. ATRA induced abundance changes of proteins that may be important for granulocyte migration, such as EGF-TM7 receptors or plexins, are interesting in connection with increased leukocyte transmigration in the differentiation syndrome in ATRA treated APL patients. Furthermore, the proteins LT4R1 and IGSF2 were detected to be less abundant in unstimulated compared with ATRA stimulated AML cells. Interestingly, levels of mRNA coding for these 2 proteins were recently described to be decreased in chronic myeloid leukemia in blast crisis.49 Protein abundances of TTYH3 and GPR97 were also detected to be increased in AML cells after ATRA stimulation, but the functions of these proteins in AML cells have not yet been clearly described and need to be analyzed in more detail. Overall, many here identified ATRA regulated membrane proteins in AML cells have been described before in the literature, indicating the robustness and reliability of phenotyping cells with the CSC technology. In addition, we identified numerous functionally uncharacterized and also putative proteins in AML cells, showing the value of the CSC technology for protein marker discovery.
Targeted MS-based phenotyping
The presented AML surface proteome illustrates the complexity of functional cell surface protein classes, which determine to a large extent the interaction capacities of AML cells with their microenvironment. Probably not only the classes of proteins present at the cell surface at a certain time are important but also their relative quantities between each other and their absolute abundances in the cell are critical to regulate cellular functions. Sensitive and accurate quantification of selected proteins, to determine protein copy numbers per cell, can now be achieved by MS workflows through the development of peptide-specific Selected Reaction Monitoring (SRM) assays, which are based on the measurement of selected fragment ions of peptides.50 The data generated in the here described experiments enable now the selection of such peptide fragment ions for targeted SRM-based strategies. The combination of the CSC technology with quantitative SRM assays will significantly contribute to the comprehensive and systematic validation of abundance changes of selected cell surface proteins. Such technologies that enable the systematic comparison of cell surface phenotypes of different cell states have the potential to create new perspectives for improved classification and targeted treatment of hematologic malignancies.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank U. Omasits, M. Claassen, L. Reiter, and J. Eng for help with data analysis and members of the Wollscheid and Aebersold group for constant help and discussions.
This project was supported by the Krebsliga Zurich, the NCCR Neural Plasticity and Repair, and the InfectX project within the Swiss Initiative in Systems Biology (SystemsX.ch). A.H. was supported in part by the Mueller Fellowship from the PhD Program in Molecular Life Sciences.
Authorship
Contribution: A.H., R.A., and B.W. designed research; A.H., B.G., A.S., T.B., and D.B. performed the research; A.H. analyzed data; and A.H. and B.W. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for B.G. is Novartis Pharma AG, Developmental and Molecular Pathways, Basel, Switzerland. The current affiliation for A.S. is Biozentrum, University of Basel, Basel, Switzerland.
Correspondence: Bernd Wollscheid, Institute of Molecular Systems Biology, HPT D77, ETH Zurich, Wolfgang-Pauli-Strasse 16, 8093 Zurich, Switzerland; e-mail: bernd.wollscheid@imsb.biol.ethz.ch.